Islet Dysfunction in a Novel Transgenic Model of T Cell Insulitis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Islet Isolation
2.3. mRNA Analysis by Quantitative Real Time PCR (qRT-PCR)
2.4. Intraventricular Perfusion
2.5. Immunofluorescence Staining (IF)
2.6. Histological Microscopy
2.7. Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) Assay
2.8. Ki67 Proliferation Assay
2.9. Peripheral Blood Serum Insulin Level Analysis
2.10. Total Pancreatic Insulin Content ELISA Analysis
2.11. Intraperitoneal Glucose Tolerance Testing (IPGTT)
2.12. Glucose Stimulated Insulin Secretion Studies In Vitro
2.13. Transgenic Splenocyte Adoptive Transfer into RAG−/− Recipients
2.14. Statistics
3. Results
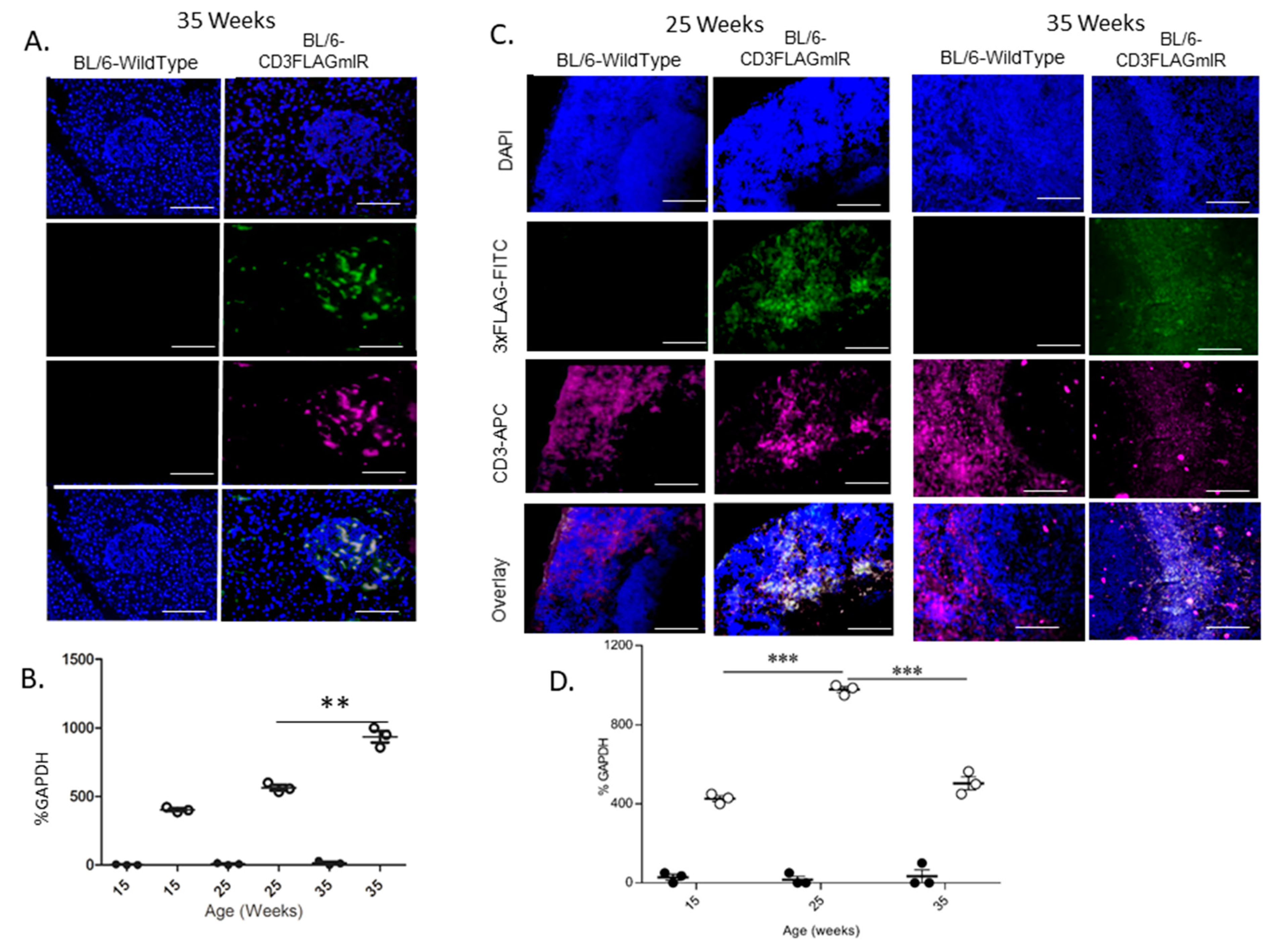
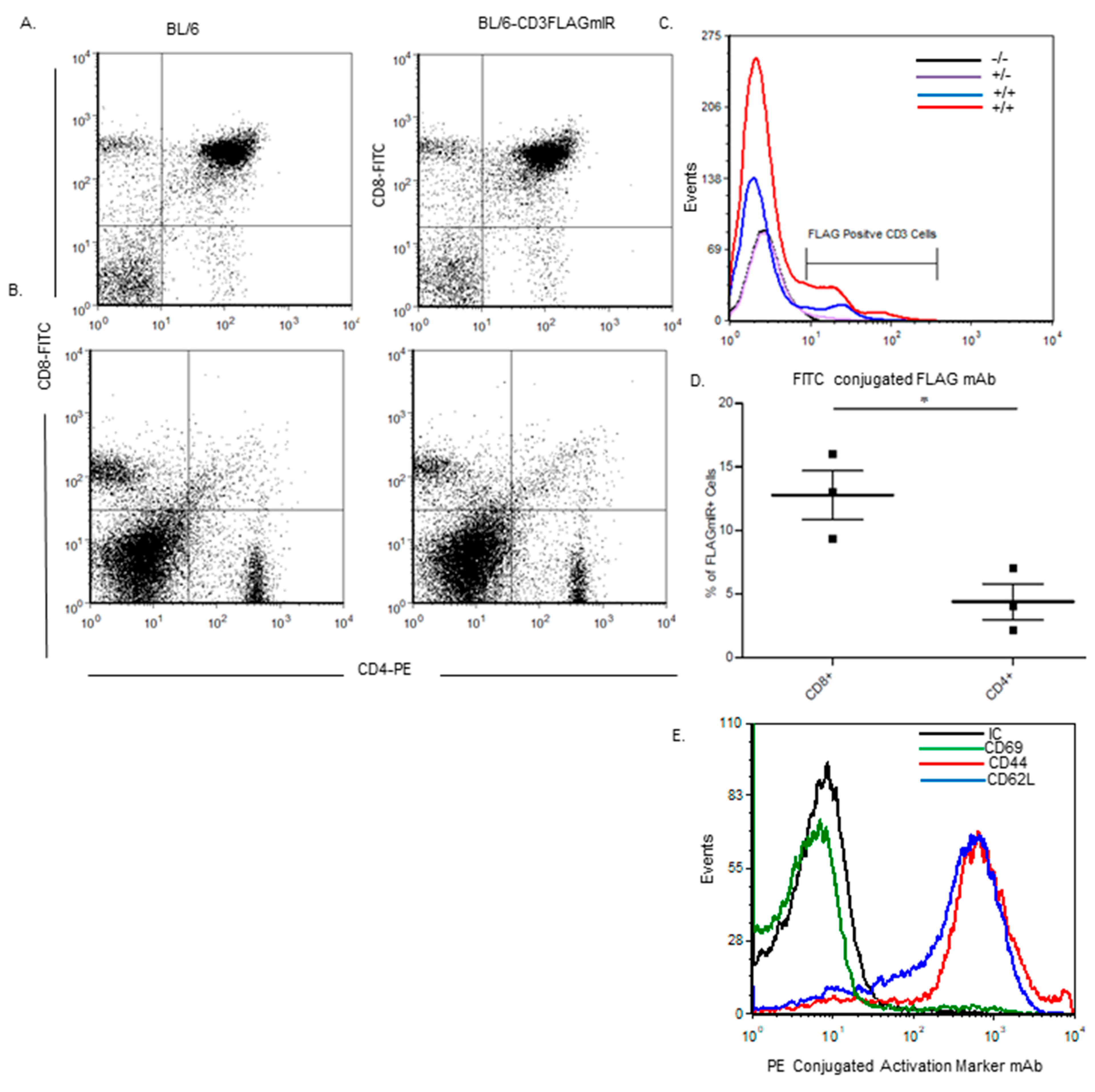
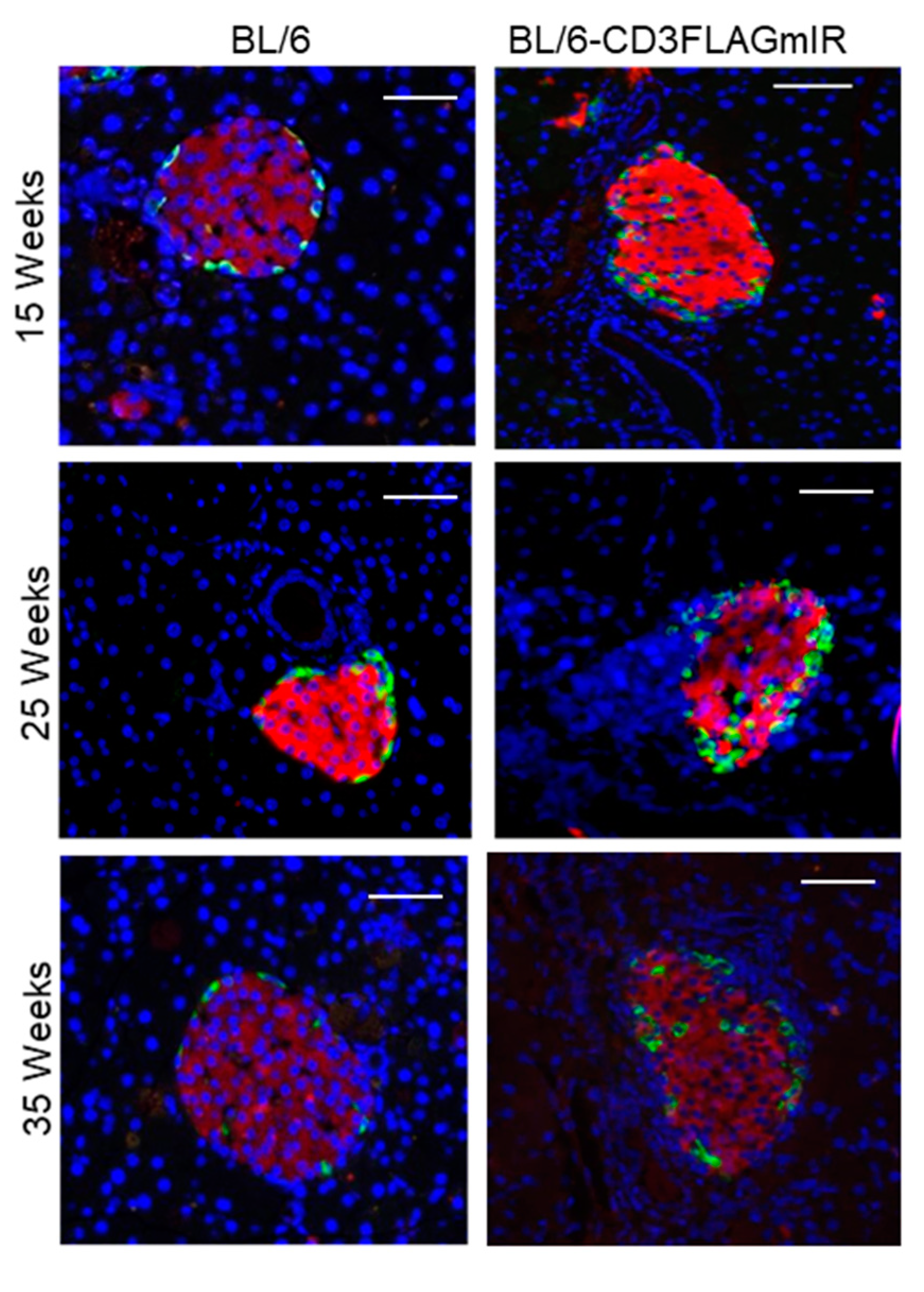
3.1. Characterization of IR+ T Cells Infiltrating into the Pancreatic Islet
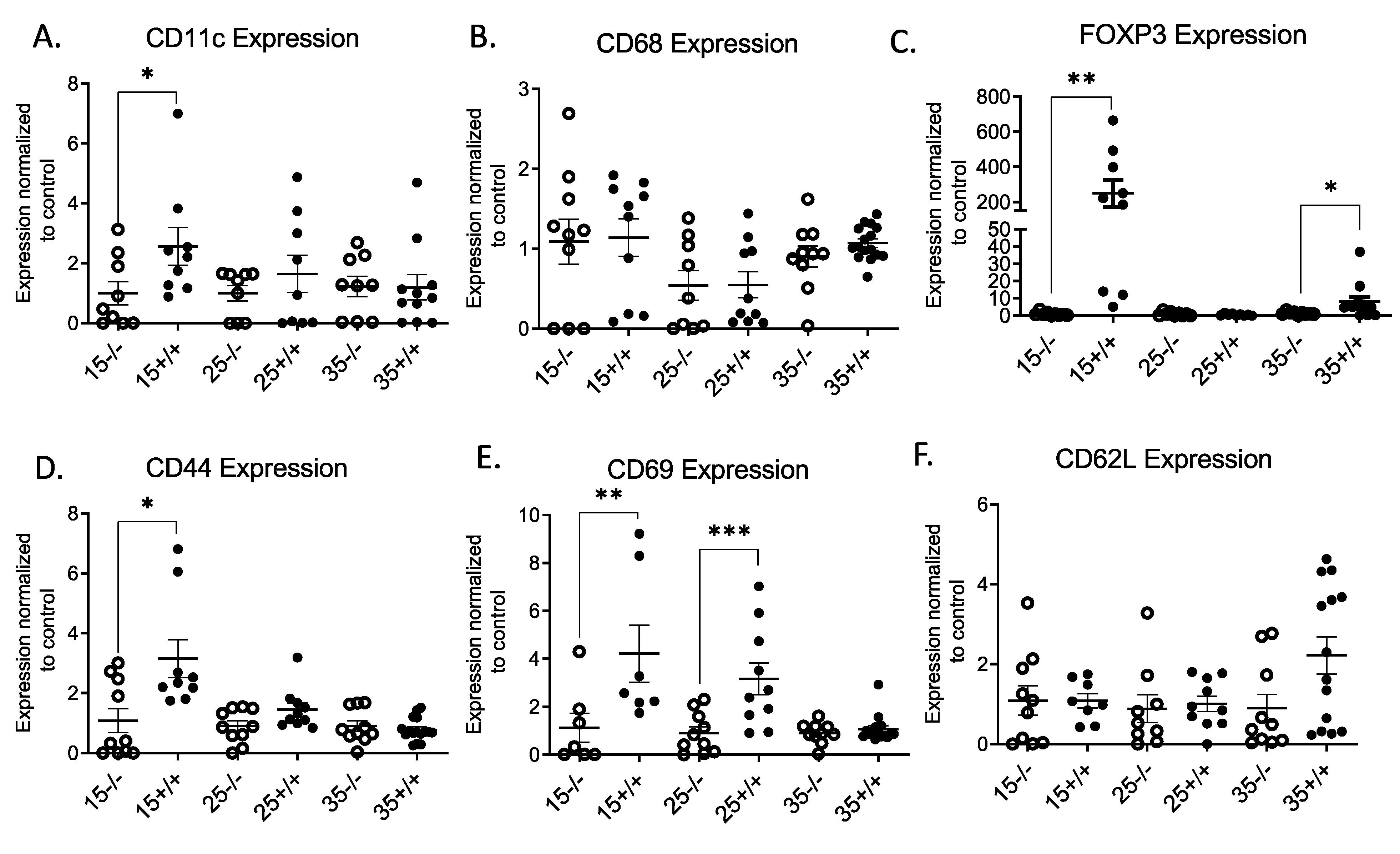
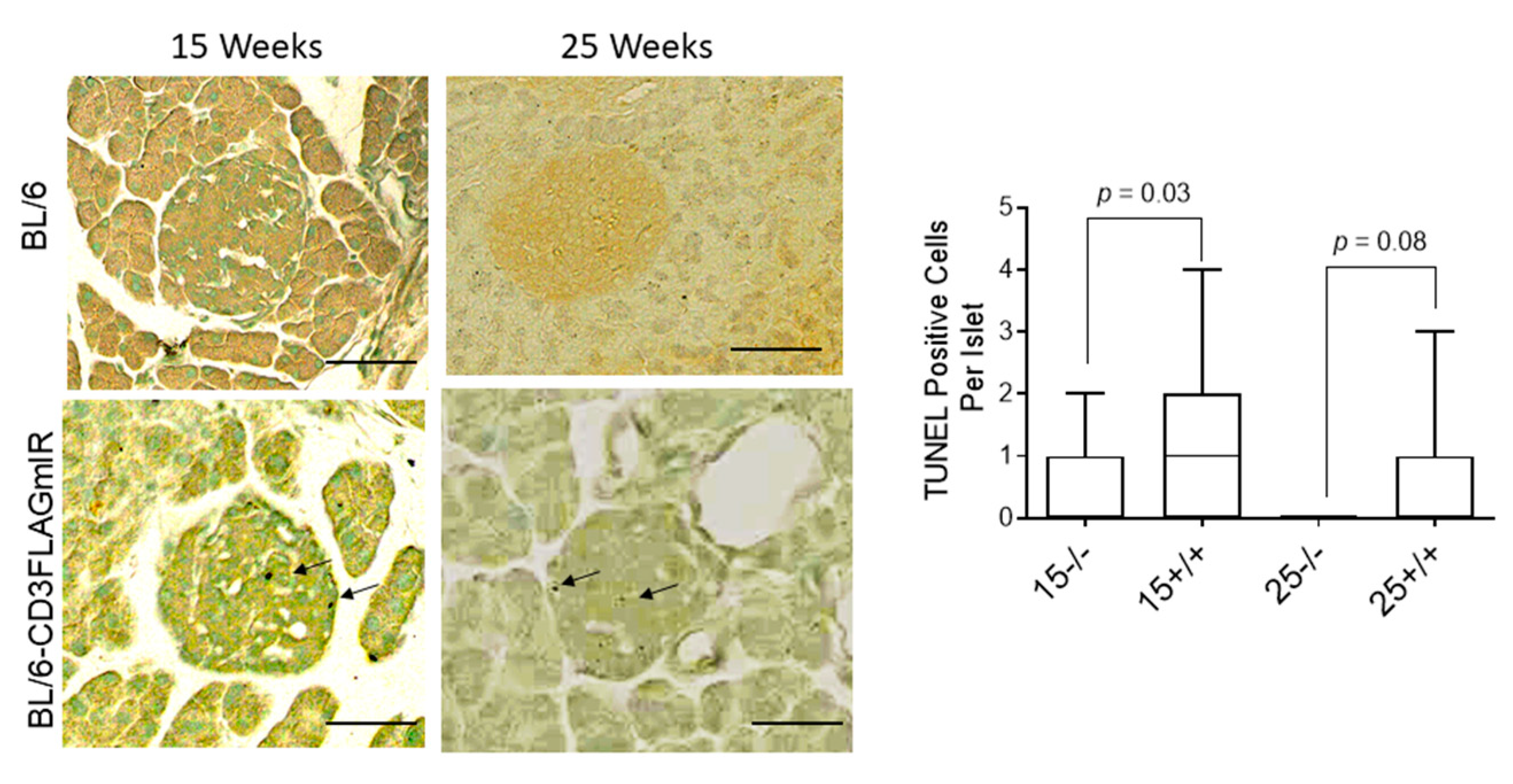
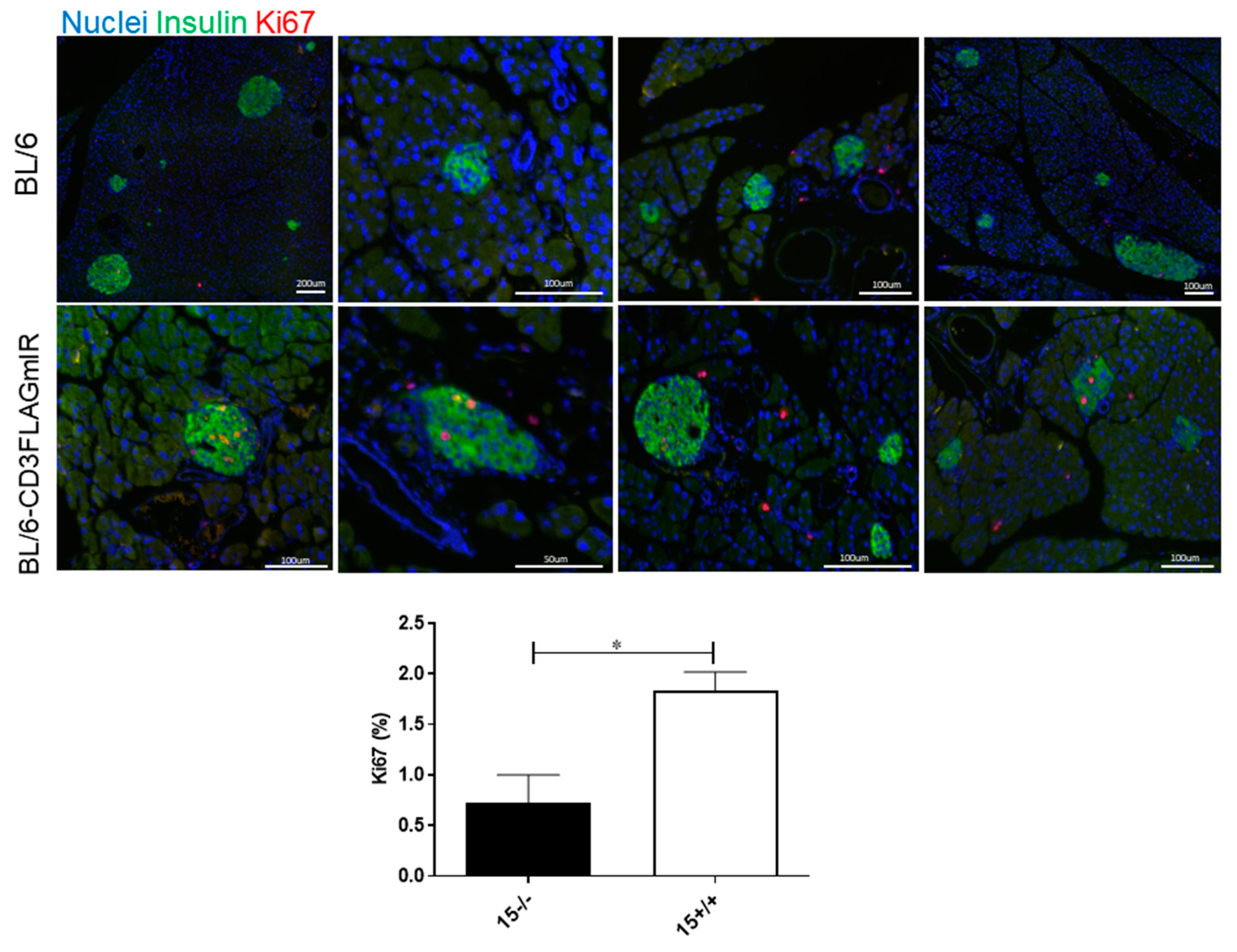
3.2. Effect of IR Expressing T Cell Infiltration into Pancreatic Islets
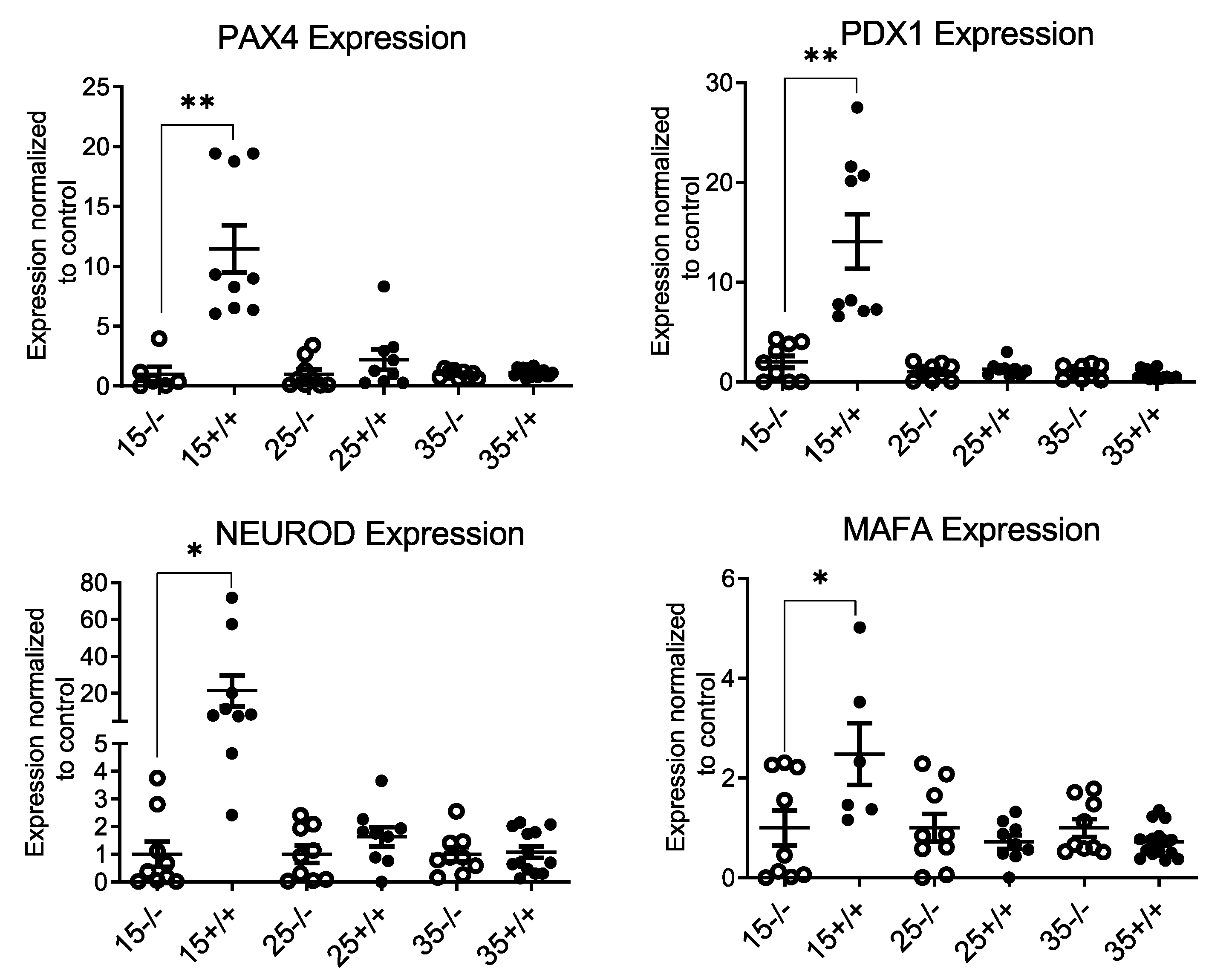
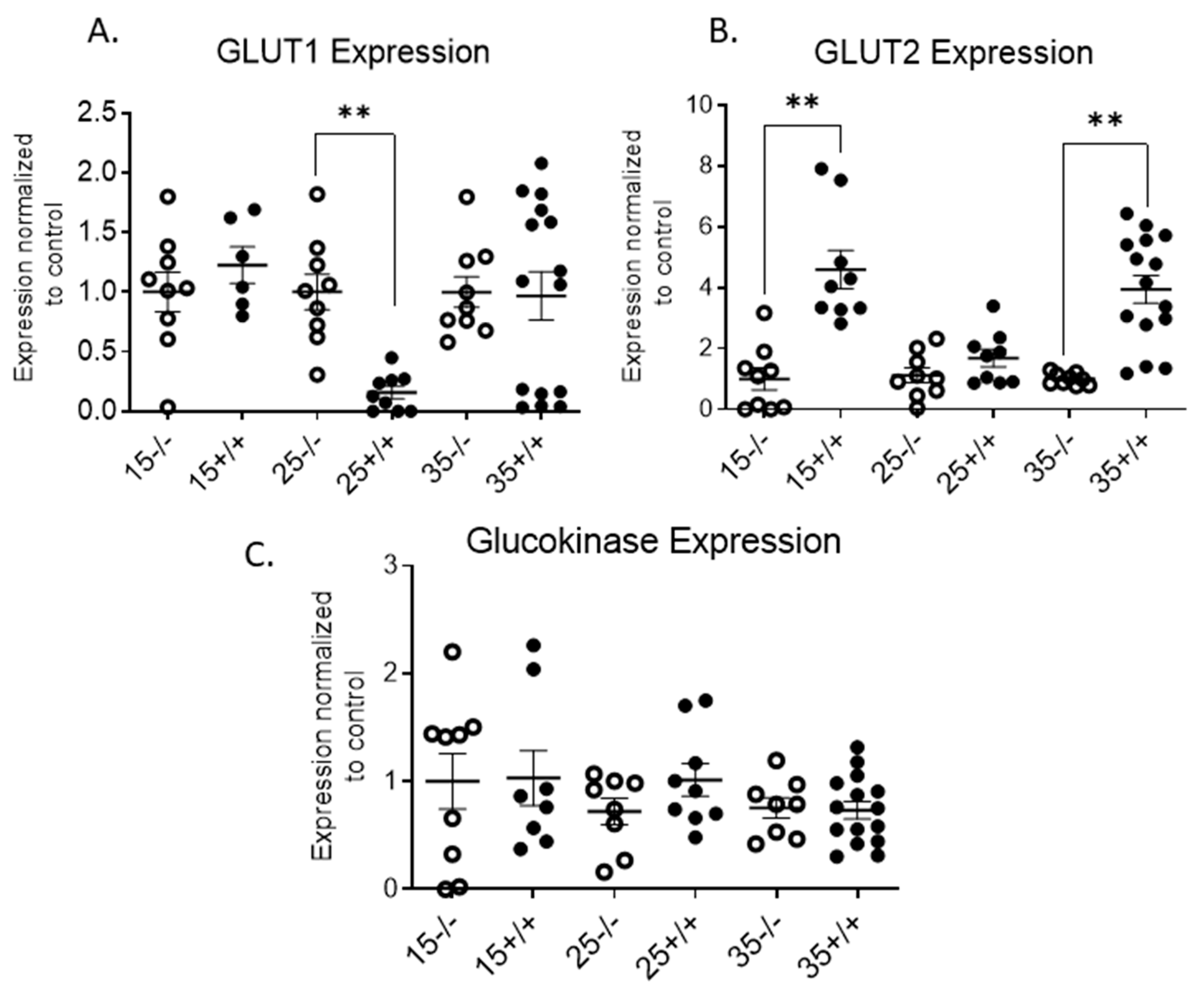
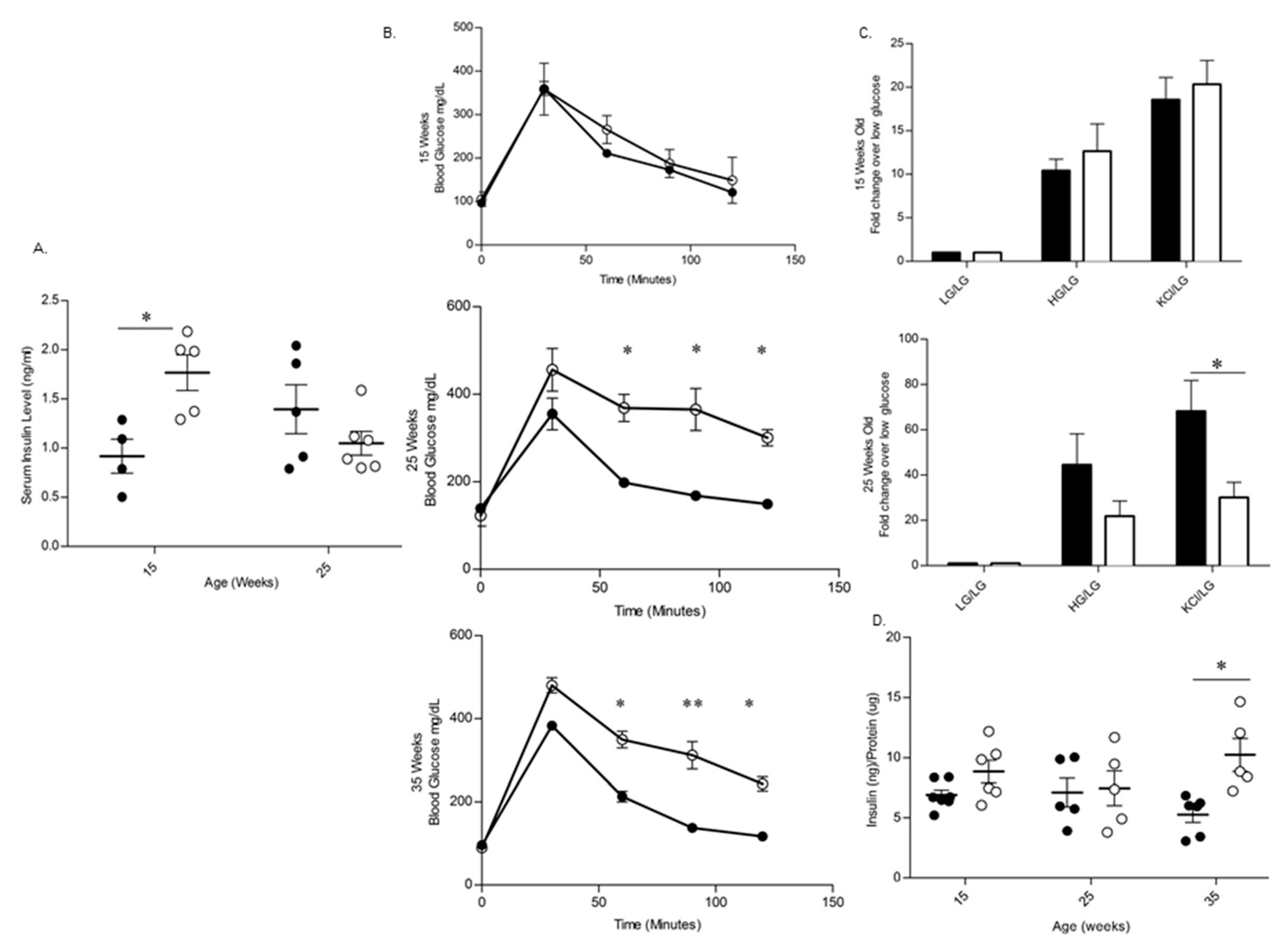
3.3. BL/6-CD3FLAGmIR Transgenic Animal Metabolic Abnormalities
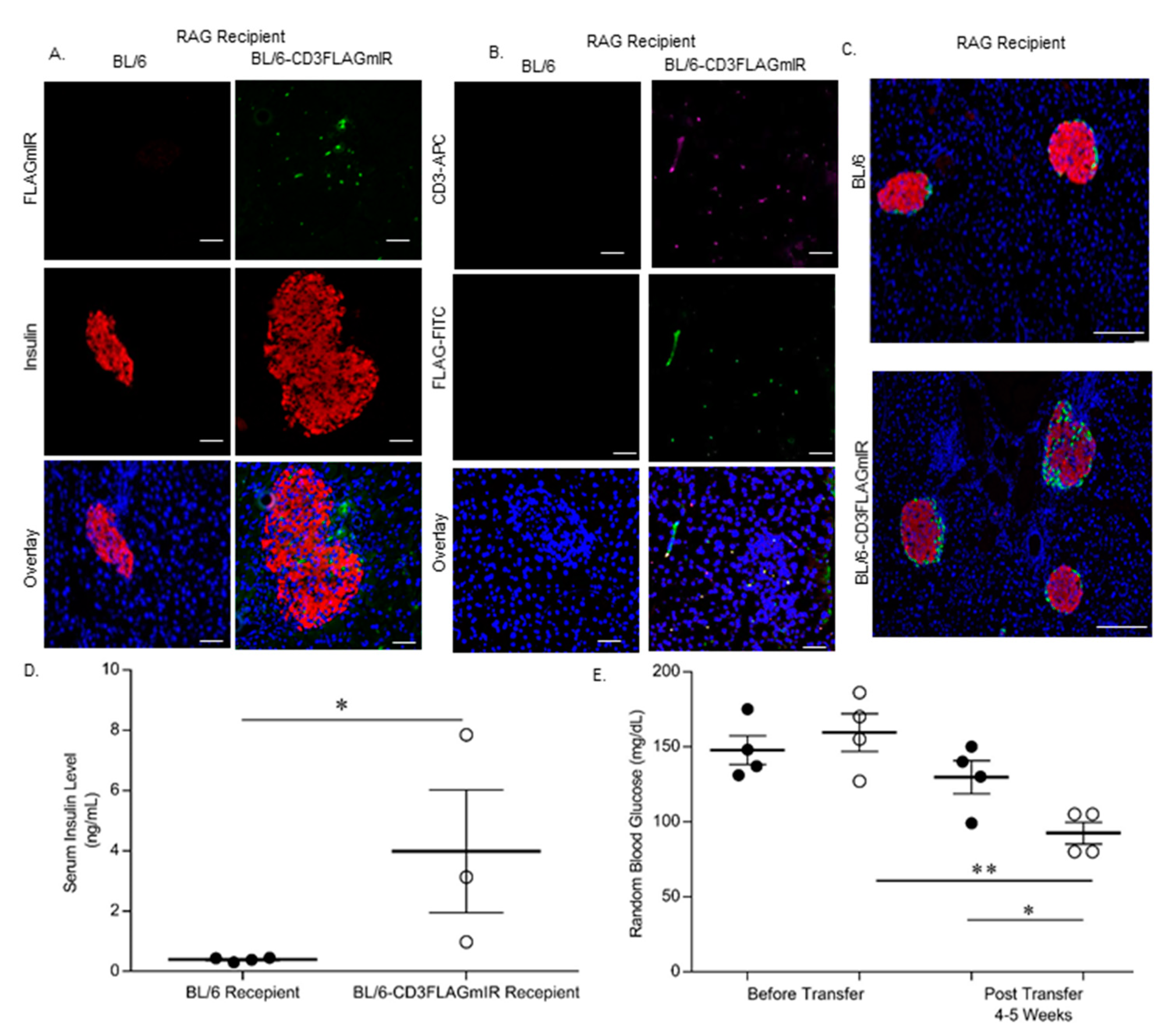
3.4. Adoptive Transfer Studies
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fischer, H.J.; Sie, C.; Schumann, E.; Witte, A.-K.; Dressel, R.; Brandt, J.V.D.; Reichardt, H.M. The Insulin Receptor Plays a Critical Role in T Cell Function and Adaptive Immunity. J. Immunol. 2017, 198, 1910–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stentz, F.B.; Kitabchi, A.E. Transcriptome and Proteome Expressions Involved in Insulin Resistance in Muscle and Activated T-Lymphocytes of Patients with Type 2 Diabetes. Genom. Proteom. Bioinform. 2007, 5, 216–235. [Google Scholar] [CrossRef] [Green Version]
- Stentz, F.B.; Kitabchi, A.E. Activated T lymphocytes in Type 2 diabetes: Implications from in vitro studies. Curr. Drug Targets 2003, 4, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.; Clemente-Casares, X.; Zhou, A.C.; Lei, H.; Ahn, J.J.; Chan, Y.T.; Choi, O.; Luck, H.; Woo, M.; Dunn, S.E.; et al. Insulin Receptor-Mediated Stimulation Boosts T Cell Immunity during Inflammation and Infection. Cell Metab. 2018, 28, 922–934.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, J.S.; Center, D.M. Chemotactic activity of porcine insulin for human T lymphocytes in vitro. J. Immunol. 1987, 138, 2100–2103. [Google Scholar]
- McInerney, M.F.; Flynn, J.C.; Goldblatt, P.J.; Najjar, S.M.; Sherwin, R.S.; Janeway, C.A. High density insulin receptor-positive T lymphocytes from nonobese diabetic mice transfer insulitis and diabetes. J. Immunol. 1996, 157, 3716–3726. [Google Scholar]
- Yenush, L.; Kundra, V.; White, M.F.; Zetter, B.R. Functional domains of the insulin receptor responsible for chemotactic signaling. J. Biol. Chem. 1994, 269, 100–104. [Google Scholar] [CrossRef]
- Knip, M.; Honkanen, J. Modulation of Type 1 Diabetes Risk by the Intestinal Microbiome. Curr. Diabetes Rep. 2017, 17, 105. [Google Scholar] [CrossRef] [Green Version]
- Foulis, A.K. Pancreatic Pathology in Type 1 Diabetes in Human. Novartis Found. Symp. 2008, 292, 2–13. [Google Scholar] [CrossRef]
- Atkinson, M.A.; Eisenbarth, G.S. Type 1 diabetes: New perspectives on disease pathogenesis and treatment. Lancet 2001, 358, 221–229. [Google Scholar] [CrossRef]
- Bluestone, J.A.; Herold, K.; Eisenbarth, G.S. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature 2010, 464, 1293–1300. [Google Scholar] [CrossRef] [Green Version]
- Makino, S.; Kunimoto, K.; Muraoka, Y.; Mizushima, Y.; Katagiri, K.; Tochino, Y. Breeding of a Non-Obese, Diabetic Strain of Mice. Jikken Dobutsu 1980, 29, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Itoh, N.; Hanafusa, T.; Miyazaki, A.; Miyagawa, J.; Yamagata, K.; Yamamoto, K.; Waguri, M.; Imagawa, A.; Tamura, S.; Inada, M. Mononuclear cell infiltration and its relation to the expression of major histocompatibility complex antigens and adhesion molecules in pancreas biopsy specimens from newly diagnosed insulin-dependent diabetes mellitus patients. J. Clin. Investig. 1993, 92, 2313–2322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katz, J.; Benoist, C.; Mathis, D. Major histocompatibility complex class I molecules are required for the development of insulitis in non-obese diabetic mice. Eur. J. Immunol. 1993, 23, 3358–3360. [Google Scholar] [CrossRef] [PubMed]
- Yagi, H.; Matsumoto, M.; Kunimoto, K.; Kawaguchi, J.; Makino, S.; Harada, M. Analysis of the roles of CD4+ and CD8+ T cells in autoimmune diabetes of NOD mice using transfer to NOD athymic nude mice. Eur. J. Immunol. 1992, 22, 2387–2393. [Google Scholar] [CrossRef] [PubMed]
- Sibley, R.K.; Sutherland, D.E.; Goetz, F.; Michael, A.F. Recurrent diabetes mellitus in the pancreas iso- and allograft. A light and electron microscopic and immunohistochemical analysis of four cases. Lab. Investig. 1985, 53, 132–144. [Google Scholar]
- Sandor, A.M.; Lindsay, R.S.; Dyjack, N.; Whitesell, J.C.; Rios, C.; Bradley, B.J.; Haskins, K.; Serreze, D.V.; Geurts, A.M.; Chen, Y.-G.; et al. CD11c+ Cells Are Gatekeepers for Lymphocyte Trafficking to Infiltrated Islets During Type 1 Diabetes. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Alejandro, R.; Shienvold, F.L.; Hajek, S.V.; Ryan, U.; Miller, J.; Mintz, D.H. Immunocytochemical Localization of HLA-DR in Human Islets of Langerhans. Diabetes 1982, 31 (Suppl. 4), 17–22. [Google Scholar] [CrossRef]
- Campbell-Thompson, M.; Fu, A.; Kaddis, J.S.; Wasserfall, C.; Schatz, D.A.; Pugliese, A.; Atkinson, M.A. Insulitis and beta-Cell Mass in the Natural History of Type 1 Diabetes. Diabetes 2016, 65, 719–731. [Google Scholar] [CrossRef] [Green Version]
- McInerney, M.F.; Rath, S.; Janeway, C.A., Jr. Exclusive expression of MHC class II proteins on CD45+ cells in pancreatic islets of NOD mice. Diabetes 1991, 40, 648–651. [Google Scholar] [CrossRef]
- Bohme, J.; Haskins, K.; Stecha, P.; Van Ewijk, W.; LeMeur, M.; Gerlinger, P.; Benoist, C.; Mathis, D. Transgenic mice with I-A on islet cells are normoglycemic but immunologically intolerant. Science 1989, 244, 1179–1183. [Google Scholar] [CrossRef]
- Hamilton-Williams, E.E.; Palmer, S.E.; Charlton, B.; Slattery, R.M. Beta cell MHC class I is a late requirement for diabetes. Proc. Natl. Acad. Sci. USA 2003, 100, 6688–6693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morran, M.P.; Al-Dieri, A.G.; Nestor-Kalinoski, A.L.; Jordan, R.K.; Gupta, N.K.; McInerney, M.F. Insulin receptor based lymphocyte trafficking in the progression of type 1 diabetes. J. Biol. Methods 2018, 5, e85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandedkar-Kulkarni, N.; Esakov, E.; Gregg, B.; Atkinson, M.A.; Rogers, D.G.; Horner, J.D.; Singer, K.; Lundy, S.K.; Felton, J.L.; Al-Huniti, T.; et al. Insulin Receptor–Expressing T Cells Appear in Individuals at Risk for Type 1 Diabetes and Can Move into the Pancreas in C57BL/6 Transgenic Mice. J. Immunol. 2021, 206, 1443–1453. [Google Scholar] [CrossRef] [PubMed]
- Szot, G.L.; Koudria, P.; Bluestone, J.A. Murine Pancreatic Islet Isolation. J. Vis. Exp. 2007, 2007, 255. [Google Scholar] [CrossRef]
- Jörns, A.; Arndt, T.; Zu Vilsendorf, A.M.; Klempnauer, J.; Wedekind, D.; Hedrich, H.-J.; Marselli, L.; Marchetti, P.; Harada, N.; Nakaya, Y.; et al. Islet infiltration, cytokine expression and beta cell death in the NOD mouse, BB rat, Komeda rat, LEW.1AR1-iddm rat and humans with type 1 diabetes. Diabetologia 2014, 57, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Ventura-Oliveira, D.; Vilella, C.; Zanin, M.; Castro, G.; Filho, D.M.; Zollner, R. Kinetics of TNF-alpha and IFN-gamma mRNA expression in islets and spleen of NOD mice. Braz. J. Med. Biol. Res. 2002, 35, 1347–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melmed, S.; Polonsky, K.S.; Burant, C.F. (Eds.) Chapter 31—Type 2 Diabetes Mellitus. In Williams Textbook of Endocrinology, 13th ed.; Elsevier: Philadelphia, PA, USA, 2016; pp. 1385–1450. [Google Scholar]
- Christoffersson, G.; Chodaczek, G.; Ratliff, S.S.; Coppieters, K.; Von Herrath, M.G. Suppression of diabetes by accumulation of non–islet-specific CD8 + effector T cells in pancreatic islets. Sci. Immunol. 2018, 3, eaam6533. [Google Scholar] [CrossRef] [Green Version]
- Magnuson, A.M.; Thurber, G.M.; Kohler, R.H.; Weissleder, R.; Mathis, D.; Benoist, C. Population dynamics of islet-infiltrating cells in autoimmune diabetes. Proc. Natl. Acad. Sci. USA 2015, 112, 1511–1516. [Google Scholar] [CrossRef] [Green Version]
- Willcox, A.; Richardson, S.J.; Bone, A.; Foulis, A.K.; Morgan, N.G. Analysis of islet inflammation in human type 1 diabetes. Clin. Exp. Immunol. 2009, 155, 173–181. [Google Scholar] [CrossRef]
- Claudine, S.; Clark, S.R.; Norman, U.M. Use of CD44 by CD4+ Th1 and Th2 lymphocytes to roll and adhere. Blood 2006, 107, 4798–4806. [Google Scholar]
- Reiser, J.; Banerjee, A. Effector, Memory, and Dysfunctional CD8+T Cell Fates in the Antitumor Immune Response. J. Immunol. Res. 2016, 2016, 8941260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samji, T.; Khanna, K.M. Understanding memory CD8 + T cells. Immunol. Lett. 2017, 185, 32–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gossel, G.; Hogan, T.; Cownden, D.; Seddon, B.; Yates, A.J. Memory CD4 T cell subsets are kinetically heterogeneous and replenished from naive T cells at high levels. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, A.B.; Pinelli, D.F.; Liu, D.; Wagener, M.; Ford, M.L. Memory T cell-mediated rejection is mitigated by FcgammaRIIB expression on CD8(+) T cells. Am. J. Transplant. 2020, 20, 2206–2215. [Google Scholar] [CrossRef]
- Garyu, J.W.; Uduman, M.; Stewart, A.; Rui, J.; Deng, S.; Shenson, J.; Staron, M.M.; Kaech, S.M.; Kleinstein, S.H.; Herold, K.C. Characterization of Diabetogenic CD8+ T Cells: Immune Therapy with Metabolic Blockade. J. Biol. Chem. 2016, 291, 11230–11240. [Google Scholar] [CrossRef] [Green Version]
- De Marquesini, L.G.; Fu, J.; Connor, K.J.; Bishop, A.J.; McLintock, N.E.; Pope, C.; Wong, F.S.; Dayan, C.M. IFN-γ and IL-10 islet-antigen-specific T cell responses in autoantibody-negative first-degree relatives of patients with type 1 diabetes. Diabetologia 2010, 53, 1451–1460. [Google Scholar] [CrossRef] [Green Version]
- Moseley, T.A.; Haudenschild, D.; Rose, L.; Reddi, A. Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev. 2003, 14, 155–174. [Google Scholar] [CrossRef]
- Fonseca, S.G.; Gromada, J.; Urano, F. Endoplasmic reticulum stress and pancreatic β-cell death. Trends Endocrinol. Metab. 2011, 22, 266–274. [Google Scholar] [CrossRef] [Green Version]
- Marhfour, I.; Lopez, X.M.; Lefkaditis, D.; Salmon, I.; Allagnat, F.; Richardson, S.J.; Morgan, N.G.; Eizirik, D.L. Expression of endoplasmic reticulum stress markers in the islets of patients with type 1 diabetes. Diabetologia 2012, 55, 2417–2420. [Google Scholar] [CrossRef]
- Leiter, E.; Atkinson, M. Nod Mice and Related Strains: Research Applications in Diabetes, AIDS, Cancer, and Other Diseases (Molecular Biology Intelligence Unit); R. G. Landes Company: Austin, TX, USA, 1998; p. 208. [Google Scholar]
- Sherry, N.A.; Kushner, J.A.; Glandt, M.; Kitamura, T.; Brillantes, A.-M.; Herold, K.C. Effects of autoimmunity and immune therapy on beta-cell turnover in type 1 diabetes. Diabetes 2006, 55, 3238–3245. [Google Scholar] [CrossRef] [Green Version]
- Brawerman, G.; Thompson, P.J. Beta Cell Therapies for Preventing Type 1 Diabetes: From Bench to Bedside. Biomolecules 2020, 10, 1681. [Google Scholar] [CrossRef] [PubMed]
- Dirice, E.; Kahraman, S.; Jiang, W.; Ouaamari, A.E.; De Jesus, D.F.; Teo, A.K.K.; Hu, J.; Kawamori, D.; Gaglia, J.L.; Mathis, D.; et al. Soluble Factors Secreted by T Cells Promote β-Cell Proliferation. Diabetes 2014, 63, 188–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Tsang, K.S.; Chan, J.C.N.; Yuan, P.; Fan, R.; Kaneto, H.; Xu, G. The Combined Expression of Pdx1 and MafA with Either Ngn3 or NeuroD Improves the Differentiation Efficiency of Mouse Embryonic Stem Cells into Insulin-Producing Cells. Cell Transplant. 2013, 22, 147–158. [Google Scholar] [CrossRef]
- Willcox, A.; Richardson, S.J.; Bone, A.; Foulis, A.K.; Morgan, N.G. Evidence of increased islet cell proliferation in patients with recent-onset type 1 diabetes. Diabetologia 2010, 53, 2020–2028. [Google Scholar] [CrossRef] [Green Version]
- Devedjian, J.C.; George, M.; Casellas, A.; Pujol, A.; Visa, J.; Pelegrin, M.; Gros, L.; Bosch, F. Transgenic mice overexpressing insulin-like growth factor-II in beta cells develop type 2 diabetes. J. Clin. Investig. 2000, 105, 731–740. [Google Scholar] [CrossRef] [Green Version]
- Thorens, B.; Guillam, M.T.; Beermann, F.; Burcelin, R.; Jaquet, M. Transgenic reexpression of GLUT1 or GLUT2 in pancreatic beta cells rescues GLUT2-null mice from early death and restores normal glucose-stimulated insulin secretion. J. Biol. Chem. 2000, 275, 23751–23758. [Google Scholar] [PubMed] [Green Version]
- Sosenko, J.M.; Palmer, J.P.; Greenbaum, C.J.; Mahon, J.; Cowie, C.; Krischer, J.P.; Chase, H.P.; White, N.H.; Buckingham, B.; Herold, K.C.; et al. Patterns of Metabolic Progression to Type 1 Diabetes in the Diabetes Prevention Trial-Type 1. Diabetes Care 2006, 29, 643–649. [Google Scholar] [CrossRef] [Green Version]
- Ramlo-Halsted, B.A.; Edelman, S.V. The Natural History of Type 2 Diabetes. Implications for Clinical Practice. Prim. Care Clin. Off. Pract. 1999, 26, 771–789. [Google Scholar] [CrossRef]




| Reagent | 20 µL Reaction Volume | 70 µL Reaction Volume |
|---|---|---|
| 2× SYBR Green Master Mix | 10 µL | 35 µL |
| Forward Primer | 1 µL | 3.5 µL |
| Reverse Primer | 1 µL | 3.5 µL |
| DI Water | 7 µL | 24.5 µL |
| cDNA (150 ng–200 ng/µL) | 1 µL | 3.5 µL |
| FOR_WFS | CCATCAACATGCTCCCGTTC | |
| REV_WFS | GGGTAGGCCTCGCCATACA | |
| FOR_BIP | TTCAGCCAATTATCAGCAAACTCT | |
| REV_BIP | TTTTCTGATGTATCCTCTTCACCAGT | |
| FOR_ERO1 | GCTCTCTCCAAAGTGCTTCCA | |
| REV_ERO1 | TGCATCCTGAACTTTATTCCCA | |
| FOR_PDX1 | GACACATCAAAATCTGGTTCCAAA | |
| REV_PDX1 | TCCCGCTACTACGTTTCTTATCTTC | |
| FOR_GAPDH | AAGGTCATCCCAGAGCTGAA | |
| REV_GAPDH | ATGTAGGCCATGAGGTCCAC | |
| FOR_FLAGmIR | CCATGTCTGCACTTCTGACTAGCTCTT | |
| REV_FLAGmIR | CCATAGACACGGAAGAGAGCAGGTAATC | |
| FOR_IFNG | CGGCACAGTCATTGAAAGCCTA | |
| REV_IFNG | GTTGCTGATGGCCTGATTGTC | |
| FOR_IL-17 | GCTCCAGAAGGCCCTCAGA | |
| REV_IL-17 | CTTTCCCTCCGCATTGACA | |
| FOR_IL-10 | CACAAAGCAGCCTTGCAGAA | |
| REV_IL-10 | AGAGCAGGCAGCATAGCAGTG | |
| FOR_TNFA | GATCTCAAAGACAACCAACATGTG | |
| REV_TNFA | CTCCAGCTGGAAGACTCCTCCCAG | |
| FOR_TGFB | CACCGGAGAGCCCTGGATA | |
| REV_TGFB | TGTACAGCTGCCGCACACA | |
| FOR_Beta-Globin | CCAATCTGCTCACACAGGATA | |
| Rev_Beta-Globin | CCTTGAGGCTGTCCAAGTGAT | |
| FOR_WFS | CCATCAACATGCTCCCGTTC | |
| REV_WFS | GGGTAGGCCTCGCCATACA | |
| FOR_BIP | TTCAGCCAATTATCAGCAAACTCT | |
| REV_BIP | TTTTCTGATGTATCCTCTTCACCAGT | |
| FOR_ERO1 | GCTCTCTCCAAAGTGCTTCCA | |
| REV_ERO1 | TGCATCCTGAACTTTATTCCCA | |
| FOR_PDX1 | GACACATCAAAATCTGGTTCCAAA | |
| REV_PDX1 | TCCCGCTACTACGTTTCTTATCTTC | |
| FOR_PAX4 | GTGTTGGCTCCAGTTCTTCC | |
| REV_PAX4 | AACCAAACCCTCACCGTGTC | |
| FOR_GLUT2 | GGCTAATTTCAGGACTGGTT | |
| REV_GLUT2 | TTTCTTTGCCCTGACTTCCT | |
| FOR_MAFA | TTCAGCAAGGAGGAGGTCAT | |
| REV_MAFA | CCGCCAACTTCTCGTATTTC | |
| FOR_GLUT1 | CATCCTTATTGCCCAGGTGTTT | |
| REV_GLUT1 | GAAGACGACACTGAGCAGCAGA | |
| FOR_NEUROD1 | TATTGCGTTGCCTTAGCACT | |
| REV_NEUROD1 | CATCCTCTTGAGTGTTATGG | |
| FOR_CD11C | ACACAGTGTGCTCCAGTATGA | |
| REV_CD11C | GCCCAGGGATATGTTGACAGC | |
| FOR_CD68 | CTTCCCACAGGCAGCACAG | |
| REV_CD68 | AATGATGAGAGGCAGCAAGAGG | |
| FOR_FOXP3 | ACTCGCATGTTCGCCTACTTCAG | |
| REV_FOXP3 | GGCGGATGGCATTCTTCCAGGT | |
| FOR_CD44 | TGAAACATGCAGGTATGGGT | |
| REV_CD44 | GCTGAGGCATTGAAGCAATA | |
| FOR_CD69 | TGGTCCTCATCACGTCCTTAATAA | |
| REV_CD69 | TCCAACTTCTCGTACAAGCCTG | |
| FOR_CD62L | CATTCCTGTAGCCGTCATGG | |
| REV_CD62L | AGGAGGAGCTGTTGGTCATG | |
| FOR_GLUCOKINASE | GAATCTTCTGTTCCACGGAG | |
| REV_GLUCOKINASE | AGTGCTCAGGATGTTAAGGA | |
| FOR_CHOP | CCACCACACCTGAAAGCAGAA | |
| REV_CHOP | AGGTGAAAGGCAGGGACTCA | |
| FOR_PAX4 | GTGTTGGCTCCAGTTCTTCC | |
| REV_PAX4 | AACCAAACCCTCACCGTGTC | |
| FOR_GLUT2 | GGCTAATTTCAGGACTGGTT | |
| REV_GLUT2 | TTTCTTTGCCCTGACTTCCT |
| Step | Temperature (°C) | Time |
|---|---|---|
| 1. Initial Denaturation | 95 | 3 min |
| 2. Template Denaturation | 95 | 30 s |
| 3. Primer Annealing | Depending upon the Forward and Reverse Primer melting temperature | 30 s |
| 4. Primer Extension | 72 | 30 s |
| 5. Cycle Repeat (steps 2–4) 39 times’ | ||
| 6. Plate Read for SYBR Green after every cycle | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esakov, E.; Nandedkar-Kulkarni, N.; Al-Dieri, A.G.; Hafner, H.; Gregg, B.; McInerney, M.F. Islet Dysfunction in a Novel Transgenic Model of T Cell Insulitis. Biomolecules 2021, 11, 552. https://doi.org/10.3390/biom11040552
Esakov E, Nandedkar-Kulkarni N, Al-Dieri AG, Hafner H, Gregg B, McInerney MF. Islet Dysfunction in a Novel Transgenic Model of T Cell Insulitis. Biomolecules. 2021; 11(4):552. https://doi.org/10.3390/biom11040552
Chicago/Turabian StyleEsakov, Emily, Neha Nandedkar-Kulkarni, Ali G. Al-Dieri, Hannah Hafner, Brigid Gregg, and Marcia F. McInerney. 2021. "Islet Dysfunction in a Novel Transgenic Model of T Cell Insulitis" Biomolecules 11, no. 4: 552. https://doi.org/10.3390/biom11040552
APA StyleEsakov, E., Nandedkar-Kulkarni, N., Al-Dieri, A. G., Hafner, H., Gregg, B., & McInerney, M. F. (2021). Islet Dysfunction in a Novel Transgenic Model of T Cell Insulitis. Biomolecules, 11(4), 552. https://doi.org/10.3390/biom11040552







