Deep Learning for Novel Antimicrobial Peptide Design
Abstract
:1. Introduction
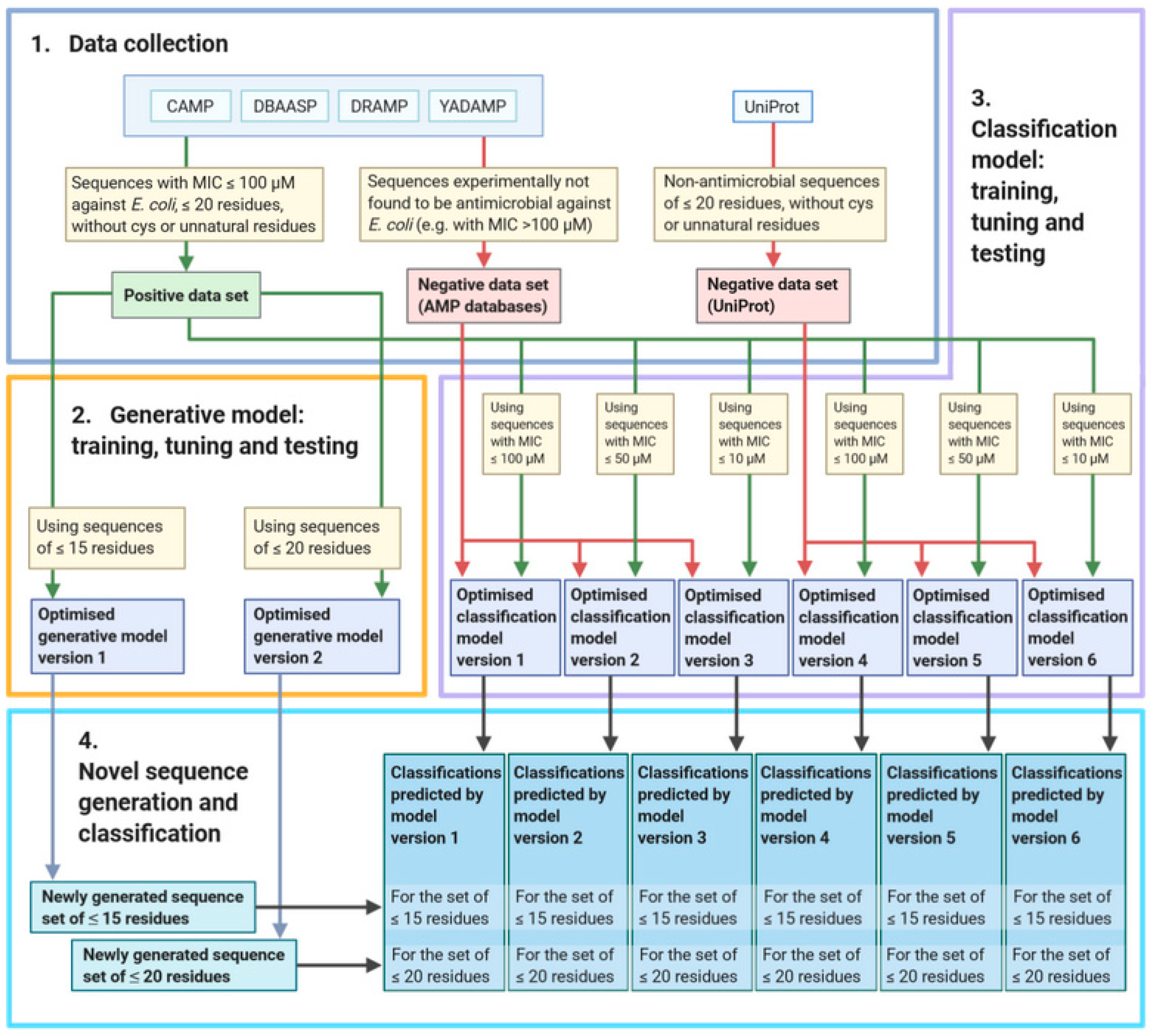
2. Materials and Methods
2.1. Data Set Collection
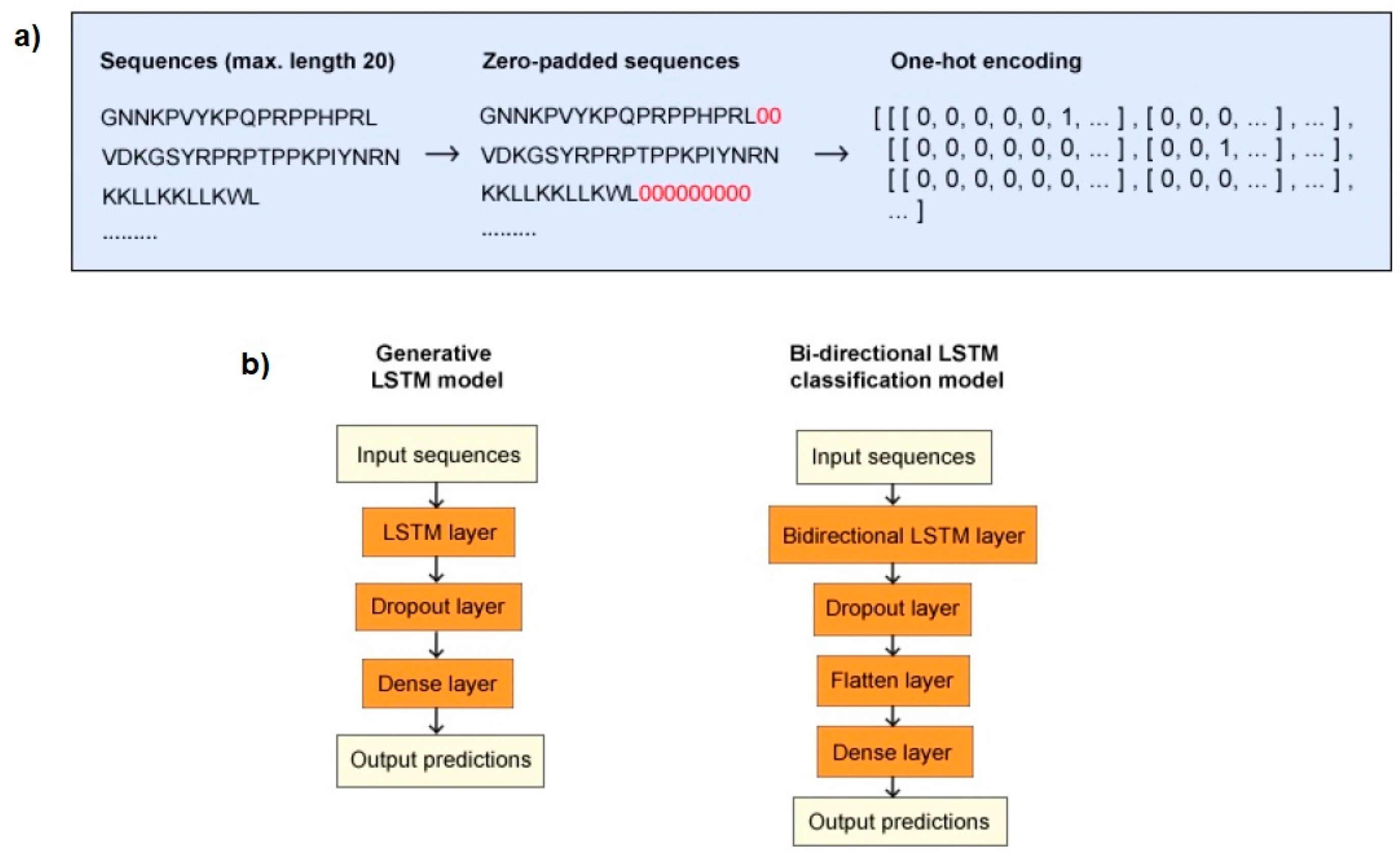
2.2. Data Processing
2.3. Model Structures and Training
2.3.1. Generative Model
2.3.2. Classification Model
2.4. Model Tuning
2.5. Model Evaluation
2.6. Data Generation and Classification
2.7. Further Analysis and 3D Structure Generation
2.8. Experimental Setup
3. Results
3.1. Data Sets
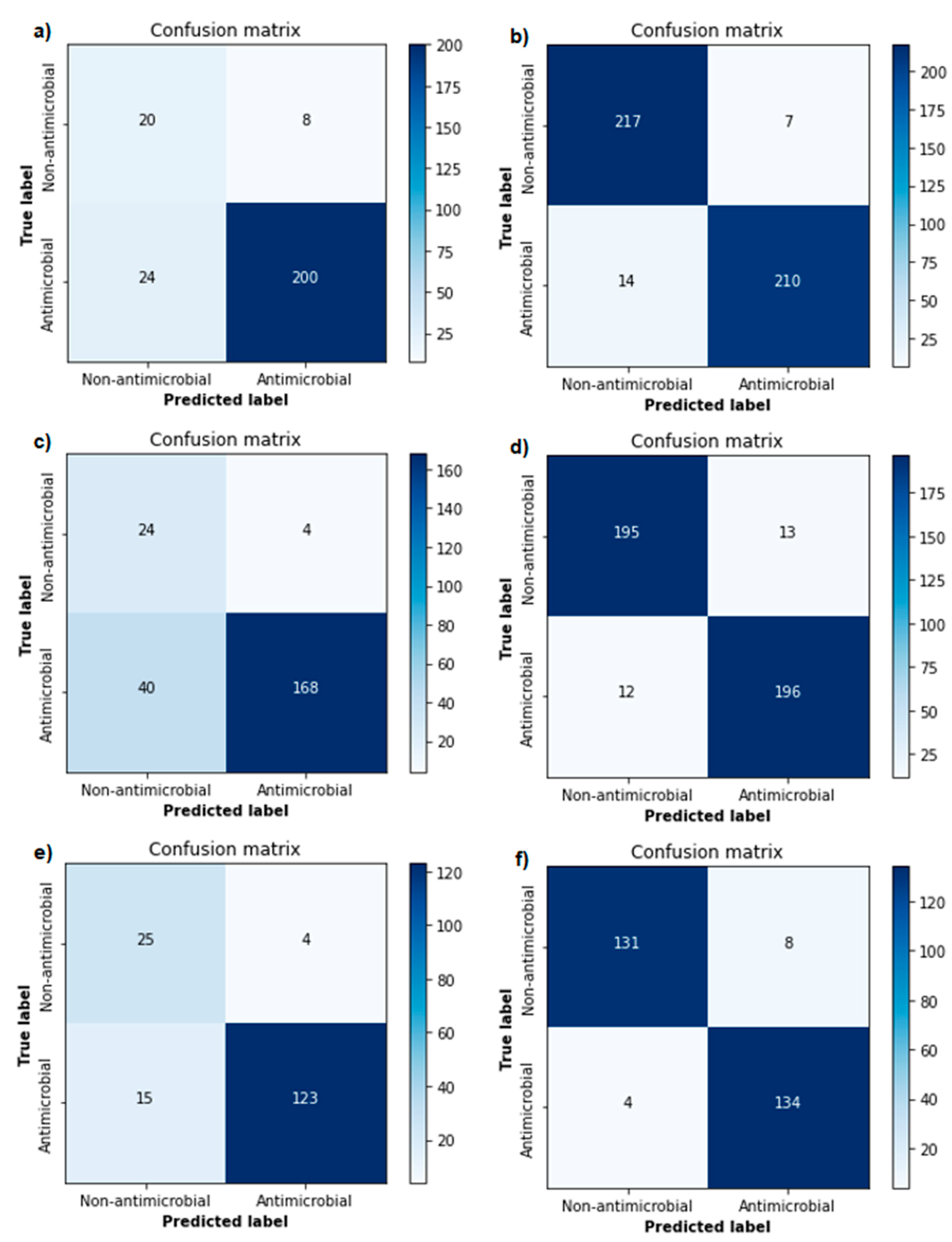
3.2. Classification Model Tuning, Evaluation and Validation
3.3. Novel Sequence Generation and Classification
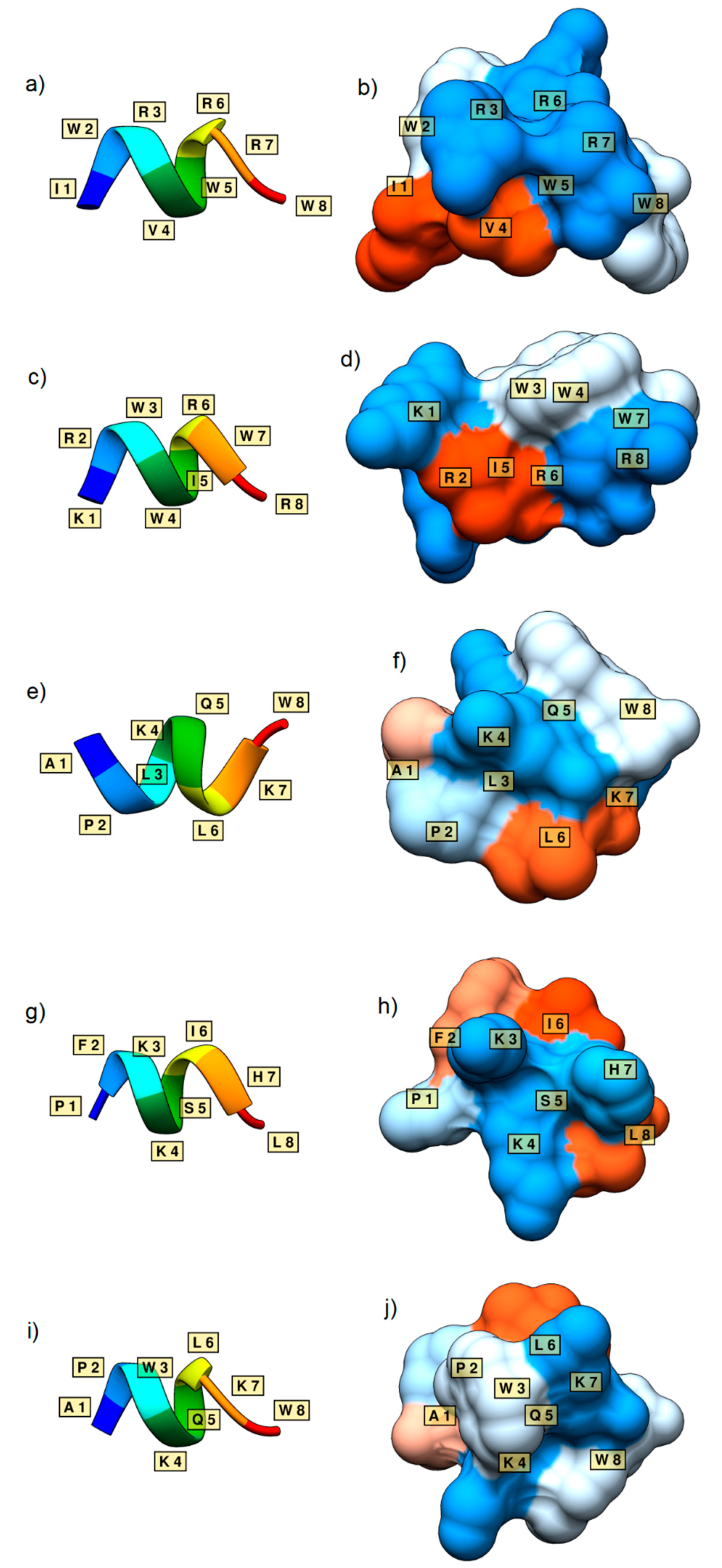
3.4. Further Analysis and Generated 3D Structures
4. Discussion
Future Perspective in Clinical Application
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Petrosillo, N. Infections: The Emergency of the New Millennium. In Nuclear Medicine in Infectious Diseases; Signore, A., Glaudemans, A.W.J.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–8. [Google Scholar]
- O’Neil, J. Tackling drug-resistant infections globally: Final report and recommendations. In Review on Antimicrobial Resistance; AMR-Review: London, UK, 2016; pp. 1–84. [Google Scholar]
- Sohrabi, C.; Alsafi, Z.; O’Neill, N.; Khan, M.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, R. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 2020, 76, 71–76. [Google Scholar] [CrossRef]
- International Severe Acute Respiratory and Emerging Infection Consortium. COVID-19 Report. 2020. Available online: https://media.tghn.org/medialibrary/2020/04/ISARIC_Data_Platform_COVID-19_Report_8APR20.pdf (accessed on 10 June 2020).
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Rawson, T.M.; Moore, L.S.P.; Castro-Sanchez, E.; Charani, E.; Davies, F.; Satta, G.; Ellington, M.J.; Holmes, A.H. COVID-19 and the potential long-term impact on antimicrobial resistance. J. Antimicrob. Chemother. 2020, 75, 1681–1684. [Google Scholar] [CrossRef]
- World Health Organization. 2019 Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moghaddam, M.M.; Mirnejad, R. Antimicrobial Peptides: Features, Action, and Their Resistance Mechanisms in Bacteria. Microb. Drug Resist. 2018, 24, 747–767. [Google Scholar] [CrossRef]
- Pushpanathan, M.; Gunasekaran, P.; Rajendhran, J. Antimicrobial Peptides: Versatile Biological Properties. Int. J. Pept. 2013, 2013, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Rational Design of α-Helical Antimicrobial Peptides with Enhanced Activities and Specificity/Therapeutic Index. J. Biol. Chem. 2005, 280, 12316–12329. [Google Scholar] [CrossRef] [Green Version]
- Passarini, I.; Rossiter, S.; Malkinson, J.; Zloh, M. In Silico Structural Evaluation of Short Cationic Antimicrobial Peptides. Pharmaceutics 2018, 10, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfalzgraff, A.; Brandenburg, K.; Weindl, G. Antimicrobial Peptides and Their Therapeutic Potential for Bacterial Skin Infections and Wounds. Front. Pharmacol. 2018, 9, 281. [Google Scholar] [CrossRef]
- Koprivnjak, T.; Peschel, A. Bacterial resistance mechanisms against host defense peptides. Cell. Mol. Life Sci. 2011, 68, 2243–2254. [Google Scholar] [CrossRef]
- Marr, A.K.; Gooderham, W.J.; Hancock, R.E. Antibacterial peptides for therapeutic use: Obstacles and realistic outlook. Curr. Opin. Pharmacol. 2006, 6, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-C.; Park, Y.; Hahm, K.-S. The Role of Antimicrobial Peptides in Preventing Multidrug-Resistant Bacterial Infections and Biofilm Formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-T.; Lee, C.-C.; Yang, J.-R.; Lai, J.Z.C.; Chang, K.Y. A Large-Scale Structural Classification of Antimicrobial Peptides. BioMed Res. Int. 2015, 2015, 1–6. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [Green Version]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides: Table 1. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirtskhalava, M.; Gabrielian, A.; Cruz, P.; Griggs, H.L.; Squires, R.B.; Hurt, D.E.; Grigolava, M.; Chubinidze, M.; Gogoladze, G.; Vishnepolsky, B.; et al. DBAASP v.2: An enhanced database of structure and antimicrobial/cytotoxic activity of natural and synthetic peptides. Nucleic Acids Res. 2016, 44, D1104–D1112. [Google Scholar] [CrossRef]
- Kang, X.; Dong, F.; Shi, C.; Liu, S.; Sun, J.; Chen, J.; Li, H.; Xu, H.; Lao, X.; Zheng, H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 2019, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Witten, J.; Witten, Z. Deep learning regression model for antimicrobial peptide design. bioRxiv 2019, 692681. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Wu, H.; Lu, H.; Li, G.; Huang, Q. LAMP: A Database Linking Antimicrobial Peptides. PLoS ONE 2013, 8, e66557. [Google Scholar] [CrossRef] [Green Version]
- Piotto, S.P.; Sessa, L.; Concilio, S.; Iannelli, P. YADAMP: Yet another database of antimicrobial peptides. Int. J. Antimicrob. Agents 2012, 39, 346–351. [Google Scholar] [CrossRef]
- Meher, P.K.; Sahu, T.K.; Saini, V.; Rao, A.R. Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC. Sci. Rep. 2017, 7, srep42362. [Google Scholar] [CrossRef]
- Lata, S.; Sharma, B.; Raghava, G. Analysis and prediction of antibacterial peptides. BMC Bioinform. 2007, 8, 263. [Google Scholar] [CrossRef] [Green Version]
- Lata, S.; Mishra, N.K.; Raghava, G.P.S. AntiBP2: Improved version of antibacterial peptide prediction. BMC Bioinform. 2010, 11 (Suppl. S1), S19. [Google Scholar] [CrossRef] [Green Version]
- Torrent, M.; Nogués, V.M.; Boix, E. A theoretical approach to spot active regions in antimicrobial proteins. BMC Bioinform. 2009, 10, 373. [Google Scholar] [CrossRef] [Green Version]
- Torrent, M.; Andreu, D.; Nogués, V.M.; Boix, E. Connecting Peptide Physicochemical and Antimicrobial Properties by a Rational Prediction Model. PLoS ONE 2011, 6, e16968. [Google Scholar] [CrossRef]
- Torrent, M.; Di Tommaso, P.; Pulido, D.; Nogués, M.V.; Notredame, C.; Boix, E.; Andreu, D. AMPA: An automated web server for prediction of protein antimicrobial regions. Bioinformatics 2012, 28, 130–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Hu, L.; Liu, G.; Jiang, N.; Chen, X.; Xu, J.; Zheng, W.; Li, L.; Tan, M.; Chen, Z.; et al. Prediction of Antimicrobial Peptides Based on Sequence Alignment and Feature Selection Methods. PLoS ONE 2011, 6, e18476. [Google Scholar] [CrossRef]
- Khosravian, M.; Faramarzi, F.K.; Beigi, M.M.; Behbahani, M.; Mohabatkar, H. Predicting Antibacterial Peptides by the Concept of Chou’s Pseudo-amino Acid Composition and Machine Learning Methods. Protein Pept. Lett. 2013, 20, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Wang, P.; Lin, W.-Z.; Jia, J.-H.; Chou, K.-C. iAMP-2L: A two-level multi-label classifier for identifying antimicrobial peptides and their functional types. Anal. Biochem. 2013, 436, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, P.; Yan, J.; Li, J.; Fong, S.; Siu, S.W.I. AmPEP: Sequence-based prediction of antimicrobial peptides using distribution patterns of amino acid properties and random forest. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Joseph, S.; Karnik, S.; Nilawe, P.; Jayaraman, V.K.; Idicula-Thomas, S. ClassAMP: A Prediction Tool for Classification of Antimicrobial Peptides. IEEE/ACM Trans. Comput. Biol. Bioinform. 2012, 9, 1535–1538. [Google Scholar] [CrossRef]
- Fjell, C.D.; Jenssen, H.; Hilpert, K.; Cheung, W.A.; Panté, N.; Hancock, R.E.W.; Cherkasov, A. Identification of Novel Antibacterial Peptides by Chemoinformatics and Machine Learning. J. Med. Chem. 2009, 52, 2006–2015. [Google Scholar] [CrossRef]
- Porto, W.F.; Pires, Á.S.; Franco, O.L. CS-AMPPred: An Updated SVM Model for Antimicrobial Activity Prediction in Cysteine-Stabilized Peptides. PLoS ONE 2012, 7, e51444. [Google Scholar] [CrossRef]
- Veltri, D.; Kamath, U.; Shehu, A. Deep learning improves antimicrobial peptide recognition. Bioinformatics 2018, 34, 2740–2747. [Google Scholar] [CrossRef] [Green Version]
- Schneider, P.; Müller, A.T.; Gabernet, G.; Button, A.L.; Posselt, G.; Wessler, S.; Hiss, J.A.; Schneider, G. Hybrid Network Model for “Deep Learning” of Chemical Data: Application to Antimicrobial Peptides. Mol. Inform. 2016, 36, 1600011. [Google Scholar] [CrossRef]
- Su, X.; Xu, J.; Yin, Y.; Quan, X.; Zhang, H. Antimicrobial peptide identification using multi-scale convolutional network. BMC Bioinform. 2019, 20, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.; Moriwaki, Y.; Li, C.; Shimizu, K. Prediction of Antifungal Peptides by Deep Learning with Character Embedding. IPSJ Trans. Bioinform. 2019, 12, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Müller, A.T.; Hiss, J.A.; Schneider, G. Recurrent Neural Network Model for Constructive Peptide Design. J. Chem. Inf. Model. 2018, 58, 472–479. [Google Scholar] [CrossRef]
- Najafabadi, M.M.; Villanustre, F.; Khoshgoftaar, T.M.; Seliya, N.; Wald, R.; Muharemagic, E. Deep learning applications and challenges in big data analytics. J. Big Data 2015, 2, 1. [Google Scholar] [CrossRef] [Green Version]
- Nagarajan, D.; Nagarajan, T.; Roy, N.; Kulkarni, O.; Ravichandran, S.; Mishra, M.; Chakravortty, D.; Chandra, N. Computational antimicrobial peptide design and evaluation against multidrug-resistant clinical isolates of bacteria. J. Biol. Chem. 2018, 293, 3492–3509. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Qi, Z.; Zheng, S.; Xu, J.; Bao, H.; Xu, B. Text Classification Improved by Integrating Bidirectional LSTM with Two-dimensional Max Pooling. arXiv 2016, arXiv:1611.06639. [Google Scholar]
- Liu, G.; Guo, J. Bidirectional LSTM with attention mechanism and convolutional layer for text classification. Neurocomputing 2019, 337, 325–338. [Google Scholar] [CrossRef]
- World Health Organization. Antibacterial Agents in Preclinical Development: An Open Access Database; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO Press: Geneva, Switzerland, 2017. [Google Scholar]
- Centers for Disease Control and Prevention CDC. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019.
- Milo, R. What is the total number of protein molecules per cell volume? A call to rethink some published values. BioEssays 2013, 35, 1050–1055. [Google Scholar] [CrossRef] [Green Version]
- Weeks, A.M.; Wells, J.A. Engineering peptide ligase specificity by proteomic identification of ligation sites. Nat. Chem. Biol. 2018, 14, 50–57. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Ghojogh, B.; Crowley, M. The Theory Behind Overfitting, Cross Validation, Regularization, Bagging, and Boosting: Tutorial. arXiv 2019, arXiv:1905.12787. [Google Scholar]
- Dwarampudi, M.; Reddy, N. Effects of padding on LSTMs and CNNs. arXiv 2019, arXiv:1903.07288. [Google Scholar]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. Open Source Drug Discovery Consortium in Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [Green Version]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Fasterde novostructure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Chollet, F. Keras. GitHub. 2015. Available online: https://github.com/fchollet/keras (accessed on 31 January 2021).
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- O’Malley, T.; Bursztein, E.; Long, J.; Chollet, F.; Jin, H.; Invernizzi, L. Keras Tuner. Github. 2019. Available online: https://github.com/keras-team/keras-tuner (accessed on 31 January 2021).
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. JMLR 2011, 12, 2825–2830. [Google Scholar]
- McKinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar]
- Passarini, I.; De Resende, P.E.; Soares, S.; Tahmasi, T.; Stapleton, P.; Malkinson, J.; Zloh, M.; Rossiter, S. Synthesis and in Silico Modelling of the Potential Dual Mechanistic Activity of Small Cationic Peptides Potentiating the Antibiotic Novobiocin against Susceptible and Multi-Drug Resistant Escherichia coli. Int. J. Mol. Sci. 2020, 21, 9134. [Google Scholar] [CrossRef]
- Pitout, J.D.D. Extraintestinal pathogenicEscherichia coli: An update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev. Anti-Infect. Ther. 2012, 10, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Roldan-Masedo, E.; Sainz, T.; Gutierrez-Arroyo, A.; Gomez-Gil, R.M.; Ballesteros, E.; Escosa, L.; Baquero-Artigao, F.; Méndez-Echevarría, A. Risk factors for gentamicin-resistant E. coli in children with community-acquired urinary tract infection. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2097–2102. [Google Scholar] [CrossRef] [PubMed]
- CDC. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Surveillance Report for 2015 (Final Report); U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2018.
- Kidsley, A.K.; Abraham, S.; Bell, J.M.; O’Dea, M.; Laird, T.J.; Jordan, D.; Mitchell, P.; McDevitt, C.A.; Trott, D.J. Antimicrobial Susceptibility of Escherichia coli and Salmonella spp. Isolates From Healthy Pigs in Australia: Results of a Pilot National Survey. Front. Microbiol. 2018, 9, 1207. [Google Scholar] [CrossRef] [Green Version]
- Krawczyk, B. Learning from imbalanced data: Open challenges and future directions. Prog. Artif. Intell. 2016, 5, 221–232. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Wong, A.K.C.; Kamel, M.S. Classification of Imbalanced Data: A Review. Int. J. Pattern Recognit. Artif. Intell. 2009, 23, 687–719. [Google Scholar] [CrossRef]
- Kotsiantis, S.; Kanellopoulos, D.; Pintelas, P. Handling imbalanced datasets: A review. GESTS Int. Tran. Comput. Sci. Eng. 2005, 30, 25–36. [Google Scholar]
- Wu, J.; Chen, X.-Y.; Zhang, H.; Xiong, L.-D.; Lei, H.; Deng, S.-H. Hyperparameter Optimization for Machine Learning Models Based on Bayesian Optimizationb. JEST 2019, 17, 26–40. [Google Scholar] [CrossRef]
- Bergstra, J.; Yamins, D.; Cox, D.D. Making a Science of Model Search: Hyperparameter Optimization in Hundreds of Dimensions for Vision Architectures. In Proceedings of the 30th International Conference on International Conference on Machine Learning (ICML’13), Atlanta, GA, USA, 17–19 June 2013. [Google Scholar]
- Feurer, M.; Hutter, F. Hyperparameter Optimization. In Automated Machine Learning; Hutter, F., Kotthoff, L., Vanschoren, J., Eds.; Springer: Cham, Switzerland, 2019; pp. 3–33. [Google Scholar]
- Tucs, A.; Duy, T.; Yumoto, A.; Ito, Y.; Uzawa, T.; Tsuda, K. Generating Ampicillin-Level Antimicrobial Peptides with Activity-Aware Generative Adversarial Networks. ACS Omega 2020, 5, 22847–22851. [Google Scholar] [CrossRef]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Transformers: State-of-the-art Natural Language Processing. arXiv 2019, arXiv:1910.03771. [Google Scholar]
- Svenson, J.; Stensen, W.; Brandsdal, B.-O.; Haug, B.E.; Monrad, J.; Svendsen, J.S. Antimicrobial Peptides with Stability toward Tryptic Degradation. Biochemistry 2008, 47, 3777–3788. [Google Scholar] [CrossRef] [PubMed]
- Whitcomb, D.C.; Lowe, M.E. Human Pancreatic Digestive Enzymes. Dig. Dis. Sci. 2007, 52, 1–17. [Google Scholar] [CrossRef] [PubMed]
- McGregor, D.P. Discovering and improving novel peptide therapeutics. Curr. Opin. Pharmacol. 2008, 8, 616–619. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.; Calderon, J.; Rondón-Villarreal, P.; Torres Sáez, R. Analysis of Structure and Hemolytic Activity Relationships of Antimicrobial Peptides (AMPs). In Advances in Computational Biology; Castillo, L.F., Cristancho, M., Isaza, G., Pinzón, A., Rodríguez, J.M., Eds.; Springer International Publishing: Cham, Switzerland, 2014; Volume 232, pp. 253–258. [Google Scholar]
- Park, Y.; Park, S.-C.; Kim, J.-Y.; Park, J.O.; Seo, C.H.; Nah, J.-W.; Hahm, K.-S. In vitro efficacy of a synthetic all-d antimicrobial peptide against clinically isolated drug-resistant strains. Int. J. Antimicrob. Agents 2010, 35, 208–209. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Nan, Y.H.; Yang, S.-T.; Kang, S.W.; Kim, Y.; Park, I.-S.; Hahm, K.-S.; Shin, S.Y. Cell selectivity and anti-inflammatory activity of a Leu/Lys-rich α-helical model antimicrobial peptide and its diastereomeric peptides. Peptides 2010, 31, 1251–1261. [Google Scholar] [CrossRef]
- Park, W.; Na, K. Advances in the synthesis and application of nanoparticles for drug delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotech. 2015, 7, 494–508. [Google Scholar] [CrossRef]
- Chongsiriwatana, N.P.; Patch, J.A.; Czyzewski, A.M.; Dohm, M.T.; Ivankin, A.; Gidalevitz, D.; Zuckermann, R.N.; Barron, A.E. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2008, 105, 2794–2799. [Google Scholar] [CrossRef] [Green Version]
- Vagner, J.; Qu, H.; Hruby, V.J. Peptidomimetics, a synthetic tool of drug discovery. Curr. Opin. Chem. Biol. 2008, 12, 292–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuppusamy, R.; Willcox, M.; Black, D.S.; Kumar, N. Short Cationic Peptidomimetic Antimicrobials. Antibiotics 2019, 8, 44. [Google Scholar] [CrossRef] [Green Version]
- Barriere, S.L. Bacterial resistance to beta-lactams, and its prevention with combination antimicrobial therapy. Pharmacotherapy 1992, 12, 397–402. [Google Scholar]
- Wu, Y.L.; Scott, E.M.; Po, A.L.W.; Tariq, V.N. Ability of azlocillin and tobramycin in combination to delay or prevent resistance development in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 1999, 44, 389–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steenbergen, J.N.; Mohr, J.F.; Thorne, G.M. Effects of daptomycin in combination with other antimicrobial agents: A review of in vitro and animal model studies. J. Antimicrob. Chemother. 2009, 64, 1130–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.; Kim, H.J.; Hahm, K.-S. Antibacterial synergism of novel antibiotic peptides with chloramphenicol. Biochem. Biophys. Res. Commun. 2004, 321, 109–115. [Google Scholar] [CrossRef]
- Park, Y.; Park, S.N.; Park, S.-C.; Shin, S.O.; Kim, J.-Y.; Kang, S.-J.; Kim, M.-H.; Jeong, C.-Y.; Hahm, K.-S. Synergism of Leu–Lys rich antimicrobial peptides and chloramphenicol against bacterial cells. Biochim. Biophys. Acta 2006, 1764, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Giacometti, A.; Cirioni, O.; Kamysz, W.; D’Amato, G.; Silvestri, C.; Del Prete, M.S.; Lukasiak, J.; Scalise, G. In vitro activity and killing effect of the synthetic hybrid cecropin A–melittin peptide CA(1–7)M(2–9)NH2 on methicillin-resistant nosocomial isolates of Staphylococcus aureus and interactions with clinically used antibiotics. Diagn. Microbiol. Infect. Dis. 2004, 49, 197–200. [Google Scholar] [CrossRef]

| Classification Models (Negative Data Set, MIC Cut-Off) | VAL ACC (%, Using 0.5 Threshold) | AUC | Optimal Threshold | SENS (%) | SPEC (%) | PREC (%) | ACC (%) |
|---|---|---|---|---|---|---|---|
| Model Version 1 (AMP, ≤100 µM) | 88.9 | 0.823 | 1.000000 | 89.3 | 71.4 | 96.2 | 87.3 |
| Model Version 2 (AMP, ≤50 µM) | 88.5 | 0.862 | 0.999985 | 80.8 | 85.7 | 97.7 | 81.4 |
| Model Version 3 (AMP, ≤10 µM) 3 | 88.0 | 0.902 | 1.000000 | 89.1 | 86.2 | 96.9 | 88.6 |
| Model Version 4 (UniProt, ≤100 µM) | 85.3 | 0.982 | 0.379858 | 93.8 | 96.9 | 96.8 | 95.3 |
| Model Version 5 (UniProt, ≤50 µM) | 82.2 | 0.981 | 0.094870 | 94.2 | 93.8 | 93.8 | 94.0 |
| Model Version 6 (UniProt, ≤10 µM) | 81.6 | 0.981 | 0.700969 | 97.1 | 94.2 | 94.4 | 95.7 |
| Classification Models (Negative Data Set, MIC Cut-Off) | Generated Set with Sequences ≤ 15 Residues (% Antimicrobial) | Generated Set with Sequences ≤ 20 Residues (% Antimicrobial) |
|---|---|---|
| Model Version 1 (AMP, ≤100 µM) | 84.2 | 79.2 |
| Model Version 2 (AMP, ≤50 µM) | 80.7 | 76.2 |
| Model Version 3 (AMP, ≤10 µM) 3 | 72.6 | 72.5 |
| Model Version 4 (UniProt, ≤100 µM) | 86.7 | 70.7 |
| Model Version 5 (UniProt, ≤50 µM) | 91.7 | 73.6 |
| Model Version 6 (UniProt, ≤10 µM) | 78.5 | 70.6 |
| Sequence | Predicted Class by AMP Scanner | Prediction by ToxinPred | Alpha-Helical Structure? | Amphipathic Surfaces? | Novel? |
|---|---|---|---|---|---|
| RIHVIRWR | AMP | Non-toxic | No | Yes | Yes |
| IWRVWRRW | AMP | Non-toxic | Yes | Yes | Yes |
| APKNQLKW | Non-AMP | - | - | - | - |
| HRWWRWWR | AMP | Non-toxic | Yes | No | Yes |
| IRRWRRIW | AMP | Non-toxic | No | No | Yes |
| PYKISIHL | Non-AMP | - | - | - | - |
| KRWWIRWR | AMP | Non-toxic | Yes | No | Yes |
| APRRNVRW | AMP | Non-toxic | No | No | Yes |
| PFKISIHH | AMP | Non-toxic | No | No | Yes |
| RRKRWWRR | AMP | Non-toxic | Yes | No | Yes |
| APLKQLKW | AMP | Non-toxic | Yes | Yes | Yes |
| PFKKSIHL | AMP | Non-toxic | Yes | Yes | Yes |
| APWKQLKW | AMP | Non-toxic | Yes | Yes | Yes |
| RRRRFRRR | AMP | Non-toxic | No | Yes | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Garlick, S.; Zloh, M. Deep Learning for Novel Antimicrobial Peptide Design. Biomolecules 2021, 11, 471. https://doi.org/10.3390/biom11030471
Wang C, Garlick S, Zloh M. Deep Learning for Novel Antimicrobial Peptide Design. Biomolecules. 2021; 11(3):471. https://doi.org/10.3390/biom11030471
Chicago/Turabian StyleWang, Christina, Sam Garlick, and Mire Zloh. 2021. "Deep Learning for Novel Antimicrobial Peptide Design" Biomolecules 11, no. 3: 471. https://doi.org/10.3390/biom11030471
APA StyleWang, C., Garlick, S., & Zloh, M. (2021). Deep Learning for Novel Antimicrobial Peptide Design. Biomolecules, 11(3), 471. https://doi.org/10.3390/biom11030471







