Comparison of SYK Signaling Networks Reveals the Potential Molecular Determinants of Its Tumor-Promoting and Suppressing Functions
Abstract
:1. Introduction
2. Materials and Methods
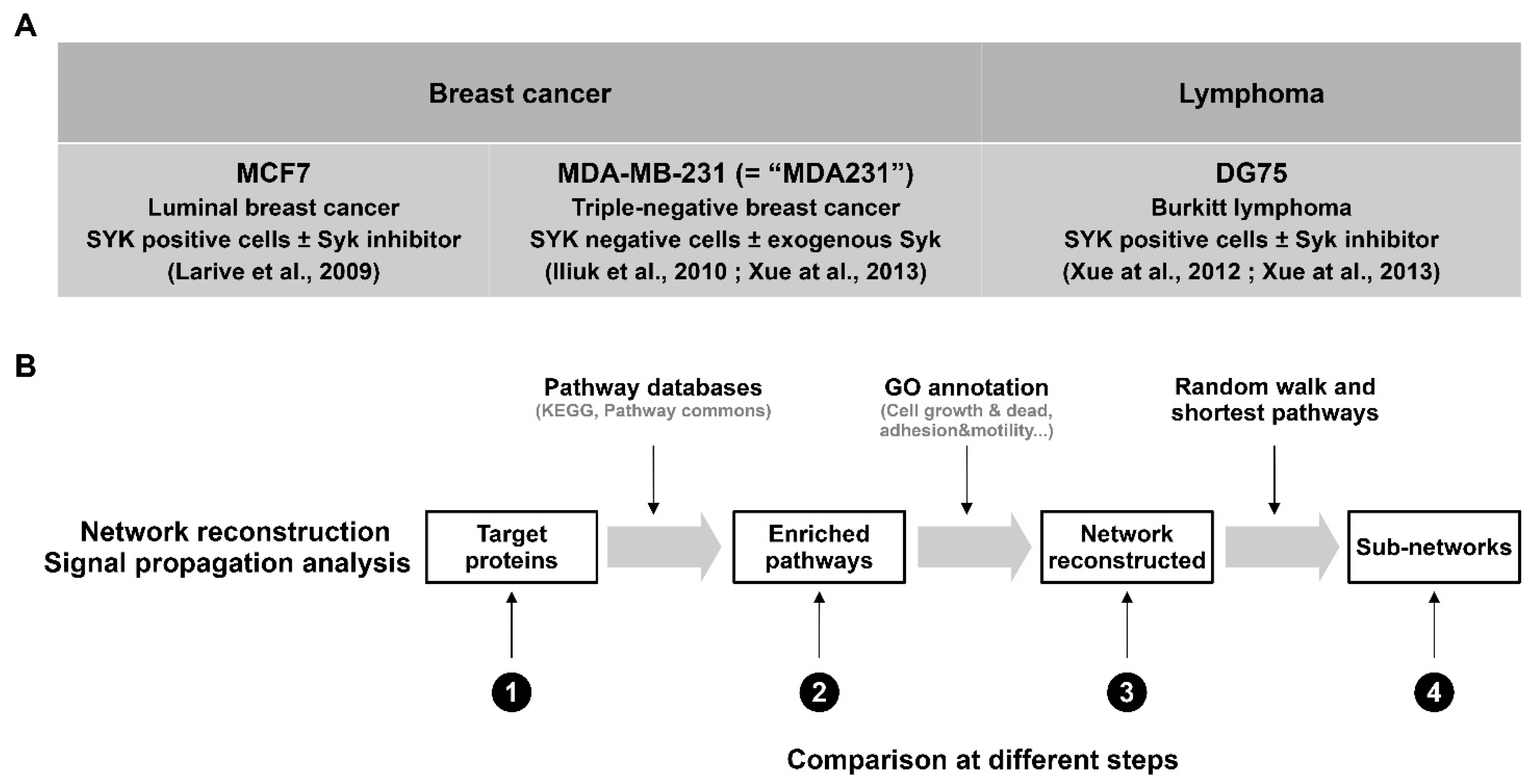
2.1. Phosphoproteomic Data for the Bioinformatic Workflow
2.2. Online Databases
2.3. Gene Ontology Annotation
2.4. Pathway Database Selection
2.5. Hierarchical Clustering
2.6. Signaling Network Comparison
2.7. Signal Propagation Analysis
3. Results
3.1. Comparison of the SYK Signaling Networks in Breast Cancer and Burkitt Lymphoma Cell Lines
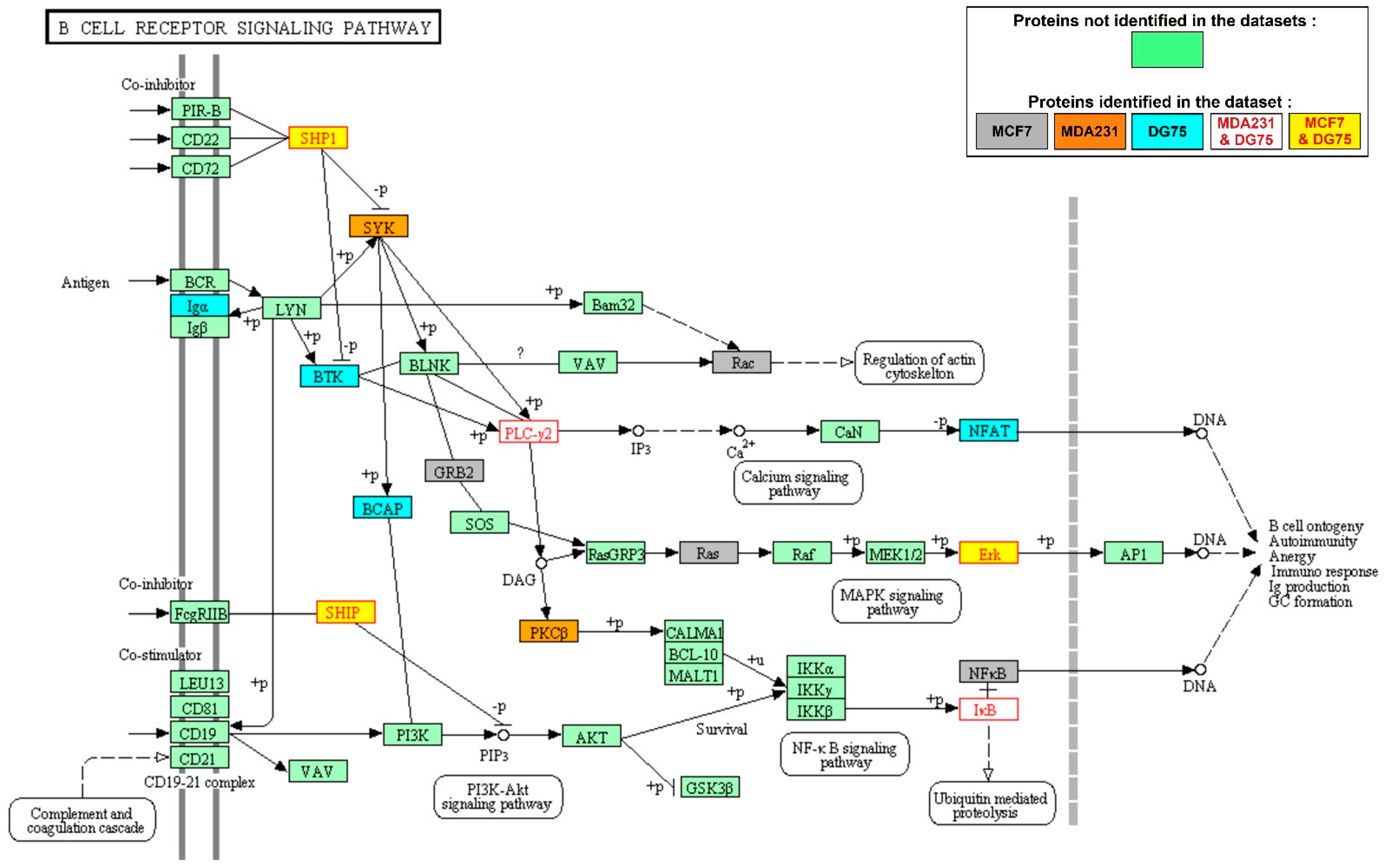
3.2. KEGG Pathways Are Differentially Enriched in SYK Targets in the Breast Cancer and Burkitt Lymphoma Cell Datasets
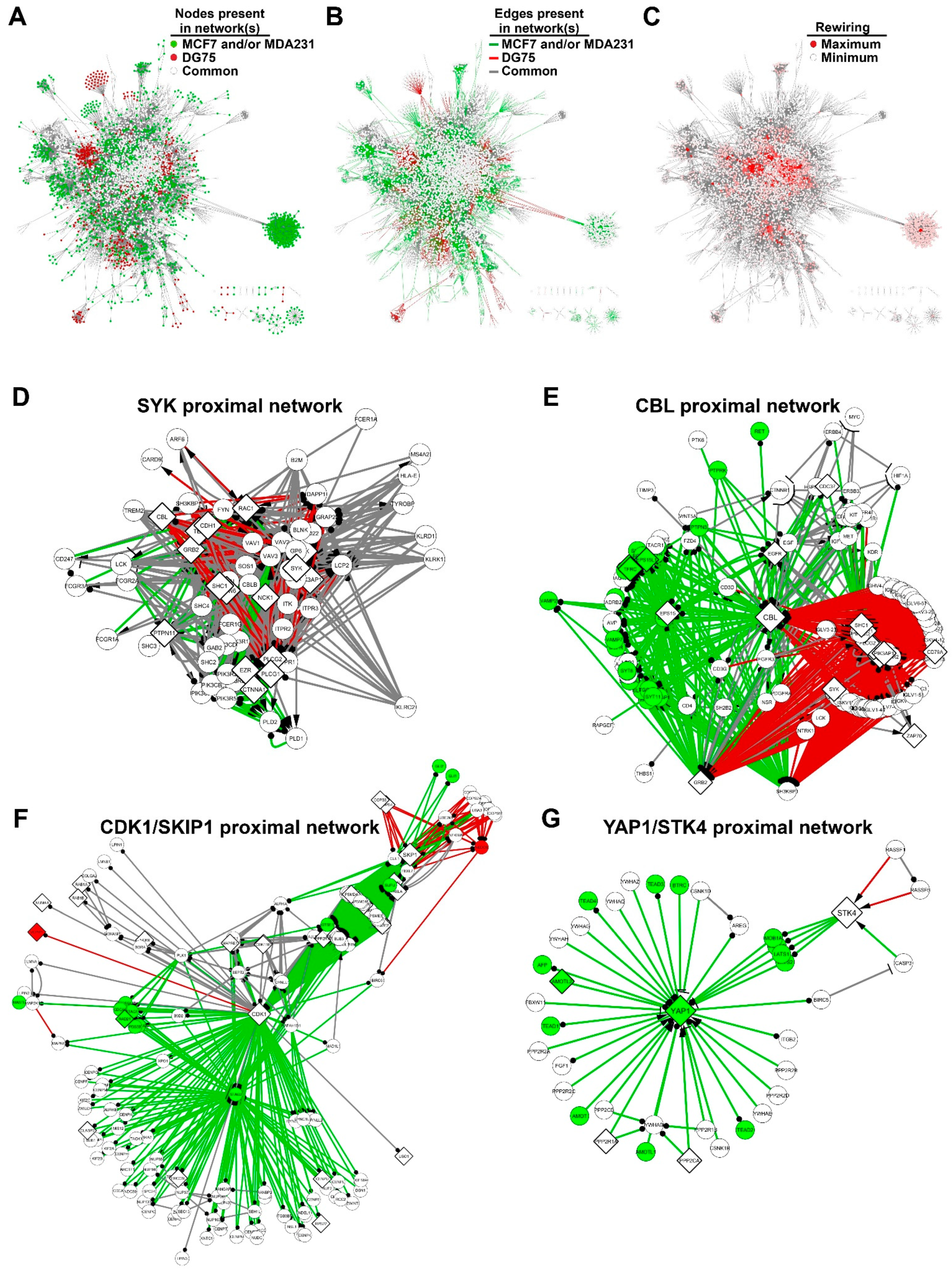
3.3. The SYK Signaling Networks Display Similar Topological Parameters
3.4. SYK Targets Are Rewired between the Breast Cancer and Burkitt Lymphoma Cell Networks
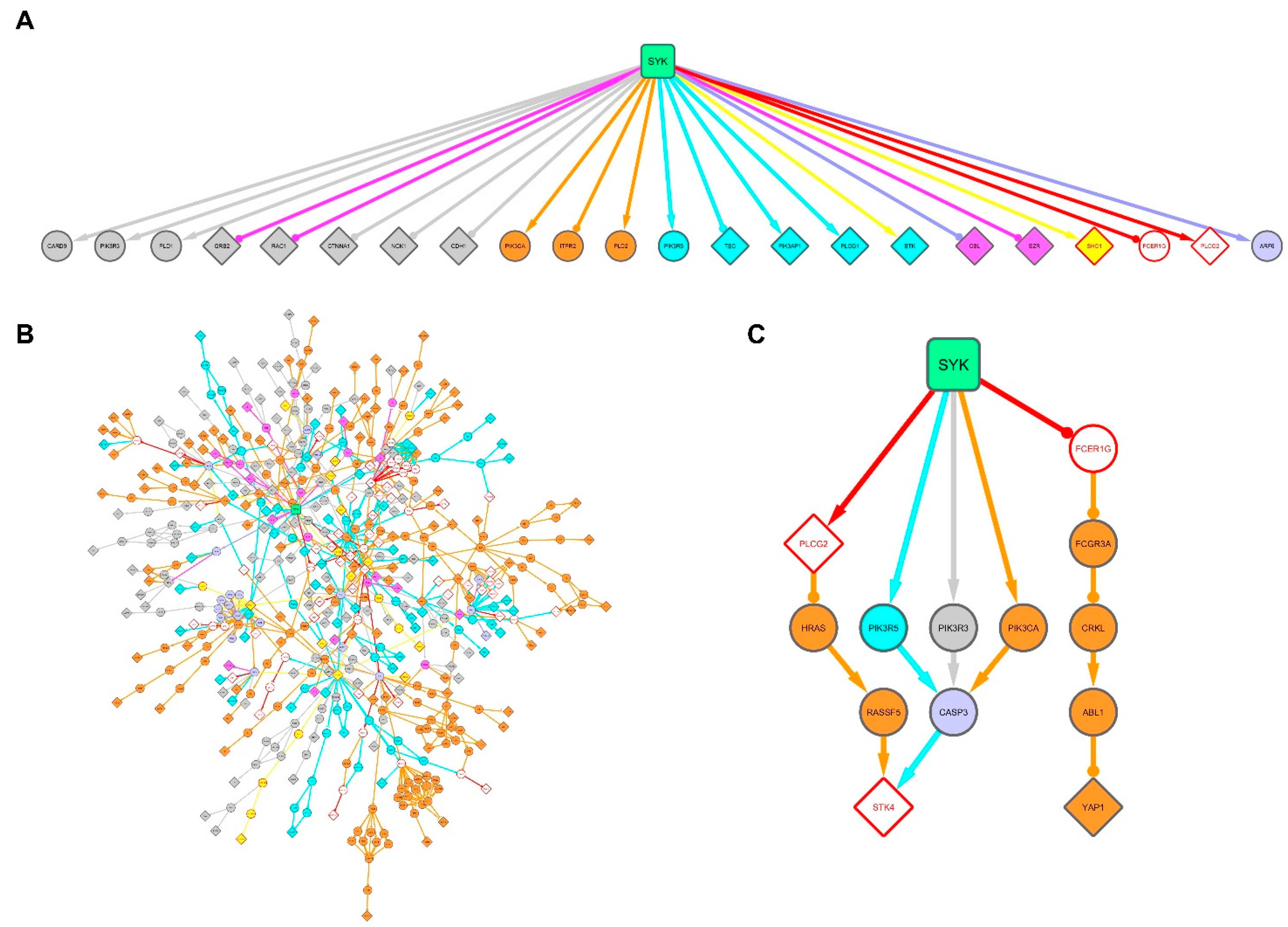
3.5. Signal Propagation from SYK to its Targets Identified in the Breast Cancer and Burkitt Lymphoma Cell Lines
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Hunter, T. Tyrosine Phosphorylation: Thirty Years and Counting. Curr. Opin. Cell Biol. 2009, 21, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Krause, D.S.; Van Etten, R.A. Tyrosine Kinases as Targets for Cancer Therapy. N. Engl. J. Med. 2005, 353, 172–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchner, M.; Fuchs, S.; Prinz, G.; Pfeifer, D.; Bartholomé, K.; Burger, M.; Chevalier, N.; Vallat, L.; Timmer, J.; Gribben, J.G.; et al. Spleen Tyrosine Kinase Is Overexpressed and Represents a Potential Therapeutic Target in Chronic Lymphocytic Leukemia. Cancer Res. 2009, 69, 5424–5432. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Monti, S.; Juszczynski, P.; Daley, J.; Chen, W.; Witzig, T.E.; Habermann, T.M.; Kutok, J.L.; Shipp, M.A. SYK-Dependent Tonic B-Cell Receptor Signaling Is a Rational Treatment Target in Diffuse Large B-Cell Lymphoma. Blood 2008, 111, 2230–2237. [Google Scholar] [CrossRef] [Green Version]
- Perova, T.; Grandal, I.; Nutter, L.M.J.; Papp, E.; Matei, I.R.; Beyene, J.; Kowalski, P.E.; Hitzler, J.K.; Minden, M.D.; Guidos, C.J.; et al. Therapeutic Potential of Spleen Tyrosine Kinase Inhibition for Treating High-Risk Precursor B Cell Acute Lymphoblastic Leukemia. Sci. Transl. Med. 2014, 6, 236ra62. [Google Scholar] [CrossRef]
- Fallah-Arani, F.; Schweighoffer, E.; Vanes, L.; Tybulewicz, V.L.J. Redundant Role for Zap70 in B Cell Development and Activation. Eur. J. Immunol. 2008, 38, 1721–1733. [Google Scholar] [CrossRef]
- Monroe, J.G. ITAM-Mediated Tonic Signalling through Pre-BCR and BCR Complexes. Nat. Rev. Immunol. 2006, 6, 283–294. [Google Scholar] [CrossRef]
- Turner, M.; Joseph Mee, P.; Costello, P.S.; Williams, O.; Price, A.A.; Duddy, L.P.; Furlong, M.T.; Geahlen, R.L.; Tybulewicz, V.L.J. Perinatal Lethality and Blocked B-Cell Development in Mice Lacking the Tyrosine Kinase Syk. Nature 1995, 378, 298–302. [Google Scholar] [CrossRef]
- Coopman, P.J.; Mueller, S.C. The Syk Tyrosine Kinase: A New Negative Regulator in Tumor Growth and Progression. Cancer Lett. 2006, 241, 159–173. [Google Scholar] [CrossRef]
- Coopman, P.J.P.; Do, M.T.H.; Barth, M.; Bowden, E.T.; Hayes, A.J.; Basyuk, E.; Blancato, J.K.; Vezza, P.R.; McLeskey, S.W.; Mangeat, P.H.; et al. The Syk Tyrosine Kinase Suppresses Malignant Growth of Human Breast Cancer Cells. Nature 2000, 406, 742–747. [Google Scholar] [CrossRef]
- Sung, Y.M.; Xu, X.; Sun, J.; Mueller, D.; Sentissi, K.; Johnson, P.; Urbach, E.; Seillier-Moiseiwitsch, F.; Johnson, M.D.; Mueller, S.C. Tumor Suppressor Function of Syk in Human MCF10A In Vitro and Normal Mouse Mammary Epithelium In Vivo. PLoS ONE 2009, 4, e7445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanagi, S.; Inatome, R.; Takano, T.; Yamamura, H. Syk Expression and Novel Function in a Wide Variety of Tissues. Biochem. Biophys. Res. Commun. 2001, 288, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Layton, T.; Stalens, C.; Gunderson, F.; Goodison, S.; Silletti, S. Syk Tyrosine Kinase Acts as a Pancreatic Adenocarcinoma Tumor Suppressor by Regulating Cellular Growth and Invasion. Am. J. Pathol. 2009, 175, 2625–2636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoeller, C.; Thallinger, C.; Pratscher, B.; Bister, M.D.; Schicher, N.; Loewe, R.; Heere-Ress, E.; Roka, F.; Sexl, V.; Pehamberger, H. The Non-Receptor-Associated Tyrosine Kinase Syk Is a Regulator of Metastatic Behavior in Human Melanoma Cells. J. Investig. Dermatol. 2005, 124, 1293–1299. [Google Scholar] [CrossRef] [Green Version]
- Mócsai, A.; Ruland, J.; Tybulewicz, V.L.J. The SYK Tyrosine Kinase: A Crucial Player in Diverse Biological Functions. Nat. Rev. Immunol. 2010, 10, 387–402. [Google Scholar] [CrossRef]
- Krisenko, M.O.; Geahlen, R.L. Calling in SYK: SYK’s Dual Role as a Tumor Promoter and Tumor Suppressor in Cancer. Biochim. Biophys. Acta BBA Mol. Cell Res. 2015, 1853, 254–263. [Google Scholar] [CrossRef] [Green Version]
- Larive, R.M.; Urbach, S.; Poncet, J.; Jouin, P.; Mascre, G.; Sahuquet, A.; Mangeat, P.H.; Coopman, P.J.; Bettache, N. Phosphoproteomic Analysis of Syk Kinase Signaling in Human Cancer Cells Reveals Its Role in Cell-Cell Adhesion. Oncogene 2009, 28, 2337–2347. [Google Scholar] [CrossRef] [Green Version]
- Iliuk, A.B.; Martin, V.A.; Alicie, B.M.; Geahlen, R.L.; Tao, W.A. In-Depth Analyses of Kinase-Dependent Tyrosine Phosphoproteomes Based on Metal Ion-Functionalized Soluble Nanopolymers. Mol. Cell. Proteom. 2010, 9, 2162–2172. [Google Scholar] [CrossRef] [Green Version]
- Xue, L.; Wang, W.-H.; Iliuk, A.; Hu, L.; Galan, J.A.; Yu, S.; Hans, M.; Geahlen, R.L.; Tao, W.A. Sensitive Kinase Assay Linked with Phosphoproteomics for Identifying Direct Kinase Substrates. Proc. Natl. Acad. Sci. USA 2012, 109, 5615–5620. [Google Scholar] [CrossRef] [Green Version]
- Naldi, A.; Larive, R.M.; Czerwinska, U.; Urbach, S.; Montcourrier, P.; Roy, C.; Solassol, J.; Freiss, G.; Coopman, P.J.; Radulescu, O. Reconstruction and Signal Propagation Analysis of the Syk Signaling Network in Breast Cancer Cells. PLoS Comput. Biol. 2017, 13, e1005432. [Google Scholar] [CrossRef] [Green Version]
- Buffard, M.; Naldi, A.; Radulescu, O.; Coopman, P.J.; Larive, R.M.; Freiss, G. Network Reconstruction and Significant Pathway Extraction Using Phosphoproteomic Data from Cancer Cells. Proteomics 2019, 19, 1800450. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Geahlen, R.L.; Tao, W.A. Identification of Direct Tyrosine Kinase Substrates Based on Protein Kinase Assay-Linked Phosphoproteomics. Mol. Cell. Proteom. 2013, 12, 2969–2980. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New Approach for Understanding Genome Variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Cerami, E.G.; Gross, B.E.; Demir, E.; Rodchenkov, I.; Babur, Ö.; Anwar, N.; Schultz, N.; Bader, G.D.; Sander, C. Pathway Commons, a Web Resource for Biological Pathway Data. Nucleic Acids Res. 2011, 39, D685–D690. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing Topological Parameters of Biological Networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Assenov, Y.; Domingues, F.S.; Albrecht, M. Topological Analysis and Interactive Visualization of Biological Networks and Protein Structures. Nat. Protoc. 2012, 7, 670–685. [Google Scholar] [CrossRef]
- Goenawan, I.H.; Bryan, K.; Lynn, D.J. DyNet: Visualization and Analysis of Dynamic Molecular Interaction Networks. Bioinformatics 2016, 32, 2713–2715. [Google Scholar] [CrossRef] [Green Version]
- Pal Singh, S.; Dammeijer, F.; Hendriks, R.W. Role of Bruton’s Tyrosine Kinase in B Cells and Malignancies. Mol. Cancer 2018, 17, 57. [Google Scholar] [CrossRef]
- Janda, E.; Palmieri, C.; Pisano, A.; Pontoriero, M.; Iaccino, E.; Falcone, C.; Fiume, G.; Gaspari, M.; Nevolo, M.; Di Salle, E.; et al. Btk Regulation in Human and Mouse B Cells via Protein Kinase C Phosphorylation of IBtkγ. Blood 2011, 117, 6520–6531. [Google Scholar] [CrossRef] [Green Version]
- Albert, R. Scale-Free Networks in Cell Biology. J. Cell Sci. 2005, 118, 4947–4957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barabási, A.-L. Scale-Free Networks: A Decade and Beyond. Science 2009, 325, 412–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barabási, A.-L.; Oltvai, Z.N. Network Biology: Understanding the Cell’s Functional Organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Watts, D.J.; Strogatz, S.H. Collective Dynamics of ‘Small-World’ Networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef]
- Freeman, L.C. Centrality in Social Networks Conceptual Clarification. Soc. Netw. 1978, 1, 215–239. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.; Blumer, A.; Lee, K. An Algorithm for Modularity Analysis of Directed and Weighted Biological Networks Based on Edge-Betweenness Centrality. Bioinformatics 2006, 22, 3106–3108. [Google Scholar] [CrossRef] [Green Version]
- Bewarder, N.; Weinrich, V.; Budde, P.; Hartmann, D.; Flaswinkel, H.; Reth, M.; Frey, J. In Vivo and in Vitro Specificity of Protein Tyrosine Kinases for Immunoglobulin G Receptor (FcgammaRII) Phosphorylation. Mol. Cell. Biol. 1996, 16, 4735–4743. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Gong, L.; Gu, H. Regulation of Immune System Development and Function by Cbl-Mediated Ubiquitination. Immunol. Rev. 2019, 291, 123–133. [Google Scholar] [CrossRef]
- Lyle, C.L.; Belghasem, M.; Chitalia, V.C. C-Cbl: An Important Regulator and a Target in Angiogenesis and Tumorigenesis. Cells 2019, 8, 498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deckert, M.; Elly, C.; Altman, A.; Liu, Y.-C. Coordinated Regulation of the Tyrosine Phosphorylation of Cbl by Fyn and Syk Tyrosine Kinases. J. Biol. Chem. 1998, 273, 8867–8874. [Google Scholar] [CrossRef] [Green Version]
- Rao, N.; Ghosh, A.K.; Ota, S.; Zhou, P.; Reddi, A.L.; Hakezi, K.; Druker, B.K.; Wu, J.; Band, H. The Non-Receptor Tyrosine Kinase Syk Is a Target of Cbl-Mediated Ubiquitylation upon B-Cell Receptor Stimulation. EMBO J. 2001, 20, 7085–7095. [Google Scholar] [CrossRef] [Green Version]
- Hussain, M.; Lu, Y.; Liu, Y.-Q.; Su, K.; Zhang, J.; Liu, J.; Zhou, G.-B. Skp1: Implications in Cancer and SCF-Oriented Anti-Cancer Drug Discovery. Pharmacol. Res. 2016, 111, 34–42. [Google Scholar] [CrossRef]
- Mueller, P.R.; Coleman, T.R.; Kumagai, A.; Dunphy, W.G. Myt1: A Membrane-Associated Inhibitory Kinase That Phosphorylates Cdc2 on Both Threonine-14 and Tyrosine-15. Science 1995, 270, 86–90. [Google Scholar] [CrossRef]
- Nurse, P. Universal Control Mechanism Regulating Onset of M-Phase. Nature 1990, 344, 503–508. [Google Scholar] [CrossRef]
- Parker, L.L.; Piwnica-Worms, H. Inactivation of the P34cdc2-Cyclin B Complex by the Human WEE1 Tyrosine Kinase. Science 1992, 257, 1955–1957. [Google Scholar] [CrossRef] [PubMed]
- Prinos, P.; Garneau, D.; Lucier, J.-F.; Gendron, D.; Couture, S.; Boivin, M.; Brosseau, J.-P.; Lapointe, E.; Thibault, P.; Durand, M.; et al. Alternative Splicing of SYK Regulates Mitosis and Cell Survival. Nat. Struct. Mol. Biol. 2011, 18, 673–679. [Google Scholar] [CrossRef]
- Uckun, F.M.; Ma, H.; Ozer, Z.; Goodman, P.; Zhang, J.; Qazi, S. A Previously Unknown Unique Challenge for Inhibitors of SYK ATP-Binding Site: Role of SYK as A Cell Cycle Checkpoint Regulator. EBioMedicine 2014, 1, 16–28. [Google Scholar] [CrossRef] [Green Version]
- Zyss, D.; Montcourrier, P.; Vidal, B.; Anguille, C.; Mérezègue, F.; Sahuquet, A.; Mangeat, P.H.; Coopman, P.J. The Syk Tyrosine Kinase Localizes to the Centrosomes and Negatively Affects Mitotic Progression. Cancer Res. 2005, 65, 10872–10880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heallen, T.; Zhang, M.; Wang, J.; Bonilla-Claudio, M.; Klysik, E.; Johnson, R.L.; Martin, J.F. Hippo Pathway Inhibits Wnt Signaling to Restrain Cardiomyocyte Proliferation and Heart Size. Science 2011, 332, 458–461. [Google Scholar] [CrossRef] [Green Version]
- Levy, D.; Adamovich, Y.; Reuven, N.; Shaul, Y. Yap1 Phosphorylation by C-Abl Is a Critical Step in Selective Activation of Proapoptotic Genes in Response to DNA Damage. Mol. Cell 2008, 29, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Mócsai, A.; Humphrey, M.B.; Van Ziffle, J.A.G.; Hu, Y.; Burghardt, A.; Spusta, S.C.; Majumdar, S.; Lanier, L.L.; Lowell, C.A.; Nakamura, M.C. The Immunomodulatory Adapter Proteins DAP12 and Fc Receptor γ-Chain (FcRγ) Regulate Development of Functional Osteoclasts through the Syk Tyrosine Kinase. Proc. Natl. Acad. Sci. USA 2004, 101, 6158–6163. [Google Scholar] [CrossRef] [Green Version]
- Ulanova, M.; Puttagunta, L.; Marcet-Palacios, M.; Duszyk, M.; Steinhoff, U.; Duta, F.; Kim, M.-K.; Indik, Z.K.; Schreiber, A.D.; Befus, A.D. Syk Tyrosine Kinase Participates in Β1-Integrin Signaling and Inflammatory Responses in Airway Epithelial Cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005, 288, L497–L507. [Google Scholar] [CrossRef] [Green Version]
- Sada, K.; Minami, Y.; Yamamura, H. Relocation of Syk Protein-Tyrosine Kinase to the Actin Filament Network and Subsequent Association with Fak. Eur. J. Biochem. 1997, 248, 827–833. [Google Scholar] [CrossRef]
- Kassouf, T.; Larive, R.M.; Morel, A.; Urbach, S.; Bettache, N.; Marcial Medina, M.C.; Mèrezègue, F.; Freiss, G.; Peter, M.; Boissière-Michot, F.; et al. The Syk Kinase Promotes Mammary Epithelial Integrity and Inhibits Breast Cancer Invasion by Stabilizing the E-Cadherin/Catenin Complex. Cancers 2019, 11, 1974. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.-H.; Childress, M.O.; Geahlen, R.L. Syk Interacts with and Phosphorylates Nucleolin To Stabilize Bcl-XL MRNA and Promote Cell Survival. Mol. Cell. Biol. 2014, 34, 3788–3799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blancato, J.; Graves, A.; Rashidi, B.; Moroni, M.; Tchobe, L.; Ozdemirli, M.; Kallakury, B.; Makambi, K.H.; Marian, C.; Mueller, S.C. SYK Allelic Loss and the Role of Syk-Regulated Genes in Breast Cancer Survival. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shrikhande, U.; Alicie, B.M.; Zhou, Q.; Geahlen, R.L. A Role for the Protein-Tyrosine Kinase, Syk, in Regulating Cell-Cell Adhesion and Motility in Breast Cancer Cells. Mol. Cancer Res. MCR 2009, 7, 634–644. [Google Scholar] [CrossRef] [Green Version]
- Cottini, F.; Hideshima, T.; Xu, C.; Sattler, M.; Dori, M.; Agnelli, L.; ten Hacken, E.; Bertilaccio, M.T.; Antonini, E.; Neri, A.; et al. Rescue of hippo co-activator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat. Med. 2014, 20, 599–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoner, S.A.; Yan, M.; Liu, K.T.H.; Arimoto, K.-I.; Shima, T.; Wang, H.-Y.; Johnson, D.T.; Bejar, R.; Jamieson, C.; Guan, K.-L.; et al. Hippo Kinase Loss Contributes to Del(20q) Hematologic Malignancies through Chronic Innate Immune Activation. Blood 2019, 134, 1730–1744. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Bhushan, B.; Mars, W.M.; Bowen, W.; Tao, J.; Orr, A.; Stoops, J.; Yu, Y.; Luo, J.; Duncan, A.W.; et al. Phosphorylated Ezrin (Thr567) Regulates Hippo Pathway and Yes-Associated Protein (Yap) in Liver. Am. J. Pathol. 2020, 190, 1427–1437. [Google Scholar] [CrossRef]
- Keshet, R.; Adler, J.; Ricardo Lax, I.; Shanzer, M.; Porat, Z.; Reuven, N.; Shaul, Y. C-Abl Antagonizes the YAP Oncogenic Function. Cell Death Differ. 2015, 22, 935–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turunen, S.P.; von Nandelstadh, P.; Öhman, T.; Gucciardo, E.; Seashore-Ludlow, B.; Martins, B.; Rantanen, V.; Li, H.; Höpfner, K.; Östling, P.; et al. FGFR4 Phosphorylates MST1 to Confer Breast Cancer Cells Resistance to MST1/2-Dependent Apoptosis. Cell Death Differ. 2019, 26, 2577–2593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlahov, N.; Scrace, S.; Soto, M.S.; Grawenda, A.M.; Bradley, L.; Pankova, D.; Papaspyropoulos, A.; Yee, K.S.; Buffa, F.; Goding, C.R.; et al. Alternate RASSF1 Transcripts Control SRC Activity, E-Cadherin Contacts, and YAP-Mediated Invasion. Curr. Biol. 2015, 25, 3019–3034. [Google Scholar] [CrossRef] [Green Version]
- Xiao, L.; Chen, D.; Hu, P.; Wu, J.; Liu, W.; Zhao, Y.; Cao, M.; Fang, Y.; Bi, W.; Zheng, Z.; et al. The C-Abl-MST1 Signaling Pathway Mediates Oxidative Stress-Induced Neuronal Cell Death. J. Neurosci. 2011, 31, 9611–9619. [Google Scholar] [CrossRef]
- Rada, M.; Barlev, N.; Macip, S. BTK: A Two-Faced Effector in Cancer and Tumour Suppression. Cell Death Dis. 2018, 9, 1–3. [Google Scholar] [CrossRef]
- Huber, C.; Sundström, C.; Nilsson, K.; Wigzell, H. Surface Receptors on Human Haematopoietic Cell Lines. Clin. Exp. Immunol. 1976, 25, 367–376. [Google Scholar]
- Katzav, S.; Schmitz, M.L. Mutations of C-Cbl in Myeloid Malignancies. Oncotarget 2015, 6, 10689–10696. [Google Scholar] [CrossRef] [Green Version]
- Kumaradevan, S.; Lee, S.Y.; Richards, S.; Lyle, C.; Zhao, Q.; Tapan, U.; Jiangliu, Y.; Ghumman, S.; Walker, J.; Belghasem, M.; et al. C-Cbl Expression Correlates with Human Colorectal Cancer Survival and Its Wnt/β-Catenin Suppressor Function Is Regulated by Tyr371 Phosphorylation. Am. J. Pathol. 2018, 188, 1921–1933. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Teng, Y.; Wu, X.; Li, Z.; Bao, B.; Liu, Y.; Qu, X.; Zhang, L. The E3 Ubiquitin Ligase Cbl-b Predicts Favorable Prognosis in Breast Cancer. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Elbæk, C.R.; Petrosius, V.; Sørensen, C.S. WEE1 Kinase Limits CDK Activities to Safeguard DNA Replication and Mitotic Entry. Mutat. Res. Mol. Mech. Mutagen. 2020, 819–820, 111694. [Google Scholar] [CrossRef]
- Schmidt, M.; Rohe, A.; Platzer, C.; Najjar, A.; Erdmann, F.; Sippl, W. Regulation of G2/M Transition by Inhibition of WEE1 and PKMYT1 Kinases. Molecules 2017, 22, 2045. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, G.; Rangaswami, H.; Jain, S.; Kundu, G.C. Hypoxia Regulates Cross-Talk between Syk and Lck Leading to Breast Cancer Progression and Angiogenesis. J. Biol. Chem. 2006, 281, 11322–11331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takada, Y.; Aggarwal, B.B. TNF Activates Syk Protein Tyrosine Kinase Leading to TNF-Induced MAPK Activation, NF-KappaB Activation, and Apoptosis. J. Immunol. Baltim. Md 1950 2004, 173, 1066–1077. [Google Scholar] [CrossRef] [Green Version]
- Markham, A. Fostamatinib: First Global Approval. Drugs 2018, 78, 959–963. [Google Scholar] [CrossRef] [PubMed]
- Rivera, J.; Colbert, R.A. Healing the Syk through Kinase Inhibitors. N. Engl. J. Med. 2010, 363, 1362–1364. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Doré, P.C.; Sewell, W.A.C. Could Oral Spleen Tyrosine Kinase Inhibitors Lead to Neoplastic Transformation? J. Allergy Clin. Immunol. 2007, 119, 1277. [Google Scholar] [CrossRef]


| Gene Symbol | KEGG Entry | UniProt Entry | MCF7 Cell Dataset (Present) | MDA231 Cell Dataset (Phosphosites) | DG75 Cell Dataset (Phosphosites) |
|---|---|---|---|---|---|
| NFKBIE | 4794 | O00221 | 155 | 155 | |
| NFATC2 | 4773 | Q13469 | 752 | ||
| PTPN6 | 5777 | P29350 | X | 536 | |
| NRAS | 4893 | P01111 | X | ||
| RAC1 | 5879 | P63000 | X | ||
| GRB2 | 2885 | P62993 | X | ||
| PLCG2 | 5336 | P16885 | 1217 | 759 * ; 550 ; 858 ; 1245 | |
| BTK | 695 | Q06187 | 551 * | ||
| MAPK3 | 5595 | P27361 | 204 | ||
| RELA | 5970 | Q04206 | X | ||
| SYK | 6850 | P43405 | 352 | ||
| INPPL1 | 3636 | O15357 | X | 1135 | |
| PRKCB | 5579 | P05771 | 507 | ||
| PIK3AP1 | 118788 | Q6ZUJ8 | 694 | ||
| CD79A | 973 | P11912 | 210 | ||
| MAPK1 | 5594 | P28482 | X | 187 |
| Gene Symbol | KEGG Entry | UniProt Entry | MCF7 Cell Dataset (Present) | MDA231 Cell Dataset (Phosphosites) | DG75 Cell Dataset (Phosphosites) |
|---|---|---|---|---|---|
| PXN | 5829 | P49023 | 409 ; 468 | 88 | |
| WASF2 | 10163 | Q9Y6W5 | X | ||
| MYL12B | 103910 | O14950 | 143 | ||
| ARHGEF12 | 23365 | Q9NZN5 | X | 1232 | |
| NRAS | 4893 | P01111 | X | ||
| ACTN1 | 87 | P12814 | X | ||
| RDX | 5962 | P35241 | |||
| PPP1R12A | 4659 | O14974 | 446 ; 446 | ||
| CFL2 | 1073 | Q9Y281 | 89 * | 89 | |
| ARHGEF7 | 8874 | Q14155 | X | ||
| PPP1CB | 5500 | P62140 | X | ||
| MAPK1 | 5594 | P28482 | X | 187 | |
| ACTB | 60 | P60709 | 240 | 240 | |
| CFL1 | 1072 | P23528 | 89 * | 68 ; 89 | |
| GIT1 | 28964 | Q9Y2X7 | 598 ; 545 | ||
| PPP1CA | 5499 | P62136 | X | ||
| RAC1 | 5879 | P63000 | X | ||
| EZR | 7430 | P15311 | X | 424 | |
| IQGAP2 | 10788 | Q13576 | X | ||
| MAPK3 | 5595 | P27361 | 204 | ||
| MYL12A | 10627 | P19105 | X | 142 | |
| EGFR | 1956 | P00533 | 1069 | ||
| ACTN4 | 81 | O43707 | X | ||
| ARHGAP35 | 2909 | Q9NRY4 | 1087 | ||
| ACTG1 | 71 | P63261 | 240 | 240 | |
| BAIAP2 | 10458 | Q9UQB8 | 505 |
| Gene Symbol | UniProt Entry | Target in Network(s) [Site(s) of Phosphorylation] | Present in Network(s) |
|---|---|---|---|
| ARFGAP1 | Q8N6T3 | MCF7 ; MDA231 [Y175] | MCF7 ; MDA231 |
| BLK | P51451 | DG75 [Y187] | MCF7 ; DG75 |
| CBL | P22681 | MCF7 ; MDA231 [Y700] | MCF7 ; MDA231 ; DG75 |
| CDK1 | P06493 | DG75 [Y15] | MCF7 ; MDA231 ; DG75 |
| CLTA | P09496 | MCF7 | MCF7 ; MDA231 ; DG75 |
| COPS4 | Q9BT78 | MCF7 | MCF7 ; DG75 |
| COPS8 | Q99627 | MCF7 | MCF7 ; DG75 |
| DNMT1 | P26358 | MDA231 [Y359 ; Y399] | MDA231 |
| EPS15L1 | Q9UBC2 | MCF7 | MCF7 ; MDA231 |
| ESPL1 | Q14674 | MDA231 [Y719] | MCF7 ; MDA231 ; DG75 |
| FIP1L1 | Q6UN15 | DG75 [Y113] | DG75 |
| GBF1 | Q92538 | MDA231 [Y377] | MDA231 |
| KPNA4 | O00629 | MDA231 [Y66] | MDA231 |
| MTMR6 | Q9Y217 | MDA231 [Y108] | MDA231 |
| NFKB2 | Q00653 | DG75 [Y39 ; Y867] | MCF7 ; MDA231 ; DG75 |
| PTDSS1 | P48651 | MDA231 [Y424] | MDA231 |
| PYGB | P11216 | MDA231 [Y263] | MCF7 ; MDA231 |
| REEP4 | Q9H6H4 | MDA231 [Y172] | MDA231 |
| RRP8 | O43159 | DG75 [Y259] | DG75 |
| SAP30BP | Q9UHR5 | MDA231 [Y46] | MDA231 |
| SF3B1 | O75533 | MDA231 [Y38 ; Y70] | MDA231 |
| SKP1 | P63208 | MCF7 | MCF7 ; DG75 |
| STAM | Q92783 | MCF7 | MCF7 ; MDA231 |
| SUPT5H | O00267 | MDA231 [Y140] | MDA231 |
| TFRC | P02786 | MDA231 [Y20] | MCF7 ; MDA231 |
| TRIP10 | Q15642 | MCF7 | MCF7 ; DG75 |
| TYMS | P04818 | MDA231 [Y153] | MCF7 ; MDA231 |
| UPF1 | Q92900 | MDA231 [Y113 ; Y1112] ; DG75 [Y113 ; Y1112] | MCF7 ; MDA231 ; DG75 |
| VAMP8 | Q9BV40 | MCF7 | MCF7 ; MDA231 |
| YAP1 | P46937 | MDA231 [Y407 ; Y391] | MCF7 ; MDA231 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buffard, M.; Naldi, A.; Freiss, G.; Deckert, M.; Radulescu, O.; Coopman, P.J.; Larive, R.M. Comparison of SYK Signaling Networks Reveals the Potential Molecular Determinants of Its Tumor-Promoting and Suppressing Functions. Biomolecules 2021, 11, 308. https://doi.org/10.3390/biom11020308
Buffard M, Naldi A, Freiss G, Deckert M, Radulescu O, Coopman PJ, Larive RM. Comparison of SYK Signaling Networks Reveals the Potential Molecular Determinants of Its Tumor-Promoting and Suppressing Functions. Biomolecules. 2021; 11(2):308. https://doi.org/10.3390/biom11020308
Chicago/Turabian StyleBuffard, Marion, Aurélien Naldi, Gilles Freiss, Marcel Deckert, Ovidiu Radulescu, Peter J. Coopman, and Romain M. Larive. 2021. "Comparison of SYK Signaling Networks Reveals the Potential Molecular Determinants of Its Tumor-Promoting and Suppressing Functions" Biomolecules 11, no. 2: 308. https://doi.org/10.3390/biom11020308






