Coupling Machine Learning and Lipidomics as a Tool to Investigate Metabolic Dysfunction-Associated Fatty Liver Disease. A General Overview
Abstract
:1. Introduction
2. NAFLD Is a Major Global Health Challenge: A Silent Pandemic
3. Lipidomics, A Latecomer Omics Technology, Is Being Consolidated
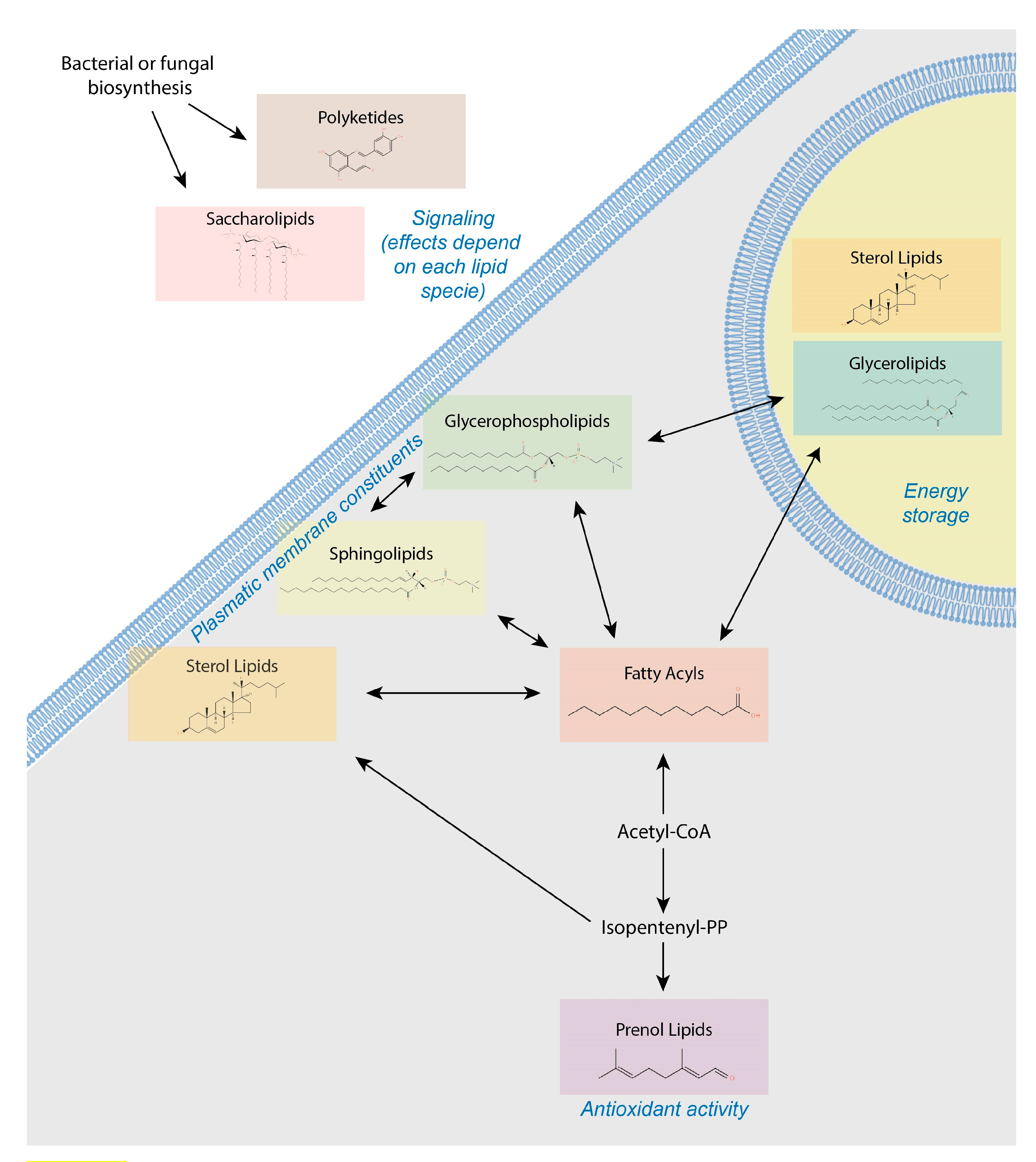
4. Lipids Form a Heterogeneous and Complex Group of Small Molecules
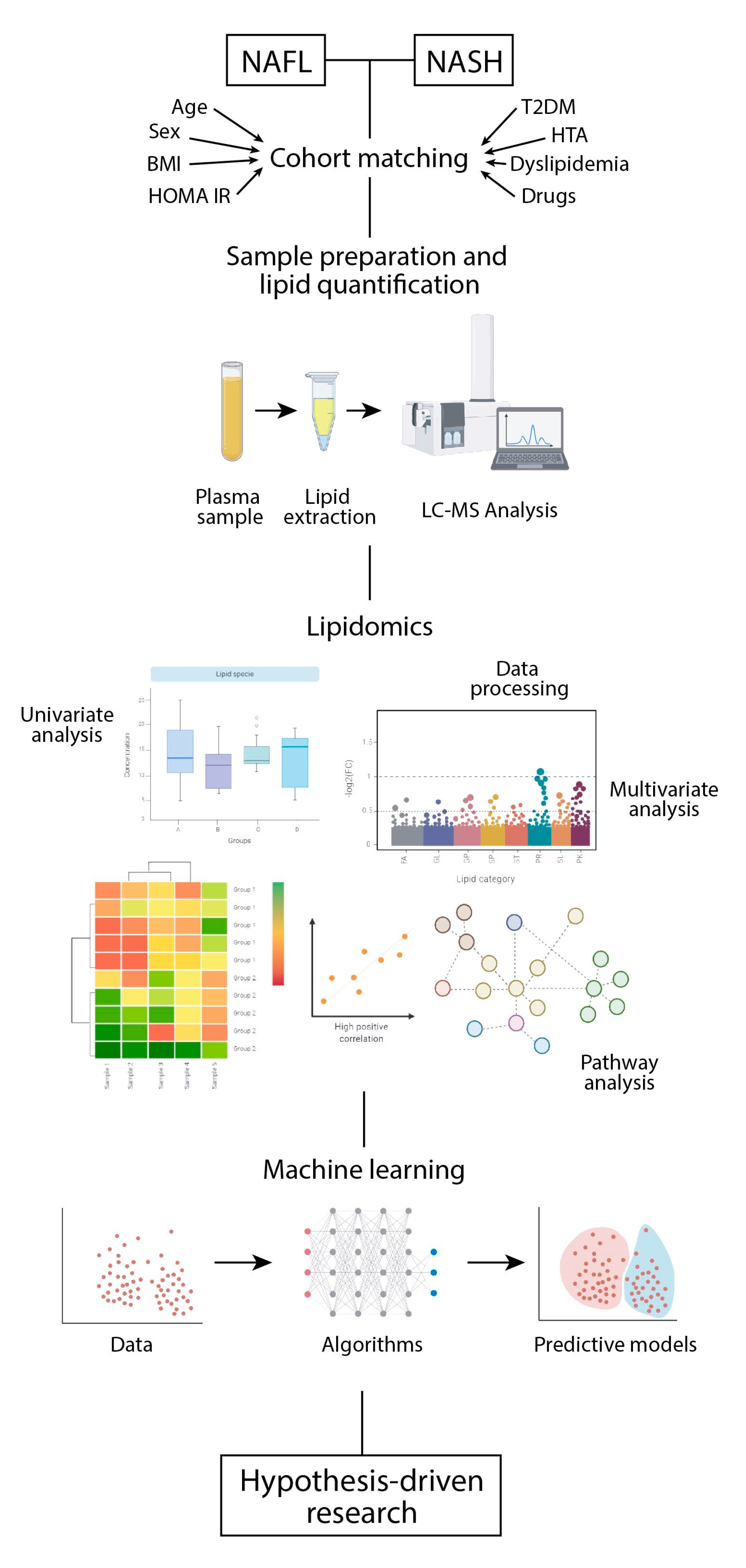
5. Sample Preparation and Systematic Error Removal
6. Addressing the Chemical Diversity of the Lipidome in a Biological System
7. Extracting the Relevant Information
8. Interorgan Communication in the Course of NAFLD
9. Can Lipidomics Provide Insights into the Pathogenesis of NAFLD?
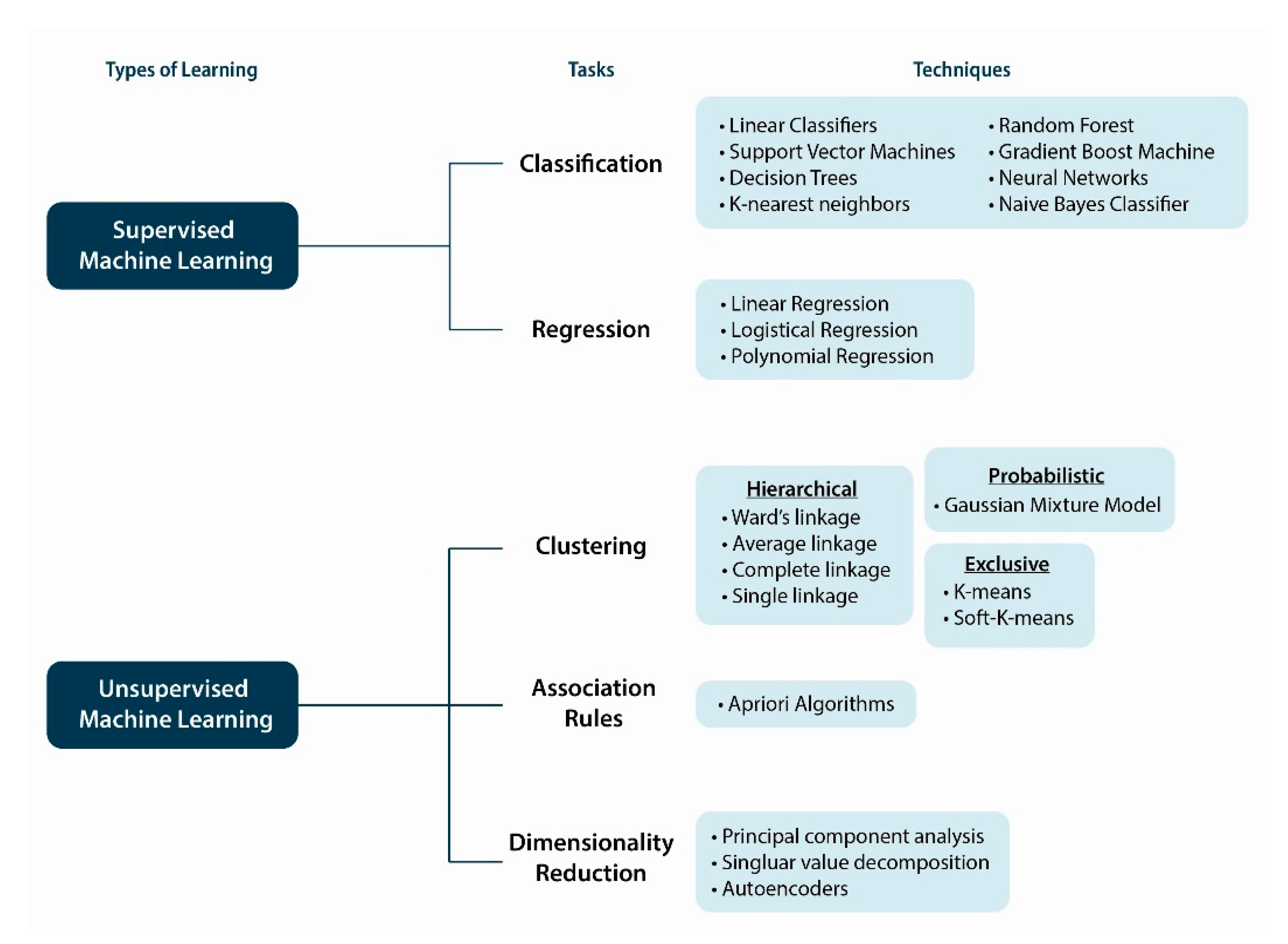
10. Machine Intelligence and Learning Approaches
11. Predicting the Risk of NASH with Lipidomics and Machine Learning
12. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Romby, P.; Charpentier, E. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell. Mol. Life Sci. 2009, 67, 217–237. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343. [Google Scholar] [CrossRef] [Green Version]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Dulai, P.S.; Singh, S.; Patel, J.; Soni, M.; Prokop, L.J.; Younossi, Z.; Loomba, R. Increased risk of mortality by fibrosis stage in non-alcoholic fatty liver disease: Systematic review and metaanalysis. Hepatology 2017, 65, 1557–1565. [Google Scholar] [CrossRef]
- Anstee, Q.M.; Targher, G.; Day, C.P. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Newsome, P.N.; Sarin, S.K.; Anstee, Q.M.; Targher, G.; Romero-Gomez, M.; Zelber-Sagi, S.; Wai-Sun Wong, V.; Dufour, J.F.; Schattenberg, J.M.; et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J. Hepatol. 2020, 73, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Metaanalytic assessment of prevalence, incidence and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [Green Version]
- McPherson, S.; Hardy, T.; Henderson, E.; Burt, A.D.; Day, C.P.; Anstee, Q.M. Evidence of NAFLD progression from steatosis to fibrosing-steatohepatitis using paired biopsies: Implications for prognosis and clinical management. J. Hepatol. 2015, 62, 1148–1155. [Google Scholar] [CrossRef] [PubMed]
- Dennis, A.; Mouchti, S.; Kelly, M.; Fallowfield, J.A.; Hirschfield, G.; Pavlides, M.; Banerjee, R. A composite biomarker using multiparametric magnetic resonance imaging and blood analytes accurately identifies patients with non-alcoholic steatohepatitis and significant fibrosis. Sci. Rep. 2020, 10, 15308. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2006, 35, D527–D532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puri, P.; Baillie, R.A.; Wiest, M.M.; Mirshahi, F.; Choudhury, J.; Cheung, O.; Sargeant, C.; Contos, M.J.; Sanyal, A.J. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007, 46, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Kartsoli, S.; Kostara, C.E.; Tsimihodimos, V.; Bairaktari, E.T.; Christodoulou, D.K. Lipidomics in non-alcoholic fatty liver disease. World J. Hepatol. 2020, 12, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Kikkawa, U. The story of PKC: A discovery marked by unexpected twists and turns. IUBMB Life 2018, 71, 697–705. [Google Scholar] [CrossRef]
- Sawada, N.; Obama, T.; Mizuno, M.; Fukuhara, K.; Iwamoto, S.; Aiuchi, T.; Makiyama, T.; Itabe, H. Transfer and enzyme-mediated metabolism of oxidized phosphatidylcholine and lysophosphatidylcholine between low and high-density lipoproteins. Antioxidants 2020, 9, 1045. [Google Scholar] [CrossRef]
- Lachkar, F.; Ferré, P.; Foufelle, F.; Papaioannou, A. Dihydroceramides: Their emerging physiological roles and functions in cancer and metabolic diseases. Am. J. Physiol. Metab. 2021, 320, E122–E130. [Google Scholar] [CrossRef]
- Pajed, L.; Wagner, C.; Taschler, U.; Schreiber, R.; Kolleritsch, S.; Fawzy, N.; Pototschnig, I.; Schoiswohl, G.; Pusch, L.-M.; Wieser, B.I.; et al. Hepatocyte-specific deletion of lysosomal acid lipase leads to cholesteryl ester but not triglyceride or retinyl ester accumulation. J. Biol. Chem. 2019, 294, 9118–9133. [Google Scholar] [CrossRef] [Green Version]
- Shannon, S.R.; Yu, J.; Defnet, A.E.; Bongfeldt, D.; Moise, A.R.; Kane, M.A.; Trainor, P.A. Identifying vitamin A signaling by visualizing gene and protein activity and by quantification of vitamin A metabolites. Methods Enzymol. 2020, 637, 367–418. [Google Scholar] [CrossRef] [PubMed]
- Kuo, T.-C.; Tseng, Y.J. Lipid Pedia: A comprehensive lipid knowledgebase. Bioinformatics 2018, 34, 2982–2987. [Google Scholar] [CrossRef] [Green Version]
- Walker, P.D.; Weir, A.N.M.; Willis, C.L.; Crump, M.P. Polyketide β-branching: Diversity, mechanism and selectivity. Nat. Prod. Rep. 2021. [Google Scholar] [CrossRef]
- Rustam, Y.H.; Reid, G.E. Analytical Challenges and Recent Advances in Mass Spectrometry Based Lipidomics. Anal. Chem. 2018, 90, 374–397. [Google Scholar] [CrossRef]
- Jurowski, K.; Kochan, K.; Walczak, J.; Barańska, M.; Piekoszewski, W.; Buszewski, B. Analytical Techniques in Lipidomics: State of the Art. Crit. Rev. Anal. Chem. 2017, 47, 418–437. [Google Scholar] [CrossRef]
- Folch, J.; Ascoli, I.; Lees, M.; Meath, J.; LeBaron, F. Preparation of lipide extracts from brain tissue. J. Biol. Chem. 1951, 191, 833–841. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satomi, Y.; Hirayama, M.; Kobayashi, H. One-step lipid extraction for plasma lipidomics analysis by liquid chromatography mass spectrometry. J. Chromatogr. B 2017, 1063, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Hájek, R.; Jirásko, R.; Lísa, M.; Cífková, E.; Holčapek, M. Hydrophilic interaction liquid chromatography-mass spectrometry characterization of gangliosides in biological samples. Anal. Chem. 2017, 89, 12425–12432. [Google Scholar] [CrossRef]
- Griffiths, W.J.; Gilmore, I.; Yutuc, E.; Abdel-Khalik, J.; Crick, P.J.; Hearn, T.; Dickson, A.; Bigger, B.W.; Wu, T.H.Y.; Goenka, A.; et al. Identification of unusual oxysterols and bile acids with 7-oxo or 3β,5α,6β-trihydroxy func-tions in human plasma by chargetagging mass spectrometry with multistage fragmentation. J. Lipid Res. 2018, 59, 1058–1070. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, W.J.; Abdel-Khalik, J.; Yutuc, E.; Morgan, A.H.; Gilmore, I.; Hearn, T.; Wang, Y. Cholesterolomics: An update. Anal. Biochem. 2017, 524, 56–67. [Google Scholar] [CrossRef] [Green Version]
- Clark, J.; Anderson, K.E.; Juvin, V.; Smith, T.S.; Karpe, F.; Wakelam, M.J.; Stephens, L.R.; Hawkins, P.T. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods 2011, 8, 267–272. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.C.; Byeon, S.K.; Moon, M.H. Relative Quantification of Phospholipids Based on Isotope-Labeled Methylation by Nanoflow Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectrometry: Enhancement in Cardiolipin Profiling. Anal. Chem. 2017, 89, 4969–4977. [Google Scholar] [CrossRef]
- Wang, M.; Wang, C.; Han, X. Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry—What, how and why? Mass Spectrom. Rev. 2017, 36, 693–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koelmel, J.P.; Cochran, J.A.; Ulmer, C.Z.; Levy, A.J.; Patterson, R.E.; Olsen, B.C.; Yost, R.A.; Bowden, J.A.; Garrett, T.J. Software tool for internal standard based normalization of lipids, and effect of data-processing strategies on resulting values. BMC Bioinform. 2019, 20, 217. [Google Scholar] [CrossRef]
- Fan, S.; Kind, T.; Cajka, T.; Hazen, S.L.; Tang, W.H.W.; Kaddurah-Daouk, R.; Irvin, M.R.; Arnett, D.K.; Barupal, D.K.; Fiehn, O. Systematic Error Removal using Random Forest for Normalizing Large-Scale Untargeted Lipidomics Data. Anal. Chem. 2019, 91, 3590–3596. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Gross, R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: A bridge to lipidomics. J. Lipid Res. 2003, 44, 1071–1079. [Google Scholar] [CrossRef] [Green Version]
- Hsu, F.-F. Mass spectrometry-based shotgun lipidomics—A critical review from the technical point of view. Anal. Bioanal. Chem. 2018, 410, 6387–6409. [Google Scholar] [CrossRef]
- Martineau, E.; Dumez, J.-N.; Giraudeau, P. Fast quantitative 2D NMR for metabolomics and lipidomics: A tutorial. Magn. Reson. Chem. 2020, 58, 390–403. [Google Scholar] [CrossRef]
- Sobczak, A.I.; Pitt, S.J.; Smith, T.K.; Ajjan, R.A.; Stewart, A.J. Lipidomic profiling of plasma free fatty acids in type-1 diabetes highlights specific changes in lipid metabolism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158823. [Google Scholar] [CrossRef]
- Züllig, T.; Trötzmüller, M.; Köfeler, H.C. Lipidomics from sample preparation to data analysis: A primer. Anal. Bioanal. Chem. 2020, 412, 2191–2209. [Google Scholar] [CrossRef] [Green Version]
- Holčapek, M.; Liebisch, G.; Ekroos, K. Lipidomic Analysis. Anal. Chem. 2018, 90, 4249–4257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lísa, M.; Cífková, E.; Khalikova, M.; Ovčačíková, M.; Holčapek, M. Lipidomic analysis of biological samples: Comparison of liquid chromatography, supercritical fluid chromatography and direct infusion mass spectrometry methods. J. Chromatogr. A. 2017, 1525, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Cífková, E.; Holčapek, M.; Lísa, M.; Ovčačíková, M.; Lyčka, A.; Lynen, F.; Sandra, P. Nontargeted quantitation of lipid classes using hydrophilic interaction liquid chromatography-electrospray ionization mass spectrometry with single internal standard and response factor approach. Anal. Chem. 2012, 84, 10064–10070. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Izumi, Y.; Takahashi, M.; Paxton, T.; Tamura, S.; Koike, T.; Yu, Y.; Kato, N.; Nagase, K.; Shiomi, M.; et al. Widely-targeted quantitative lipidomics method by supercritical fluid chromatography triple quadrupole mass spectrometry. J. Lipid Res. 2018, 59, 1283–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, W.; Wang, L.; Xuan, Q.; Zhao, X.; Liu, X.; Shi, X.; Xu, G. Pseudotargeted Method Based on Parallel Column Two-Dimensional Liquid Chromatography-Mass Spectrometry for Broad Coverage of Metabolome and Lipidome. Anal. Chem. 2020, 92, 6043–6050. [Google Scholar] [CrossRef] [PubMed]
- Contrepois, K.; Mahmoudi, S.; Ubhi, B.K.; Papsdorf, K.; Hornburg, D.; Brunet, A.; Snyder, M. Cross-Platform Comparison of Untargeted and Targeted Lipidomics Approaches on Aging Mouse Plasma. Sci. Rep. 2018, 8, 17747. [Google Scholar] [CrossRef] [PubMed]
- Hinz, C.; Liggi, S.; Mocciaro, G.; Jung, S.M.; Induruwa, I.; Pereira, M.C.; Bryant, C.E.; Meckelmann, S.W.; O’Donnell, V.B.; Farndale, R.W.; et al. A Comprehensive UHPLC Ion Mobility Quadrupole Time-of-Flight Method for Profiling and Quantification of Eicosanoids, Other Oxylipins, and Fatty Acids. Anal. Chem. 2019, 91, 8025–8035. [Google Scholar] [CrossRef]
- Zhou, Z.; Shen, X.; Chen, X.; Tu, J.; Xiong, X.; Zhu, Z.-J. LipidIMMS Analyzer: Integrating multi-dimensional information to support lipid identification in ion mobility-mass spectrometry based lipidomics. Bioinformatics 2019, 35, 698–700. [Google Scholar] [CrossRef]
- Poad, B.L.J.; Zheng, X.; Mitchell, T.W.; Smith, R.D.; Baker, E.S.; Blanksby, S.J. Online ozonolysis combined with ion mobility-mass spectrometry provides a new platform for lipid isomer analyses. Anal. Chem. 2018, 90, 1292–1300. [Google Scholar] [CrossRef] [Green Version]
- Poad, B.L.J.; Green, M.R.; Kirk, J.M.; Tomczyk, N.; Mitchell, T.W.; Blanksby, S.J. High-Pressure Ozone-Induced Dissociation for Lipid Structure Elucidation on Fast Chromatographic Timescales. Anal. Chem. 2017, 89, 4223–4229. [Google Scholar] [CrossRef] [Green Version]
- Leaptrot, K.L.; May, J.C.; Dodds, J.N.; McLean, J.A. Ion mobility conformational lipid atlas for high confidence lipidomics. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Kyle, J.E.; Zhang, X.; Weitz, K.K.; Monroe, M.E.; Ibrahim, Y.M.; Moore, R.J.; Cha, J.; Sun, X.; Lovelace, E.S.; Wagoner, J.; et al. Uncovering biologically significant lipid isomers with liquid chromatography, ion mobility spectrometry and mass spectrometry. Analyst 2016, 141, 1649–1659. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Smith, R.D.; Baker, E.S. Recent advances in lipid separations and structural elucidation using mass spectrometry combined with ion mobility spectrometry, ion-molecule reactions and fragmentation approaches. Curr. Opin. Chem. Biol. 2018, 42, 111–118. [Google Scholar] [CrossRef]
- Ma, X.; Xia, Y. Pinpointing Double Bonds in Lipids by Paternò-Büchi Reactions and Mass Spectrometry. Angew. Chem. Int. Ed. 2014, 53, 2592–2596. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Chong, L.; Tian, R.; Shi, R.; Hu, T.Y.; Ouyang, Z.; Xia, Y. Identification and quantitation of lipid C=C location isomers: A shotgun lipidomics approach enabled by photochemical reaction. Proc. Natl. Acad. Sci. USA 2016, 113, 2573–2578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfadda, A.A.; Benabdelkamel, H.; Fathaddin, A.A.; Alanazi, I.O.; Lauzon, N.; Chaurand, P.; Masood, A. A matrix-assisted laser desorption/ionization imaging mass spectrometric approach to study weight-related changes within thyroid tissue. J. Mass Spectrom. 2021, 56, 4671. [Google Scholar] [CrossRef]
- Pittenauer, E.; Allmaier, G. The renaissance of high-energy CID for structural elucidation of complex lipids: MAL-DI-TOF/RTOF-MS of alkali cationized triacylglycerols. J. Am. Soc. Mass Spectrom. 2009, 20, 1037–1047. [Google Scholar] [CrossRef] [Green Version]
- Liebisch, G.; Ekroos, K.; Hermansson, M.; Ejsing, C.S. Reporting of lipidomics data should be standardized. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Hunter, A.; Dayalan, S.; De Souza, D.; Power, B.; Lorrimar, R.; Szabo, T.; Nguyen, T.; O’Callaghan, S.; Hack, J.; Pyke, J.; et al. MASTR-MS: A web-based collaborative laboratory information management system (LIMS) for metabolomics. Metabolomics 2017, 13, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Foster, J.M.; Moreno, P.; Fabregat, A.; Hermjakob, H.; Steinbeck, C.; Apweiler, R.; Wakelam, M.J.O.; Vizcaíno, J.A. LipidHome: A Database of Theoretical Lipids Optimized for High Throughput Mass Spectrometry Lipidomics. PLoS ONE 2013, 8, e61951. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Allison, P.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Koelmel, J.P.; Kroeger, N.M.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Cochran, J.A.; Beecher, C.W.W.; Garrett, T.J.; Yost, R.A. LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinform. 2017, 18, 1–11. [Google Scholar] [CrossRef]
- Djoumbou-Feunang, Y.; Pon, A.; Karu, N.; Zheng, J.; Li, C.; Arndt, D.; Gautam, M.; Allen, F.; Wishart, D.S. CFM-ID 3.0: Sig-nificantly Improved ESI-MS/MS prediction and compound identification. Metabolites 2019, 9, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afgan, E.; Baker, D.; Beek, M.V.D.; Blankenberg, D.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolstencroft, K.; Haines, R.; Fellows, D.; Williams, A.; Withers, D.; Owen, S.; Soiland-Reyes, S.; Dunlop, I.; Nenadic, A.; Fisher, P.; et al. The Taverna workflow suite: Designing and executing workflows of Web Services on the desktop, web or in the cloud. Nucleic Acids Res. 2013, 41, W557–W561. [Google Scholar] [CrossRef]
- Fillbrunn, A.; Dietz, C.; Pfeuffer, J.; Rahn, R.; Landrum, G.A.; Berthold, M.R. KNIME for reproducible cross-domain analysis of life science data. J. Biotechnol. 2017, 261, 149–156. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciucci, S.; Ge, Y.; Durán, C.; Palladini, A.; Jiménez-Jiménez, V.; Martínez-Sánchez, L.M.; Wang, Y.; Sales, S.; Shevchenko, A.; Poser, S.W.; et al. Enlightening discriminative network functional modules behind Principal Component Analysis separation in differentialomic science studies. Sci. Rep. 2017, 7, srep43946. [Google Scholar] [CrossRef] [Green Version]
- Wheelock, C.E.; Wheelock, A.M.; Kawashima, S.; Diez, D.; Kanehisa, M.; Van Erk, M.; Kleemann, R.; Haeggström, J.Z.; Goto, S. Systems biology approaches and pathway tools for investigating cardiovascular disease. Mol. BioSyst. 2009, 5, 588–602. [Google Scholar] [CrossRef] [Green Version]
- Owen, B.M.; Ding, X.; Morgan, D.A.; Coate, K.C.; Bookout, A.L.; Rahmouni, K.; Kliewer, S.A.; Mangelsdorf, D.J. FGF21 Acts Centrally to Induce Sympathetic Nerve Activity, Energy Expenditure, and Weight Loss. Cell Metab. 2014, 20, 670–677. [Google Scholar] [CrossRef] [Green Version]
- Simcox, J.; Geoghegan, G.; Maschek, J.A.; Bensard, C.L.; Pasquali, M.; Miao, R.; Lee, S.; Jiang, L.; Huck, I.; Kershaw, E.E.; et al. Global Analysis of Plasma Lipids Identifies Liver-Derived Acylcarnitines as a Fuel Source for Brown Fat Thermogenesis. Cell Metab. 2017, 26, 509–522. [Google Scholar] [CrossRef] [Green Version]
- Flier, J.S.; Maratos-Flier, E. Leptin’s Physiologic Role: Does the Emperor of Energy Balance Have No Clothes? Cell Metab. 2017, 26, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.V.; Scherer, P.E. Adiponectin, the past two decades. J. Mol. Cell Biol. 2016, 8, 93–100. [Google Scholar] [CrossRef]
- Cao, H.; Sekiya, M.; Ertunc, M.E.; Burak, M.F.; Mayers, J.R.; White, A.; Inouye, K.; Rickey, L.M.; Ercal, B.C.; Furuhashi, M.; et al. Adipocyte Lipid Chaperone aP2 is a Secreted Adipokine Regulating Hepatic Glucose Production. Cell Metab. 2013, 17, 768–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rancoule, C.; Dusaulcy, R.; Treguer, K.; Grès, S.; Attané, C.; Saulnier-Blache, J.S. Involvement of autotaxin/lysophosphatidic acid signaling in obesity and impaired glucose homeostasis. Biochimie 2014, 96, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Yore, M.M.; Syed, I.; Moraes-Vieira, P.M.; Zhang, T.; Herman, M.A.; Homan, E.A.; Patel, R.T.; Lee, J.; Chen, S.; Peroni, O.D.; et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and antiinflammatory effects. Cell 2014, 159, 318–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rada, P.; González-Rodríguez, Á.; García-Monzón, C.; Valverde, Á.M. Understanding lipotoxicity in NAFLD pathogenesis: Is CD36 a key driver? Cell Death Dis. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- McCullough, A.J. The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin. Liver Dis. 2004, 8, 521–533. [Google Scholar] [CrossRef]
- Adams, L.A.; Ratziu, V. Non-alcoholic fatty liver—Perhaps not so benign. J. Hepatol. 2015, 62, 1002–1004. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Allen, A.M.; Wang, Z.; Prokop, L.J.; Murad, M.H.; Loomba, R. Fibrosis Progression in Nonalcoholic Fatty Liver vs Nonalcoholic Steatohepatitis: A Systematic Review and Meta-analysis of Paired-Biopsy Studies. Clin. Gastroenterol. Hepatol. 2015, 13, 643–654. [Google Scholar] [CrossRef] [Green Version]
- Ekstedt, M.; Hagström, H.; Nasr, P.; Fredrikson, M.; Stål, P.; Kechagias, S.; Hultcrantz, R. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology 2015, 61, 1547–1554. [Google Scholar] [CrossRef] [Green Version]
- Argo, C.K.; Northup, P.G.; Al-Osaimi, A.M.; Caldwell, S.H. Systematic review of risk factors for fibrosis progression in non-alcoholic steatohepatitis. J. Hepatol. 2009, 51, 371–379. [Google Scholar] [CrossRef]
- Batterham, R.L.; Cummings, D.E. Mechanisms of Diabetes Improvement Following Bariatric/Metabolic Surgery. Diabetes Care 2016, 39, 893–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabré, N.; Luciano-Mateo, F.; Fernández-Arroyo, S.; Baiges-Gayà, G.; Hernández-Aguilera, A.; Fibla, M.; Fernández-Julià, R.; París, M.; Sabench, F.; Del Castillo, D.; et al. Laparoscopic sleeve gastrectomy reverses non-alcoholic fatty liver disease modulating oxidative stress and inflammation. Metabolism 2019, 99, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Quezada, N.; Maturana, G.; Irarrázaval, M.J.; Muñoz, R.; Morales, S.; Achurra, P.; Azócar, C.; Crovari, F. Bariatric Surgery in Cirrhotic Patients: A Matched Case-Control Study. Obes. Surg. 2020, 30, 4724–4731. [Google Scholar] [CrossRef]
- Tan, C.H.; Al-Kalifah, N.; Ser, K.-H.; Lee, Y.-C.; Chen, J.-C.; Lee, W.-J. Long-term effect of bariatric surgery on resolution of nonalcoholic steatohepatitis (NASH): An external validation and application of a clinical NASH score. Surg. Obes. Relat. Dis. 2018, 14, 1600–1606. [Google Scholar] [CrossRef] [PubMed]
- Calvo, N.; Beltrán-Debón, R.; Rodríguez-Gallego, E.; Hernández-Aguilera, A.; Guirro, M.; MarinéCasadó, R.; Millá, L.; Alegret, J.M.; Sabench, F.; Del Castillo, D.; et al. Liver fat deposition and mitochondrial dysfunction in morbid obesity: An approach combining metabolomics with liver imaging and histology. World J. Gastroenterol. 2015, 21, 7529–7544. [Google Scholar] [CrossRef]
- Rodríguez-Calvo, R.; Samino, S.; Girona, J.; Martínez-Micaelo, N.; Ràfols, P.; García-Altares, M.; Guaita-Esteruelas, S.; Junza, A.; Heras, M.; Yanes, O.; et al. Hepatic Lipidomics and Molecular Imaging in a Murine Non-Alcoholic Fatty Liver Disease Model: Insights into Molecular Mechanisms. Biomolecules 2020, 10, 1275. [Google Scholar] [CrossRef] [PubMed]
- Anjani, K.; Lhomme, M.; Sokolovska, N.; Poitou, C.; Aron-Wisnewsky, J.; Bouillot, J.-L.; Lesnik, P.; Bedossa, P.; Kontush, A.; Clement, K.; et al. Circulating phospholipid profiling identifies portal contribution to NASH signature in obesity. J. Hepatol. 2015, 62, 905–912. [Google Scholar] [CrossRef] [Green Version]
- Jadhav, K.; Cohen, T.S. Can You Trust Your Gut? Implicating a Disrupted Intestinal Microbiome in the Progression of NAFLD/NASH. Front. Endocrinol. 2020, 11, 592157. [Google Scholar] [CrossRef]
- Araya, J.; Rodrigo, R.; Videla, L.A.; Thielemann, L.; Orellana, M.; Pettinelli, P.; Poniachik, J. Increase in long-chain polyun-saturated fatty acid n-6/n-3 ratio in relation to hepatic steatosis in patients with non-alcoholic fatty liver disease. Clin. Sci. 2004, 106, 635–643. [Google Scholar] [CrossRef] [Green Version]
- Allard, J.P.; Aghdassi, E.; Mohammed, S.; Raman, M.; Avand, G.; Arendt, B.M.; Jalali, P.; Kandasamy, T.; Prayitno, N.; Sherman, M.; et al. Nutritional assessment and hepatic fatty acid composition in non-alcoholic fatty liver disease (NAFLD): A cross-sectional study. J. Hepatol. 2008, 48, 300–307. [Google Scholar] [CrossRef]
- Chiappini, F.; Coilly, A.; Kadar, H.; Gual, P.; Tran, A.; Desterke, C.; Samuel, D.; Duclos-Vallée, J.C.; Touboul, D.; Bertrand-Michel, J.; et al. Metabolism dysregulation induces a specific lipid signature of nonal-coholic steatohepatitis in patients. Sci. Rep. 2017, 7, 46658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puri, P.; Wiest, M.M.; Cheung, O.; Mirshahi, F.; Sargeant, C.; Min, H.-K.; Contos, M.J.; Sterling, R.K.; Fuchs, M.; Zhou, H.; et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology 2009, 50, 1827–1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.-S.; Xu, A.; Huang, T.; Yu, X.; Li, D. Low Docosahexaenoic Acid Content in Plasma Phospholipids is Associated with Increased Non-alcoholic Fatty Liver Disease in China. Lipids 2012, 47, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Loomba, R.; Quehenberger, O.; Armando, A.; Dennis, E.A. Polyunsaturated fatty acid metabolites as novel lipidomic bi-omarkers for noninvasive diagnosis of nonalcoholic steatohepatitis. J. Lipid Res. 2015, 56, 185–192. [Google Scholar] [CrossRef] [Green Version]
- Walle, P.; Takkunen, M.; Männistö, V.; Vaittinen, M.; Lankinen, M.; Kärjä, V.; Käkelä, P.; Ågren, J.; Tiainen, M.; Schwab, U.; et al. Fatty acid metabolism is altered in non-alcoholic steatohepatitis independent of obesity. Metabolism 2016, 65, 655–666. [Google Scholar] [CrossRef]
- Tiwari-Heckler, S.; Gan-Schreier, H.; Stremmel, W.; Chamulitrat, W.; Pathil, A. Circulating Phospholipid Patterns in NAFLD Patients Associated with a Combination of Metabolic Risk Factors. Nutrients 2018, 10, 649. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.W.L.; Arendt, B.M.; Hillyer, L.M.; Fung, S.K.; McGilvray, I.; Guindi, M.; Allard, J.P. Plasma phospholipids and fatty acid composition differ between liver biopsy-proven nonalcoholic fatty liver disease and healthy subjects. Nutr. Diabetes 2016, 6, e220. [Google Scholar] [CrossRef] [Green Version]
- Gorden, D.L.; Myers, D.S.; Ivanova, P.T.; Fahy, E.; Maurya, M.R.; Gupta, S.; Min, J.; Spann, N.J.; McDonald, J.G.; Kelly, S.L.; et al. Biomarkers of NAFLD progression: A lipidomics approach to an epidemic. J. lipid Res. 2015, 56, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Peng, K.Y.; Watt, M.J.; Rensen, S.; Greve, J.W.; Huynh, K.; Jayawardana, K.S.; Meikle, P.J.; Meex, R.C.R. Mitochondrial dys-function-related lipid changes occur in nonalcoholic fatty liver disease progression. J. Lipid Res. 2018, 59, 1977–1986. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Alvarez, M.I.; Sebastián, D.; Vives, S.; Ivanova, S.; Bartoccioni, P.; Kakimoto, P.; Plana, N.; Veiga, S.R.; Hernández, V.; Vasconcelos, N.; et al. Deficient endoplasmic reticulum-mitochondrial phosphatidylserine transfer causes liver disease. Cell 2019, 177, 881–895. [Google Scholar] [CrossRef]
- Hernández-Aguilera, A.; Rull, A.; Rodríguez-Gallego, E.; Riera-Borrull, M.; Luciano-Mateo, F.; Camps, J.; Menéndez, J.A.; Joven, J. Mitochondrial Dysfunction: A Basic Mechanism in Inflammation-Related Non-Communicable Diseases and Therapeutic Opportunities. Mediat. Inflamm. 2013, 2013, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotronen, A.; Yki-Järvinen, H.; Sevastianova, K.; Bergholm, R.; Hakkarainen, A.; Pietiläinen, K.H.; Juurinen, L.; Lundbom, N.; Sørensen, T.I. Comparison of the Relative Contributions of Intra-Abdominal and Liver Fat to Components of the Metabolic Syndrome. Obesity 2011, 19, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Pietiläinen, K.H.; Sysi-Aho, M.; Rissanen, A.; Seppänen-Laakso, T.; Yki-Jarvinen, H.; Kaprio, J.; Oresic, M. Acquired Obesity is Associated with Changes in the Serum Lipidomic Profile Independent of Genetic Effects—A Monozygotic Twin Study. PLoS ONE 2007, 2, e218. [Google Scholar] [CrossRef]
- Graessler, J.; Schwudke, D.; Schwarz, P.E.H.; Herzog, R.; Shevchenko, A.; Bornstein, S.R. Top-Down Lipidomics Reveals Ether Lipid Deficiency in Blood Plasma of Hypertensive Patients. PLoS ONE 2009, 4, e6261. [Google Scholar] [CrossRef] [PubMed]
- Kotronen, A.; Velagapudi, V.R.; Yetukuri, L.; Westerbacka, J.; Bergholm, R.; Ekroos, K.; Makkonen, J.; Taskinen, M.-R.; Oresic, M.; Yki-Järvinen, H. Serum saturated fatty acids containing triacylglycerols are better markers of insulin resistance than total serum triacylglycerol concentrations. Diabetologia 2009, 52, 684–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016, 12, 668–679. [Google Scholar] [CrossRef]
- Kiebish, M.A.; Bell, R.; Yang, K.; Phan, T.; Zhao, Z.; Ames, W.; Seyfried, T.N.; Gross, R.W.; Chuang, J.H.; Han, X. Dynamic simulation of cardiolipin remodeling: Greasing the wheels for an interpretative approach to lipidomics. J. Lipid Res. 2010, 51, 2153–2170. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Díaz-Díaz, N.; Zarringhalam, K.; Hermansson, M.; Somerharju, P.; Chuang, J. Dynamics of the ethanolamine glycerophospholipid remodeling network. PLoS ONE 2012, 7, e50858. [Google Scholar] [CrossRef]
- Wakelam, M.J.; Pettitt, T.R.; Postle, A.D. Lipidomic Analysis of Signaling Pathways. Methods Enzymol. 2007, 432, 233–246. [Google Scholar] [CrossRef]
- Osman, C.; Voelker, D.R.; Langer, T. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 2011, 192, 7–16. [Google Scholar] [CrossRef] [Green Version]
- Bechmann, L.P.; Hannivoort, R.A.; Gerken, G.; Hotamisligil, G.S.; Trauner, M.; Canbay, A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 2012, 56, 952–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietiläinen, K.H.; Róg, T.; Seppänen-Laakso, T.; Virtue, S.; Gopalacharyulu, P.; Tang, J.; Rodriguez-Cuenca, S.; Maciejewski, A.; Naukkarinen, J.; Ruskeepää, A.L.; et al. Association of lipidome remodeling in the adipocyte membrane with acquired obesity in humans. PLoS Biol. 2011, 9, e1000623. [Google Scholar] [CrossRef] [Green Version]
- Gross, R.W.; Han, X. Lipidomics in Diabetes and the Metabolic Syndrome. Methods Enzym. 2007, 433, 73–90. [Google Scholar] [CrossRef]
- Meikle, P.J.; Christopher, M.J. Lipidomics is providing new insight into the metabolic syndrome and its sequelae. Curr. Opin. Lipidol. 2011, 22, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Martel, C.; Degli Esposti, D.; Bouchet, A.; Brenner, C.; Lemoine, A. Non-alcoholic steatohepatitis: New insights from OMICS studies. Curr. Pharm. Biotechnol. 2012, 13, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Ferré, N.; Martínez-Clemente, M.; López-Parra, M.; González-Périz, A.; Horrillo, R.; Planagumà, A.; Camps, J.; Joven, J.; Tres, A.; Guardiola, F.; et al. Increased susceptibility to exacerbated liver injury in hypercholesterolemic ApoE-deficient mice: Potential involvement of oxysterols. Am. J. Physiol. Liver Physiol. 2009, 296, G553–G562. [Google Scholar] [CrossRef]
- Lopategi, A.; López-Vicario, C.; Alcaraz-Quiles, J.; García-Alonso, V.; Rius, B.; Titos, E.; Clària, J. Role of bioactive lipid me-diators in obese adipose tissue inflammation and endocrine dysfunction. Mol. Cell Endocrinol. 2016, 419, 44–59. [Google Scholar] [CrossRef] [Green Version]
- Debnath, M.; Agrawal, S.; Agrawal, A.; Dubey, G.P. Metaflammatory responses during obesity: Pathomechanism and treat-ment. Obes. Res. Clin. Pract. 2016, 10, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Caussy, C.; Chuang, J.-C.; Billin, A.; Hu, T.; Wang, Y.; Subramanian, G.M.; Djedjos, C.S.; Myers, R.P.; Dennis, E.A.; Loomba, R. Plasma eicosanoids as noninvasive biomarkers of liver fibrosis in patients with nonalcoholic steatohepatitis. Ther. Adv. Gastroenterol. 2020, 13. [Google Scholar] [CrossRef]
- Li, Q.; Rempel, J.D.; Ball, T.B.; Aukema, H.; Minuk, G.Y. Plasma Oxylipins Levels in Nonalcoholic Fatty Liver Disease. Dig. Dis. Sci. 2020, 65, 3605–3613. [Google Scholar] [CrossRef] [PubMed]
- Musso, G.; Gambino, R.; Cassader, M.; Paschetta, E.; Sircana, A. Specialized Proresolving Mediators: Enhancing Nonalcoholic Steatohepatitis and Fibrosis Resolution. Trends Pharmacol. Sci. 2018, 39, 387–401. [Google Scholar] [CrossRef]
- Rodríguez-Gallego, E.; Guirro, M.; Riera-Borrull, M.; Hernández-Aguilera, A.; Mariné-Casadó, R.; Fernández-Arroyo, S.; Beltrán-Debón, R.; Sabench, F.; Hernández, M.; Del Castillo, D.; et al. Mapping of the circulating metabolome reveals α-ketoglutarate as a predictor of morbid obesity-associated non-alcoholic fatty liver disease. Int. J. Obes. 2015, 39, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Forrester, J.S.; Milne, S.B.; Ivanova, P.T.; Brown, H.A. Computational Lipidomics: A Multiplexed Analysis of Dynamic Changes in Membrane Lipid Composition during Signal Transduction. Mol. Pharmacol. 2004, 65, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Zarringhalam, K.; Zhang, L.; Kiebish, M.A.; Yang, K.; Han, X.; Gross, R.W.; Chuang, J. Statistical Analysis of the Processes Controlling Choline and Ethanolamine Glycerophospholipid Molecular Species Composition. PLoS ONE 2012, 7, e37293. [Google Scholar] [CrossRef] [Green Version]
- Han, R.H.; Wang, M.; Fang, X.; Han, X. Simulation of triacylglycerol ion profiles: Bioinformatics for interpretation of triacyl-glycerol biosynthesis. J. Lipid Res. 2013, 54, 1023–1032. [Google Scholar] [CrossRef] [Green Version]
- Goecks, J.; Jalili, V.; Heiser, L.M.; Gray, J.W. How Machine Learning Will Transform Biomedicine. Cell 2020, 181, 92–101. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. Review the Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Współczesna Onkologia 2015, 1, 68–77. [Google Scholar] [CrossRef]
- Zhang, J.; Bajari, R.; Andric, D.; Gerthoffert, F.; Lepsa, A.; Nahal-Bose, H.; Stein, L.D.; Ferretti, V. The International Cancer Genome Consortium Data Portal. Nat. Biotechnol. 2019, 37, 367–369. [Google Scholar] [CrossRef]
- Iorio, F.; Bernardo-Faura, M.; Gobbi, A.; Cokelaer, T.; Jurman, G.; Saez-Rodriguez, J. Efficient randomization of biological networks while preserving functional characterization of individual nodes. BMC Bioinform. 2016, 17, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Sen, P.; Lamichhane, S.; Mathema, V.B.; McGlinchey, A.; Dickens, A.M.; Khoomrung, S.; Orešič, M. Deep learning meets metabolomics: A methodological perspective. Briefings Bioinform. 2020, 204. [Google Scholar] [CrossRef]
- Melnikov, A.D.; Tsentalovich, Y.P.; Yanshole, V.V. Deep Learning for the Precise Peak Detection in High-Resolution LC–MS Data. Anal. Chem. 2019, 92, 588–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Risum, A.B.; Bro, R. Using deep learning to evaluate peaks in chromatographic data. Talanta 2019, 204, 255–260. [Google Scholar] [CrossRef]
- Hyötyläinen, T.; Orešič, M. Optimizing the lipidomics workflow for clinical studies-practical considerations. Anal. Bioanal. Chem. 2015, 407, 4973–4993. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.A.; Lamichhane, S.; Dickens, A.; McGlinchey, A.; Ribeiro, H.C.; Sen, P.; Wei, F.; Hyötyläinen, T.; Orešič, M. Systems biology approaches to study lipidomes in health and disease. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2021, 1866, 158857. [Google Scholar] [CrossRef] [PubMed]
- Saeys, Y.; Inza, I.; Larrañaga, P. A review of feature selection techniques in bioinformatics. Bioinformatics 2007, 23, 2507–2517. [Google Scholar] [CrossRef] [Green Version]
- Mendez, K.M.; Broadhurst, D.I.; Reinke, S.N. Migrating from partial least squares discriminant analysis to artificial neural networks: A comparison of functionally equivalent visualisation and feature contribution tools using jupyter notebooks. Metabolomics 2020, 16, 17. [Google Scholar] [CrossRef] [Green Version]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis—A marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, K.; Misra, B.B. Software tools, databases and resources in metabolomics: Updates from 2018–2019. Metabolomics 2020, 16, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tan, J.; Han, D.; Zhu, H. From machine learning to deep learning: Progress in machine intelligence for rational drug discovery. Drug Discov. Today 2017, 22, 1680–1685. [Google Scholar] [CrossRef] [PubMed]
- Pomyen, Y.; Wanichthanarak, K.; Poungsombat, P.; Fahrmann, J.; Grapov, D.; Khoomrung, S. Deep metabolome: Applications of deep learning in metabolomics. Comput. Struct. Biotechnol. J. 2020, 18, 2818–2825. [Google Scholar] [CrossRef]
- Grapov, D.; Fahrmann, J.; Wanichthanarak, K.; Khoomrung, S. Rise of deep learning for genomic, proteomic, and etabolomic Data integration in precision medicine. OMICS 2018, 22, 630–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendez, K.M.; Reinke, S.N.; Broadhurst, D.I. A comparative evaluation of the generalised predictive ability of eight machine learning algorithms across ten clinical metabolomics data sets for binary classification. Metabolomics 2019, 15, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-generation machine learning for biological net-works. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting Network Activity from High Throughput Metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Akwaa, F.M.; Yunits, B.; Huang, S.; Alhajaji, H.; Garmire, L.X. Lilikoi: An R package for personalized pathway-based classification modeling using metabolomics data. GigaScience 2018, 7, 136. [Google Scholar] [CrossRef] [Green Version]
- Palsson, B.; Zengler, K. The challenges of integrating multiomic data sets. Nat. Chem. Biol. 2010, 6, 787–789. [Google Scholar] [CrossRef]
- Manica, M.; Oskooei, A.; Born, J.; Subramanian, V.; Sáez-Rodríguez, J.; Martínez, M.R. Toward Explainable Anticancer Compound Sensitivity Prediction via Multimodal Attention-Based Convolutional Encoders. Mol. Pharm. 2019, 16, 4797–4806. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2017, 24, 1248–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Rai, N.; Zorraquino, V.; Tagkopoulos, I. Multiomics integration accurately predicts cellular state in unexplored conditions for Escherichia coli. Nat. Commun. 2016, 7, 13090. [Google Scholar] [CrossRef] [Green Version]
- Castera, L.; Friedrich-Rust, M.; Loomba, R. Noninvasive Assessment of Liver Disease in Patients with Nonalcoholic Fatty Liver Disease. Gastroenterology 2019, 156, 1264–1281. [Google Scholar] [CrossRef] [Green Version]
- Sorino, P.; Caruso, M.G.; Misciagna, G.; Bonfiglio, C.; Campanella, A.; Mirizzi, A.; Franco, I.; Bianco, A.; Buongiorno, C.; Liuzzi, R.; et al. Selecting the best machine learning algorithm to support the diagnosis of Non-Alcoholic Fatty Liver Disease: A meta learner study. PLoS ONE 2020, 15, e0240867. [Google Scholar] [CrossRef]
- Chalasani, N.; Younossi, Z.; LaVine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Dhyani, M.; Grajo, J.R.; Sirlin, C.E.; Samir, A. Current status of imaging in nonalcoholic fatty liver disease. World J. Hepatol. 2018, 10, 530–542. [Google Scholar] [CrossRef]
- Perakakis, N.; Polyzos, S.A.; Yazdani, A.; Sala-Vila, A.; Kountouras, J.; Anastasilakis, A.D.; Mantzoros, C.S. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: A proof of concept study. Metabolism 2019, 101, 154005. [Google Scholar] [CrossRef]
- Perakakis, N.; Yazdani, A.; Karniadakis, G.E.; Mantzoros, C. Omics, big data and machine learning as tools to propel under-standing of biological mechanisms and to discover novel diagnostics and therapeutics. Metabolism 2018, 87, A1–A9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acharjee, A.; Ament, Z.; West, J.A.; Stanley, E.; Griffin, J.L. Integration of metabolomics, lipidomics and clinical data using a machine learning method. BMC Bioinform. 2016, 17, 37–49. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Chaudhary, K.; Garmire, L.X. More is Better: Recent Progress in Multi-Omics Data Integration Methods. Front. Genet. 2017, 8, 84. [Google Scholar] [CrossRef] [Green Version]
- Song, M.; Greenbaum, J.; Luttrell, J.; Zhou, W.; Wu, C.; Shen, H.; Gong, P.; Zhang, C.; Deng, H.-W. A Review of Integrative Imputation for Multi-Omics Datasets. Front. Genet. 2020, 11, 570255. [Google Scholar] [CrossRef]



| Frameworks | Programming Languages | Features |
|---|---|---|
| Apache Spark | Java, R, Python, Scala | Structured data processing for machine learning and graph processing. |
| Caffe | C++, Python | Supports different deep learning architectures like CNN or RNN. |
| Chainer | Python | Provides a flexible, intuitive and high performance of deep learning models, such as RNN and autoencoders. |
| Deeplearning4j | Java | Works with different data types, such as images, CSV, plain text, audio and video to build a full range of deep neural network. |
| h2o.ai | Java, R, Python, Scala | Provides fast and scalable machine learning and predictive analysis platform. |
| Keras | Python | It is a deep learning API that works with machine learning platform TensorFlow. |
| Neon | Python | Artificial intelligence platform that works with images and videos. |
| Pytorch | C++, Python | It is a Python library for deep learning that provides fast and flexible framework to build dynamic neural network. |
| Scikit-learn | C, C++, Python, Cython | It is library for machine learning and statistical modeling that supports supervised and unsupervised learning. |
| TensorFlow | C++, Python | Machine learning platform that builds API for implementing machine learning, deep learning and science computing models. |
| Theano | Python | It is a Python library that provide train deep neural networks algorithms. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castañé, H.; Baiges-Gaya, G.; Hernández-Aguilera, A.; Rodríguez-Tomàs, E.; Fernández-Arroyo, S.; Herrero, P.; Delpino-Rius, A.; Canela, N.; Menendez, J.A.; Camps, J.; et al. Coupling Machine Learning and Lipidomics as a Tool to Investigate Metabolic Dysfunction-Associated Fatty Liver Disease. A General Overview. Biomolecules 2021, 11, 473. https://doi.org/10.3390/biom11030473
Castañé H, Baiges-Gaya G, Hernández-Aguilera A, Rodríguez-Tomàs E, Fernández-Arroyo S, Herrero P, Delpino-Rius A, Canela N, Menendez JA, Camps J, et al. Coupling Machine Learning and Lipidomics as a Tool to Investigate Metabolic Dysfunction-Associated Fatty Liver Disease. A General Overview. Biomolecules. 2021; 11(3):473. https://doi.org/10.3390/biom11030473
Chicago/Turabian StyleCastañé, Helena, Gerard Baiges-Gaya, Anna Hernández-Aguilera, Elisabet Rodríguez-Tomàs, Salvador Fernández-Arroyo, Pol Herrero, Antoni Delpino-Rius, Nuria Canela, Javier A. Menendez, Jordi Camps, and et al. 2021. "Coupling Machine Learning and Lipidomics as a Tool to Investigate Metabolic Dysfunction-Associated Fatty Liver Disease. A General Overview" Biomolecules 11, no. 3: 473. https://doi.org/10.3390/biom11030473
APA StyleCastañé, H., Baiges-Gaya, G., Hernández-Aguilera, A., Rodríguez-Tomàs, E., Fernández-Arroyo, S., Herrero, P., Delpino-Rius, A., Canela, N., Menendez, J. A., Camps, J., & Joven, J. (2021). Coupling Machine Learning and Lipidomics as a Tool to Investigate Metabolic Dysfunction-Associated Fatty Liver Disease. A General Overview. Biomolecules, 11(3), 473. https://doi.org/10.3390/biom11030473









