The Controversial Role of Autophagy in Ewing Sarcoma Pathogenesis—Current Treatment Options
Abstract
:1. Introduction
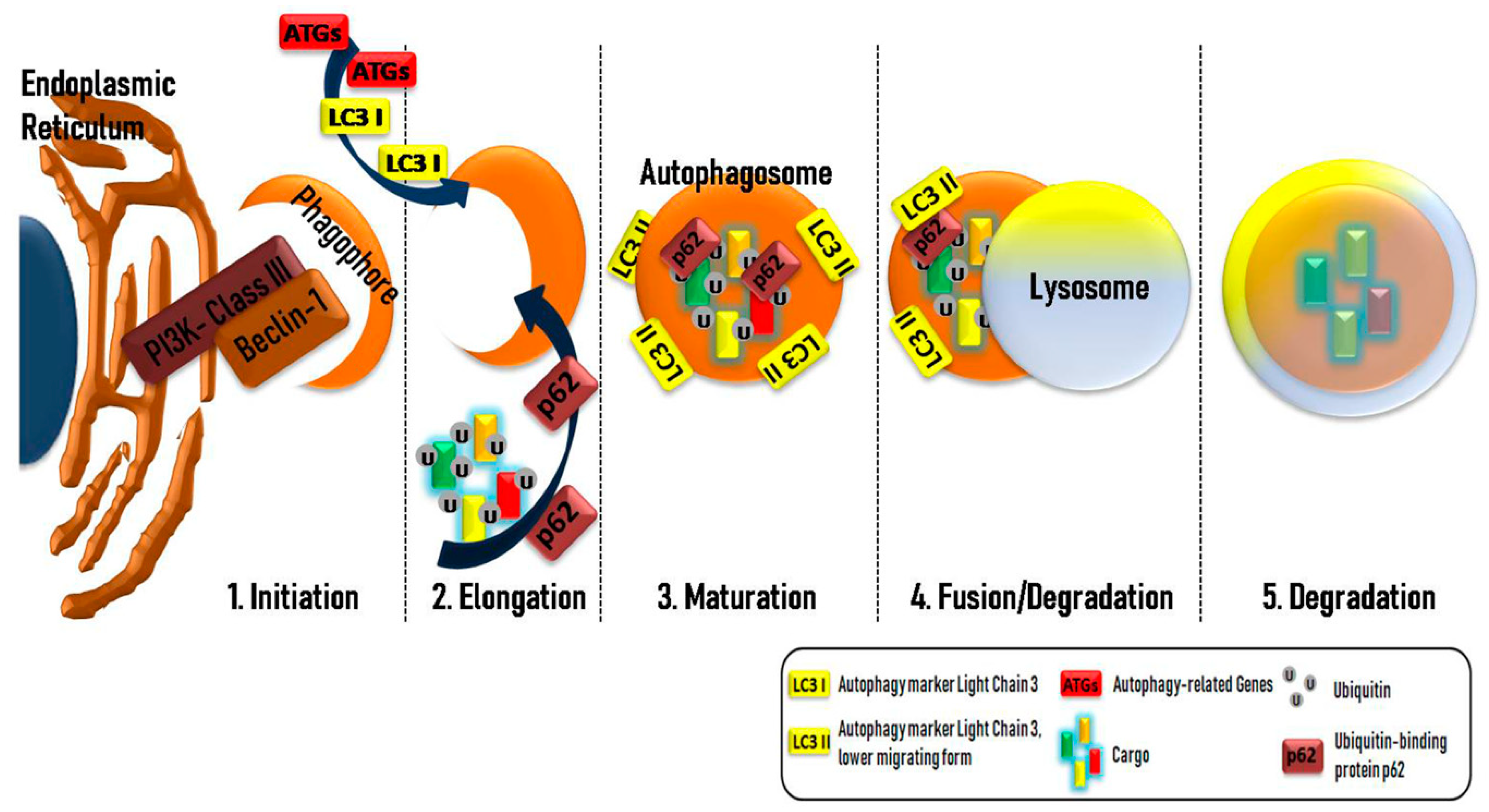
2. The Complex Mechanism of Autophagy
2.1. The Key Components in Autophagy
2.2. The Controversial Role of Autophagy in Human Cancer
3. The Impact of Autophagy in Ewing Sarcoma
4. Targeting Autophagy in Clinical Practice—A Promising Anti-Cancer Strategy for Ewing Sarcoma
4.1. The Clinical Impact of Autophagy Inhibition in Ewing Sarcoma
4.2. The Clinical Impact of Autophagy Induction in Ewing Sarcoma
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaspar, N.; Hawkins, D.S.; Dirksen, U.; Lewis, I.J.; Ferrari, S.; Le Deley, M.C.; Kovar, H.; Grimer, R.; Whelan, J.; Claude, L.; et al. Ewing sarcoma: Current manage-ment and future approaches through collaboration. J. Clin. Oncol. 2015, 33, 3036–3046. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ge, Y.; Guo, L.; Huang, L. Potential approaches to the treatment of Ewing’s sarcoma. Oncotarget 2016, 8, 5523–5539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delattre, O.; Zucman, J.; Plougastel, B.; Desmaze, C.; Melot, T.; Peter, M.; Kovar, H.; Joubert, I.; De Jong, P.; A Rouleau, G.; et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992, 359, 162–165. [Google Scholar] [CrossRef]
- Tanaka, K.; Iwakuma, T.; Harimaya, K.; Sato, H.; Iwamoto, Y. EWS-Fli1 antisense oligodeoxynucleotide inhibits prolifer-ation of human Ewing’s sarcoma and primitive neuroectodermal tumor cells. J. Clin. Investig. 1997, 99, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.; Christensen, L.; Warner, K.; States, L.; Kang, H.-G.; Vo, K.; Lawlor, E.R.; May, W.A. GLI1 Is a Central Mediator of EWS/FLI1 Signaling in Ewing Tumors. PLoS ONE 2009, 4, e7608. [Google Scholar] [CrossRef]
- Mateo-Lozano, S.; Gokhale, P.C.; Soldatenkov, V.A.; Dritschilo, A.; Tirado, O.M.; Notario, V. Combined Transcriptional and Translational Targeting of EWS/FLI-1 in Ewing’s Sarcoma. Clin. Cancer Res. 2006, 12, 6781–6790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Nguyen, P.T.; Shim, H.S.; Hyeon, S.J.; Im, H.; Choi, M.-H.; Chung, S.; Kowall, N.W.; Lee, S.B.; Ryu, H. EWSR1, a multifunctional protein, regulates cellular function and aging via genetic and epigenetic pathways. Biochim. et Biophys. Acta (BBA) Mol. Basis Dis. 2019, 1865, 1938–1945. [Google Scholar] [CrossRef]
- Scott-Browne, J.P.; Lio, C.W.J.; Rao, A. TET proteins in natural and induced differentiation. Curr. Opin. Genet. Dev. 2017, 46, 202–208. [Google Scholar] [CrossRef]
- Andersson, M.K.; Ståhlberg, A.; Arvidsson, Y.; Olofsson, A.; Semb, H.; Stenman, G.; Nilsson, O.; Aman, P. The multifunction-al FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008, 9, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahlendorf, D.E.; Staege, M.S. Characterization of Ewing sarcoma associated cancer/testis antigens. Cancer Biol. Ther. 2013, 14, 254–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, A.Y.; Manley, J.L. The TET Family of Proteins: Functions and Roles in Disease. J. Mol. Cell Biol. 2009, 1, 82–92. [Google Scholar] [CrossRef] [Green Version]
- Paronetto, M.P. Ewing Sarcoma Protein: A Key Player in Human Cancer. Int. J. Cell Biol. 2013, 2013, 1–12. [Google Scholar] [CrossRef]
- Morohoshi, F.; Ootsuka, Y.; Arai, K.; Ichikawa, H.; Mitani, S.; Munakata, N.; Ohki, M. Genomic structure of the human RBP56/hTAFII68 and FUS/TLS genes. Gene 1998, 221, 191–198. [Google Scholar] [CrossRef]
- Eskelinen, E.-L. Autophagy: Supporting cellular and organismal homeostasis by self-eating. Int. J. Biochem. Cell Biol. 2019, 111, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Yun, C.W.; Lee, S.H. The Roles of Autophagy in Cancer. Int. J. Mol. Sci. 2018, 19, 3466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burman, C.; Ktistakis, N.T. Autophagosome formation in mammalian cells. Semin. Immunopathol. 2010, 32, 397–413. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; White, E. Role of Autophagy in Cancer: Management of Metabolic Stress. Autophagy 2007, 3, 28–31. [Google Scholar] [CrossRef] [Green Version]
- Lippai, M.; Szatmári, Z. Autophagy—from molecular mechanisms to clinical relevance. Cell Biol. Toxicol. 2017, 33, 145–168. [Google Scholar] [CrossRef] [PubMed]
- Menon, M.B.; Dhamija, S. Beclin 1 Phosphorylation—at the Center of Autophagy Regulation. Front. Cell Dev. Biol. 2018, 6, 137. [Google Scholar] [CrossRef] [Green Version]
- Dunlop, E.A.; Tee, A.R. mTOR and autophagy: A dynamic relationship governed by nutrients and energy. Semin. Cell Dev. Biol. 2014, 36, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef] [PubMed]
- Chun, Y.; Kim, J. Autophagy: An Essential Degradation Program for Cellular Homeostasis and Life. Cells 2018, 7, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.E.; Tee, A.R. Exploiting cancer vulnerabilities: mTOR, autophagy, and homeostatic imbalance. Essays Biochem. 2017, 61, 699–710. [Google Scholar] [CrossRef]
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; et al. Autophagosomes form at ER–mitochondria contact sites. Nat. Cell Biol. 2013, 495, 389–393. [Google Scholar] [CrossRef]
- Munson, M.J.; Ganley, I.G. MTOR, PIK3C3, and autophagy: Signaling the beginning from the end. Autophagy 2015, 11, 2375–2376. [Google Scholar] [CrossRef]
- Kihara, A.; Noda, T.; Ishihara, N.; Ohsumi, Y. Two Distinct Vps34 Phosphatidylinositol 3–Kinase Complexes Function in Autophagy and Carboxypeptidase Y Sorting inSaccharomyces cerevisiae. J. Cell Biol. 2001, 152, 519–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arakawa, S.; Honda, S.; Yamaguchi, H.; Shimizu, S. Molecular mechanisms and physiological roles of Atg5/Atg7-independent alternative autophagy. Proc. Jpn. Acad. Ser. B 2017, 93, 378–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, J. Atg7 in development and disease: Panacea or Pandora’s Box? Protein Cell 2015, 6, 722–734. [Google Scholar] [CrossRef] [Green Version]
- Recchi, C.; Seabra, M.C. Novel functions for Rab GTPases in multiple aspects of tumour progression. Biochem. Soc. Trans. 2012, 40, 1398–1403. [Google Scholar] [CrossRef] [Green Version]
- Bucci, C.; Thomsen, P.; Nicoziani, P.; McCarthy, J.; Van Deurs, B. Rab7: A Key to Lysosome Biogenesis. Mol. Biol. Cell 2000, 11, 467–480. [Google Scholar] [CrossRef]
- Schmitz, K.J.; Ademi, C.; Bertram, S.; Schmid, K.W.; Baba, H.A. Prognostic relevance of autophagy-related markers LC3, p62/sequestosome 1, Beclin-1 and ULK1 in colorectal cancer patients with respect to KRAS mutational status. World J. Surg. Oncol. 2016, 14, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamb, C.A.; Dooley, H.C.; Tooze, S.A. Endocytosis and autophagy: Shared machinery for degradation. BioEssays 2012, 35, 34–45. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Arozena, A.A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [Green Version]
- Amaravadi, R.; Kimmelman, A.C.; White, E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016, 30, 1913–1930. [Google Scholar] [CrossRef]
- Levy, J.M.M.; Thorburn, A. Autophagy in cancer: Moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 2020, 27, 843–857. [Google Scholar] [CrossRef]
- Kimmelman, A.C. The dynamic nature of autophagy in cancer. Genes Dev. 2011, 25, 1999–2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhutia, S.K.; Mukhopadhyay, S.; Sinha, N.; Das, D.N.; Panda, P.K.; Patra, S.K.; Maiti, T.K.; Mandal, M.; Dent, P.; Wang, X.Y.; et al. Autophagy: Cancer’s friend or foe? Adv. Cancer Res. 2013, 118, 61–95. [Google Scholar]
- Hu, Y.-L.; Jahangiri, A.; DeLay, M.; Aghi, M.K. Tumor Cell Autophagy as an Adaptive Response Mediating Resistance to Treatments Such as Antiangiogenic Therapy: Figure. Cancer Res. 2012, 72, 4294–4299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, A.; Kimura, T.; Takabatake, Y.; Namba, T.; Kaimori, J.; Kitamura, H.; Matsui, I.; Niimura, F.; Matsusaka, T.; Fujita, N. Autophagy guards against cispla-tin-induced acute kidney injury. Am. J. Pathol. 2012, 180, 517–525. [Google Scholar] [CrossRef]
- Carew, J.S.; Nawrocki, S.T.; Kahue, C.N.; Zhang, H.; Yang, C.; Chung, L.; Houghton, J.A.; Huang, P.; Giles, F.J.; Cleveland, J.L. Targeting autophagy augments the anti-cancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood 2007, 110, 313–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Yu, D.-D.; Yan, F.; Jing, Y.-Y.; Han, Z.-P.; Sun, K.; Liang, L.; Hou, J.; Wei, L.-X. The role of autophagy induced by tumor microenvironment in different cells and stages of cancer. Cell Biosci. 2015, 5, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levine, B.; Kroemer, G. Autophagy in the Pathogenesis of Disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Zacharek, A.; Salkowski, A.; Grignon, D.J.; Sakr, W.; Porter, A.T.; Honn, K.V. Loss of heterozygosity of the BRCA1 and other loci on chromosome 17q in human prostate cancer. Cancer Res. 1995, 55, 1002–1005. [Google Scholar] [PubMed]
- Zhang, M.Y.; Wang, L.Y.; Zhao, S.; Guo, X.C.; Xu, Y.Q.; Zheng, Z.H.; Lu, H.; Zheng, H.C. Effects of Beclin 1 overexpres-sion on aggressive phenotypes of colon cancer cells. Oncol Lett. 2019, 17, 2441–2450. [Google Scholar]
- McKnight, N.C.; Yue, Z. Beclin 1, an Essential Component and Master Regulator of PI3K-III in Health and Disease. Curr. Pathobiol. Rep. 2013, 1, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Baehrecke, E.H. Autophagy, cell death, and cancer. Trends Stem Cell Prolif. Cancer Res. 2015, 2.3, e985913. [Google Scholar] [CrossRef] [Green Version]
- Takamura, A.; Komatsu, M.; Hara, T.; Sakamoto, A.; Kishi, C.; Waguri, S.; Eishi, Y.; Hino, O.; Tanaka, K.; Mizushima, N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011, 25, 795–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koustas, E.; Sarantis, P.; Kyriakopoulou, G.; Papavassiliou, A.G.; Karamouzis, M.V. The Interplay of Autophagy and Tumor Microenvironment in Colorectal Cancer—Ways of Enhancing Immunotherapy Action. Cancers 2019, 11, 533. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Wang, L.; Ou, J.-H.J. Autophagy, a double-edged sword in hepatocarcinogenesis. Mol. Cell. Oncol. 2015, 2, e1004968. [Google Scholar] [CrossRef] [Green Version]
- Koustas, E.; Sarantis, P.; Papavassiliou, A.G.; Karamouzis, M.V. Upgraded role of autophagy in colorectal carcinomas. World J. Gastrointest. Oncol. 2018, 10, 367–369. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhou, Y.; Sun, Q.; Zhou, J.; Pan, H.; Sui, X. Regulation of Autophagy by MiRNAs and Their Emerging Roles in Tumorigenesis and Cancer Treatment. Int. Rev. Cell Mol. Biol. 2017, 334, 1–26. [Google Scholar] [CrossRef]
- Kim, Y.; Kang, Y.S.; Lee, N.Y.; Kim, K.Y.; Hwang, Y.J.; Kim, H.W.; Rhyu, I.J.; Her, S.; Jung, M.K.; Kim, S.; et al. Uvrag targeting by Mir125a and Mir351 modu-lates autophagy associated with Ewsr1 deficiency. Autophagy 2015, 11, 796–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; He, Y.; Qiu, X.; Yang, W.; Liu, W.; Li, X.; Li, Y.; Shen, H.-M.; Wang, R.; Yue, Z.; et al. Targeting the potent Beclin 1–UVRAG coiled-coil interaction with designed peptides enhances autophagy and endolysosomal trafficking. Proc. Natl. Acad. Sci. USA 2018, 115, E5669–E5678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, C.; Yu, X.; Liu, X.; Zhan, P.; Nie, T.; Guo, R.; Liu, H.; Dai, M.; Zhang, B. Beclin-1 knockdown decreases proliferation, invasion and migra-tion of ewing sarcoma SK-ES-1 cells via inhibition of MMP-9. Oncol. Lett. 2018, 15, 3221–3225. [Google Scholar]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef]
- Lu, Q.; Zhang, Y.; Ma, L.; Li, D.; Li, M.; Li, J.; Liu, P. EWS-FLI1 positively regulates autophagy by increasing ATG4B ex-pression in Ewing sarcoma cells. Int. J. Mol. Med. 2017, 40, 1217–1225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iurlaro, R.; Muñoz-Pinedo, C. Cell death induced by endoplasmic reticulum stress. FEBS J. 2016, 283, 2640–2652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldstein, J.C.; Muñoz-Pinedo, C.; Ricci, J.E.; Adams, S.R.; Kelekar, A.; Schuler, M.; Tsien, R.Y.; Green, D.R. Cytochrome c is released in a single step during apoptosis. Cell Death Differ. 2005, 12, 453–462. [Google Scholar] [CrossRef]
- Nandy, A.; Lin, L.; Velentzas, P.D.; Wu, L.P.; Baehrecke, E.H.; Silverman, N. The NF-κB Factor Relish Regulates Atg1 Expression and Controls Autophagy. Cell Rep. 2018, 25, 2110–2120. [Google Scholar] [CrossRef] [Green Version]
- Trocoli, A.; Djavaheri-Mergny, M. The complex interplay between autophagy and NF-κB signaling pathways in cancer cells. Am. J. Cancer Res. 2011, 1, 629–649. [Google Scholar] [PubMed]
- Djavaheri-Mergny, M.; Amelotti, M.; Mathieu, J.; Besançon, F.; Bauvy, C.; Souquère, S.; Pierron, G.; Codogno, P. NF-κB activation represses tumor necrosis factor-α-induced autophagy. J. Biol. Chem. 2006, 281, 30373–30382. [Google Scholar] [CrossRef] [Green Version]
- Paquette, M.; El-Houjeiri, L.; Pause, A. mTOR Pathways in Cancer and Autophagy. Cancers 2018, 10, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noorolyai, S.; Shajari, N.; Baghbani, E.; Sadreddini, S.; Baradaran, B. The relation between PI3K/AKT signalling pathway and cancer. Gene 2019, 698, 120–128. [Google Scholar] [CrossRef]
- Rodgers, S.J.; Ferguson, D.T.; Mitchell, C.A.; Ooms, L.M. Regulation of PI3K effector signalling in cancer by the phospho-inositide phosphatases. Biosci. Rep. 2017, 37, BSR20160432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, M.; Gomez, N.C.; McFadden, A.W.; Moats-Staats, B.M.; Wu, S.; Rojas, A.; Sapp, T.; Simon, J.M.; Smith, S.V.; Kaiser-Rogers, K.; et al. PTEN deficiency mediates a reciprocal response to IGFI and mTOR inhibition. Mol. Cancer Res. 2014, 12, 1610–1620. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.; Zhang, Y.; Ma, L.; Li, D.; Li, M.; Liu, P.; Li, J. TRIM3 Negatively Regulates Autophagy Through Promoting Degradation of Beclin1 in Ewing Sarcoma Cells. OncoTargets Ther. 2019, 12, 11587–11595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorin, S.; Borges, A.; Dos Santos, L.R.; Souquère, S.; Pierron, G.; Ryan, K.M.; Codogno, P.; Djavaheri-Mergny, M. c-Jun NH2-terminal kinase activation is essential for DRAM-dependent induction of autophagyand apoptosis in 2-methoxyestradiol-treated ewing sarcoma cells. Cancer Res. 2009, 69, 6924–6931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vakifahmetoglu-Norberg, H.; Xia, H.-G.; Yuan, J. Pharmacologic agents targeting autophagy. J. Clin. Investig. 2015, 125, 5–13. [Google Scholar] [CrossRef] [Green Version]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef]
- Janji, B.; Berchem, G.; Chouaib, S. Targeting autophagy in the tumor microenvironment: New challenges and opportuni-ties for regulating tumor immunity. Front. Immunol. 2018, 9, 887. [Google Scholar] [CrossRef]
- Onorati, A.V.; Dyczynski, M.; Ojha, R.; Amaravadi, R.K. Targeting autophagy in cancer. Cancer 2018, 124, 3307–3318. [Google Scholar] [CrossRef] [Green Version]
- Veldhoen, R.A.; Banman, S.L.; Hemmerling, D.R.; Odsen, R.; Simmen, T.; Simmonds, A.J.; Underhill, D.A.; Goping, I.S. The chemotherapeutic agent paclitaxel inhibits autophagy through two distinct mechanisms that regulate apoptosis. Oncogene 2012, 32, 736–746. [Google Scholar] [CrossRef] [Green Version]
- Moreno, L.; Casanova, M.; Chisholm, J.C.; Berlanga, P.; Chastagner, P.B.; Baruchel, S.; Amoroso, L.; Melcón, S.G.; Gerber, N.U.; Bisogno, G.; et al. Phase I results of a phase I/II study of weekly nab-paclitaxel in paediatric patients with recurrent/refractory solid tumours: A collaboration with inno-vative therapies for children with cancer. Eur. J. Cancer 2018, 100, 27–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koustas, E.; Sarantis, P.; Theoharis, S.; Saetta, A.A.; Chatziandreou, I.; Kyriakopoulou, G.; Giannopoulou, I.; Michelli, M.; Schizas, D.; Papavassiliou, A.G.; et al. Autophagy-related Pro-teins as a Prognostic Factor of Patients with Colorectal Cancer. Am. J. Clin. Oncol. 2019, 42, 767–776. [Google Scholar] [CrossRef] [Green Version]
- Rosenfeld, M.R.; Ye, X.; Supko, J.G.; Desideri, S.; Grossman, S.A.; Brem, S.; Mikkelson, T.; Wang, D.; Chang, Y.C.; Hu, J.; et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed gli-oblastoma multiforme. Autophagy 2014, 10, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Thornton, K.A.; Chen, A.R.; Trucco, M.M.; Shah, P.; Wilky, B.A.; Gul, N.; Carrera-Haro, M.A.; Ferreira, M.F.; Shafique, U.; Powell, J.D.; et al. A Dose Finding Study of Temsirolimus and Liposomal Doxorubicin for Patients with Recurrent and Refractory Bone and Soft Tissue Sarcoma. Int. J. Cancer 2013, 133, 997–1005. [Google Scholar] [CrossRef] [Green Version]
- Trucco, M.M.; Meyer, C.F.; Thornton, K.A.; Shah, P.; Chen, A.R.; Wilky, B.A.; Carrera-Haro, M.A.; Boyer, L.C.; Ferreira, M.F.; Shafique, U.; et al. A phase II study of temsirolimus and liposomal doxorubicin for patients with recurrent and refractory bone and soft tissue sarcomas. Clin. Sarcoma Res. 2018, 8, 21. [Google Scholar] [CrossRef] [PubMed]
| Number of Study | Intervention/Treatment | Autophagy Modulator and Target | Phase of Study | Other Cancers |
|---|---|---|---|---|
| NCT03275818 | Nab-paclitaxel | nab paclitaxel: Inhibit phosphorylation of VPS34 | II | NCT03344172: Pancreatic cancer, gemcitabine, nab-paclitaxel, HCQ and avelumab |
| NCT02945800 | Nab-paclitaxel+gemcitabine | nab paclitaxel: Inhibit phosphorylation of VPS34 | II | NCT01649947: NSCLC, Drug: paclitaxel, carboplatin, HCQ, bevacizumab |
| NCT01962103 | Nab-paclitaxel | nab paclitaxel: Inhibit phosphorylation of VPS34 | I/II | NCT00728845: NSCLC, Biological: bevacizumab drug: carboplatin, HCQ, paclitaxel |
| NCT03507491 | Nab-paclitaxel+gemcitabine | nab paclitaxel: Inhibit phosphorylation of VPS34 | I | |
| NCT00002854 | etoposide, cisplatin, and cyclophosphamide followed by ifosfamide, carboplatin, and paclitaxel | nab paclitaxel: Inhibit phosphorylation of VPS34 | I | |
| NCT03190174 | Nab-rapamycin | nab rapamycin: Inhibitor of mTORC1 | I | NCT01396200: Myeloma, drug: HCQ, rapamycin |
| NCT03245151 | Everolimus | everolimus: inhibitor of mTORC1 | II | NCT01510119: Renal cell carcinoma, Drug: HCQ RAD001 (everolimus) |
| NCT00949325 | Temsirolimus + lip. doxorubicin | temsirolimus: inhibitor of mTORC1 | I/II | NCT00909831: Metastatic solid tumors, drug: HCQ, temsirolimus |
| NCT01016769: Squamous cell cancer, head and neck cancer, drug: temsirolimus, paclitaxel, carboplatin |
| Agents | Mechanism | Target |
|---|---|---|
| Chloroquine (CQ) | Neutralizes the acidic pH of intracellular vesicles | Lysosome |
| Hydroxy-chloroquine (HCQ) | CQ derivative | Lysosome |
| 3-Methyladenine (3-MA) | Inhibitor of PI3K Class I and III | Autophagosome formation |
| Wortmannin | Inhibitor of PI3K Class I and III | Autophagosome formation |
| LY294002 | PI3-kinase inhibitor | Autophagosome formation |
| LY3023414 | PI3-kinase and mTOR inhibitor | Autophagosome formation |
| SAR405 | (Vps18 and Vps34) inhibitor | Autophagosome formation |
| SB203580 | Inhibitor of p38α and p38β. p38α inhibits trafficking of Atg9 | Autophagosome formation |
| Bafilomycin A1 | Inhibition of lysosomal acidification | Lysosome |
| Concanamycin A | Inhibition of lysosomal acidification | Lysosome |
| Azithromycin | Inhibition of lysosomal acidification | Lysosome |
| Paclitaxel | Microtubule stabilizer inhibits phosphorylation of VPS34 | Autophagosome formation |
| SAHA | Inhibit autophagosome–lysosome fusion | Autophagosome formation |
| Monensin | Inhibit autophagosome–lysosome fusion | Autophagosome formation |
| Sputin-1 | (USP10) and (USP13) inhibitor | Autophagosome formation |
| NSC185058 | Inhibitor of ATG4B | Autophagosome formation |
| Verteporfin | Alter acidification of lysosomes | Autophagosome formation |
| Agents | Mechanism | Target |
|---|---|---|
| Rapamycin | Inhibitor of mTORC1 | Autophagosome formation |
| Temsirolimus | Inhibitor of mTORC1 | Autophagosome formation |
| Deforolimus | Inhibitor of mTORC1 | Autophagosome formation |
| Everolimus | Inhibitor of mTORC1 | Autophagosome formation |
| Metformin | AMPK activator | Autophagosome formation |
| GDC-0980 | PI3K and mTORC1 inhibitor | Autophagosome formation |
| GDC-0941 | Inhibitor of PI3K Class I | Autophagosome formation |
| fluspirilene | Antagonists of L-type Ca2+ channels | Lysosome |
| Perifosine | AKT inhibitior | Autophagosome formation |
| Tat–Beclin 1 peptide | Releases Beclin-1 into cytoplasm | Autophagosome formation |
| isoliensinine | Natural alkaloid | Autophagic flux |
| cepharanthine | Natural alkaloid | Autophagic flux |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koustas, E.; Sarantis, P.; Karamouzis, M.V.; Vielh, P.; Theocharis, S. The Controversial Role of Autophagy in Ewing Sarcoma Pathogenesis—Current Treatment Options. Biomolecules 2021, 11, 355. https://doi.org/10.3390/biom11030355
Koustas E, Sarantis P, Karamouzis MV, Vielh P, Theocharis S. The Controversial Role of Autophagy in Ewing Sarcoma Pathogenesis—Current Treatment Options. Biomolecules. 2021; 11(3):355. https://doi.org/10.3390/biom11030355
Chicago/Turabian StyleKoustas, Evangelos, Panagiotis Sarantis, Michalis V. Karamouzis, Philippe Vielh, and Stamatios Theocharis. 2021. "The Controversial Role of Autophagy in Ewing Sarcoma Pathogenesis—Current Treatment Options" Biomolecules 11, no. 3: 355. https://doi.org/10.3390/biom11030355
APA StyleKoustas, E., Sarantis, P., Karamouzis, M. V., Vielh, P., & Theocharis, S. (2021). The Controversial Role of Autophagy in Ewing Sarcoma Pathogenesis—Current Treatment Options. Biomolecules, 11(3), 355. https://doi.org/10.3390/biom11030355










