Phosphorylation, Mg-ADP, and Inhibitors Differentially Shape the Conformational Dynamics of the A-Loop of Aurora-A
Abstract
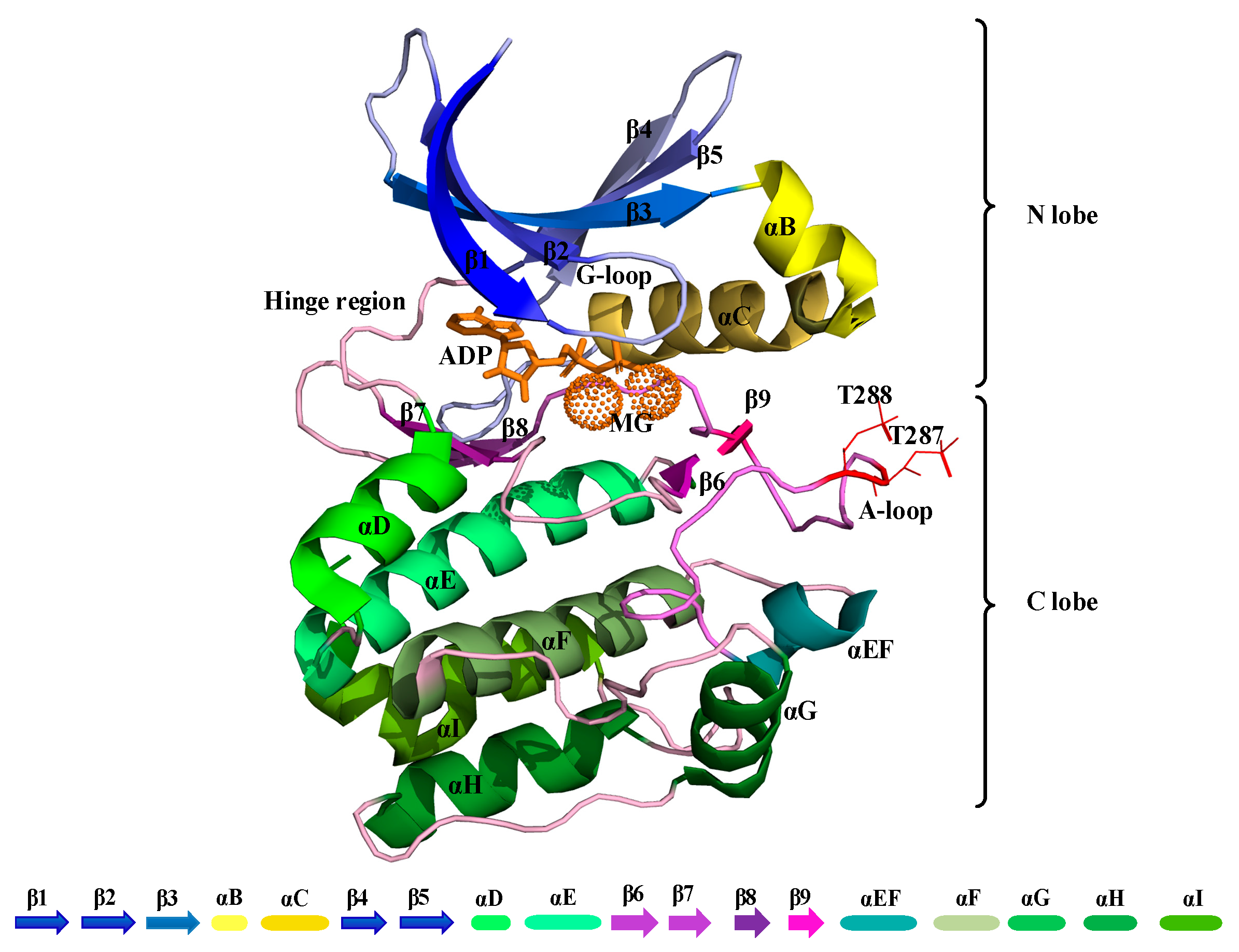
1. Introduction
2. Materials and Methods
2.1. MD Simulations
2.2. Principal Component Analysis
2.3. Cluster Analysis
2.4. Inter-Lobe Angle Analysis
3. Results
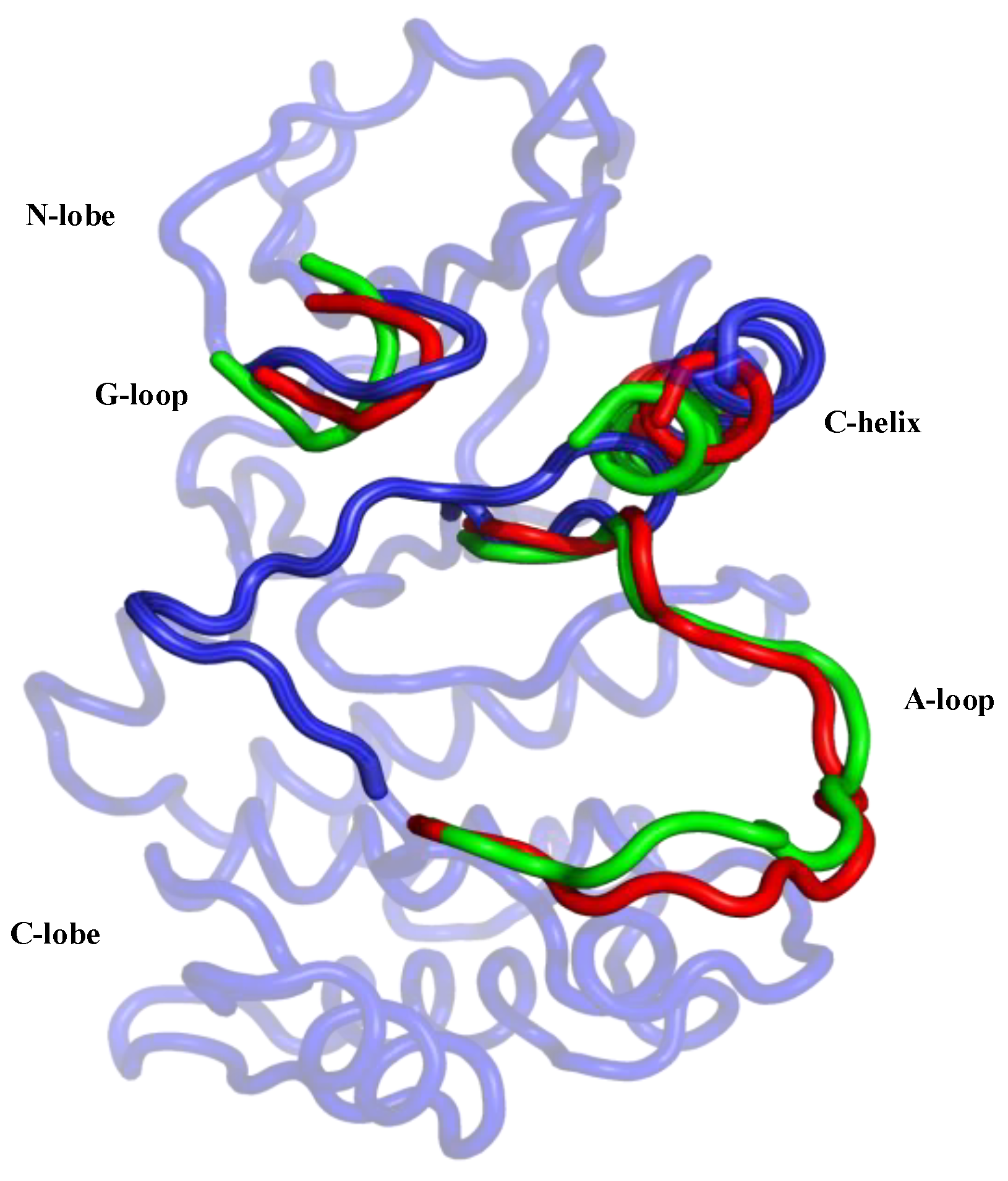
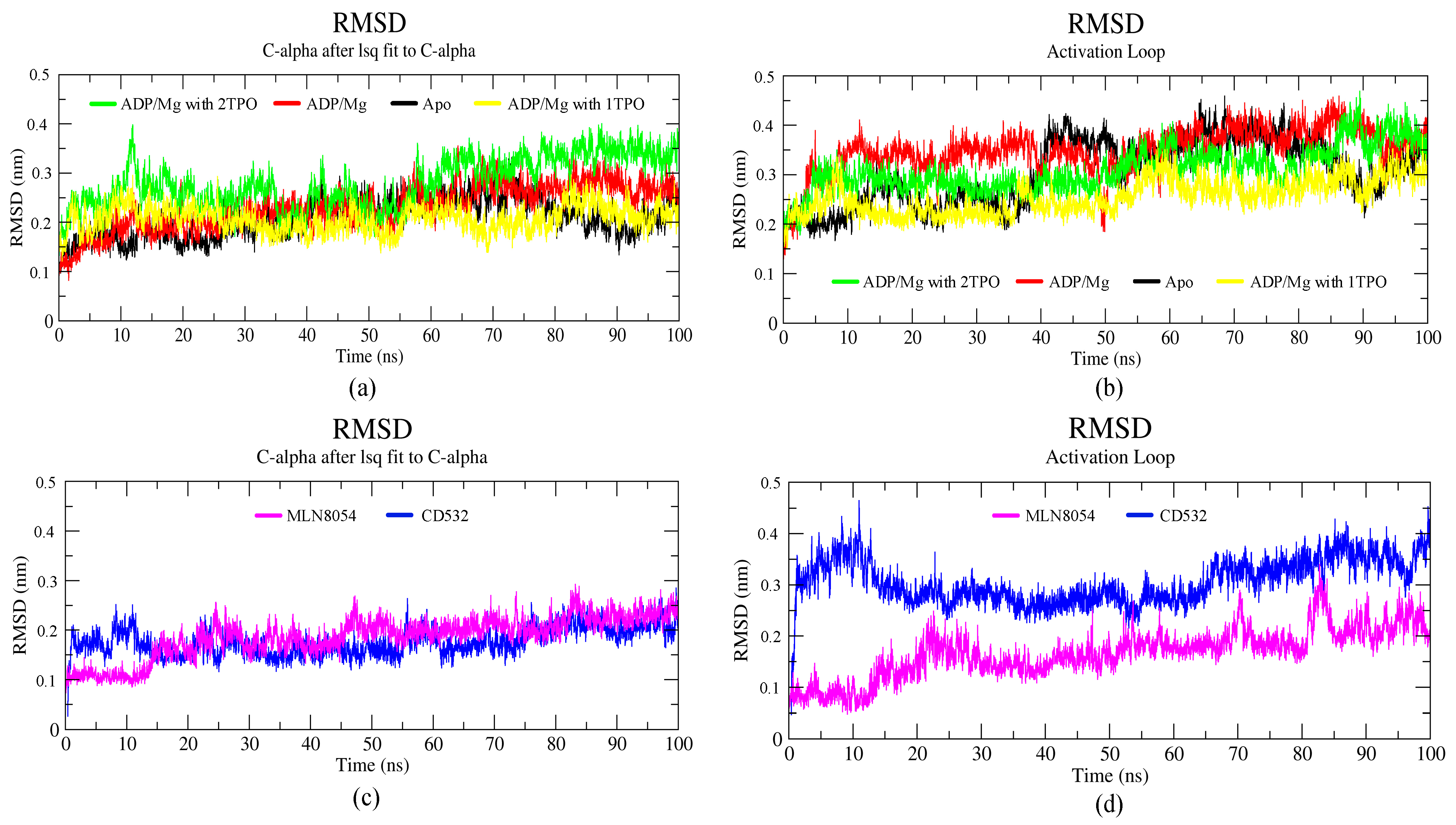
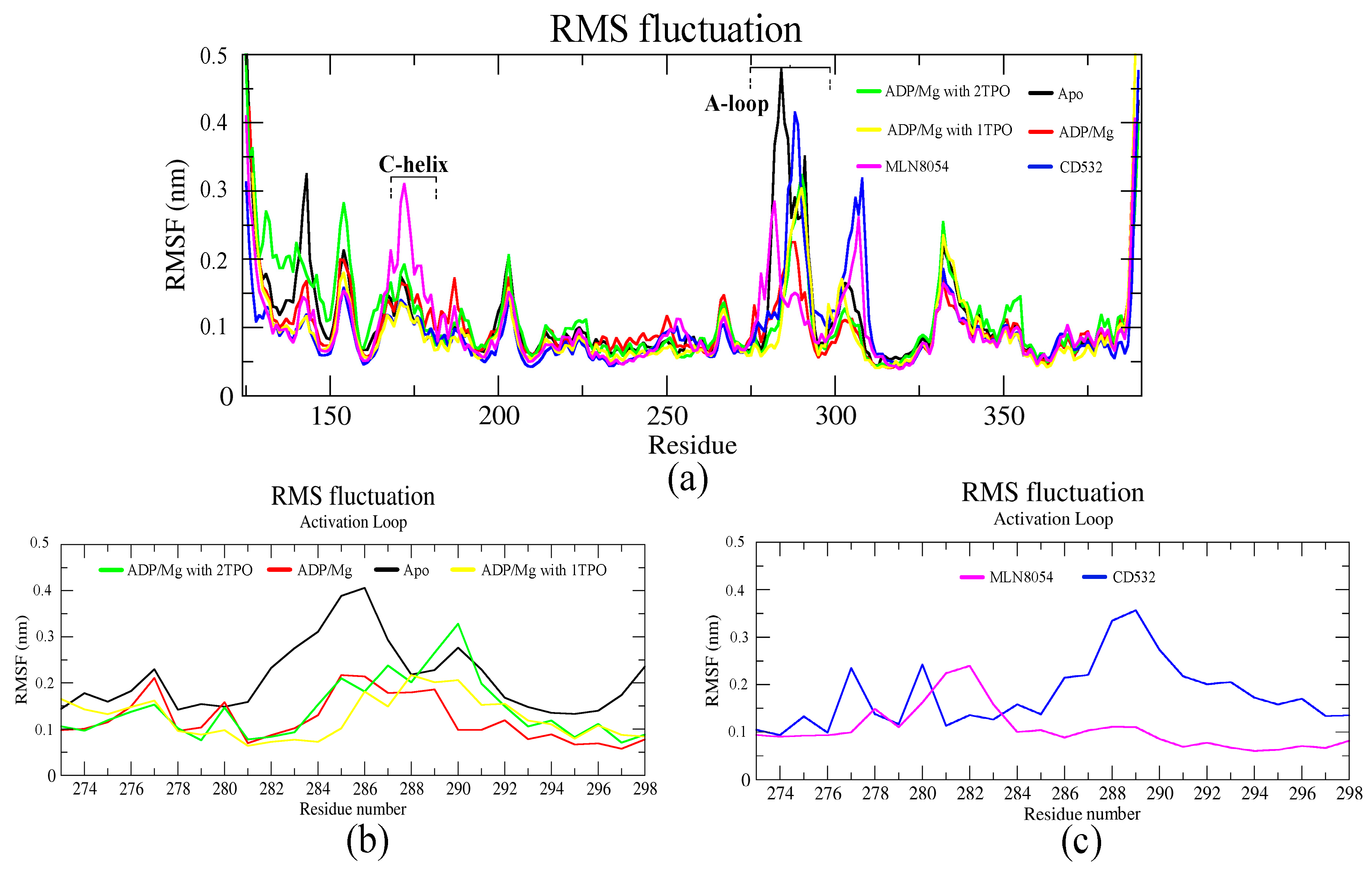
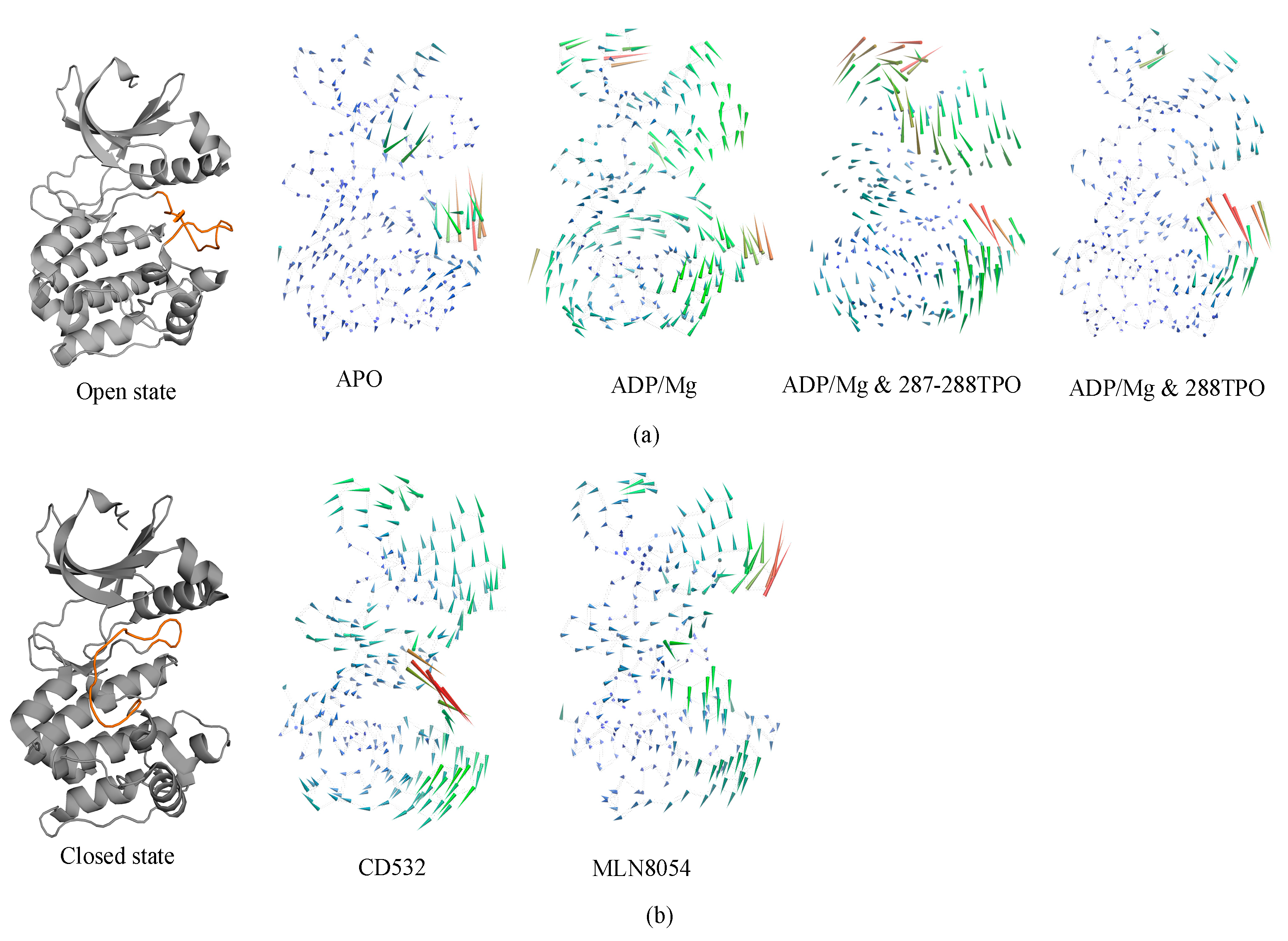
3.1. Collective Motions
3.2. Characterizations of Global and Local Concerted Motions in Aurora-A
3.3. Ligand Interactions and Inter-Lobe Motions
3.4. Angle Analysis
3.5. Protein-Ligand Contacts Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nigg, E.A. Cell division: Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001, 2, 21. [Google Scholar] [CrossRef]
- Jeng, Y.M.; Peng, S.Y.; Lin, C.Y.; Hsu, H.C. Overexpression and amplification of Aurora-A in hepatocellular carcinoma. Clin. Cancer Res. 2004, 10, 2065–2071. [Google Scholar] [CrossRef]
- Lee, S.Y.; Lee, G.R.; Woo, D.H.; Park, N.H.; Cha, H.J.; Moon, Y.H.; Han, I.S. Depletion of Aurora-A leads to upregulation of FoxO1 to induce cell cycle arrest in hepatocellular carcinoma cells. Cell Cycle 2013, 12, 67–75. [Google Scholar] [CrossRef][Green Version]
- Herberg, F.W.; Doyle, M.L.; Cox, S.; Taylor, S.S. Dissection of the nucleotide and metal− phosphate binding sites in cAMP-dependent protein kinase. Biochemistry 1999, 38, 6352–6360. [Google Scholar] [CrossRef]
- Zhao, Z.S.; Lim, J.P.; Ng, Y.W.; Lim, L.; Manser, E. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol. Cell 2005, 20, 237–249. [Google Scholar] [CrossRef]
- Nikonova, A.S.; Astsaturov, I.; Serebriiskii, I.G.; Dunbrack, R.L.; Golemis, E.A. Aurora-A kinase (AURKA) in normal and pathological cell division. Cell. Mol. Life Sci. 2013, 70, 661–687. [Google Scholar] [CrossRef]
- Walter, A.O.; Seghezzi, W.; Korver, W.; Sheung, J.; Lees, E. The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene 2000, 19, 4906. [Google Scholar] [CrossRef]
- Richards, M.W.; Burgess, S.G.; Poon, E.; Carstensen, A.; Eilers, M.; Chesler, L.; Bayliss, R. Structural basis of N-Myc binding by Aurora-A and its destabilization by kinase inhibitors. Proc. Natl. Acad. Sci. USA 2016, 113, 13726–13731. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Hu, G.; Wei, G.; Cui, K.; Yamane, A.; Resch, W.; Wang, R.; Green, D.R.; Tessarollo, L.; Casellas, R.; et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012, 151, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Lovén, J.; Rahl, P.B.; Paranal, R.M.; Burge, C.B.; Bradner, J.E.; Lee, T.I.; Young, R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 2012, 151, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Brockmann, M.; Poon, E.; Berry, T.; Carstensen, A.; Deubzer, H.E.; Rycak, L.; Jamin, Y.; Thway, K.; Robinson, S.P.; Roels, F.; et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell 2013, 24, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, W.C.; Meyerowitz, J.G.; Nekritz, E.A.; Chen, J.; Benes, C.; Charron, E.; Simonds, E.F.; Seeger, R.; Matthay, K.K.; Hertz, N.T.; et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell 2014, 26, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Dodson, C.A.; Kosmopoulou, M.; Richards, M.W.; Atrash, B.; Bavetsias, V.; Blagg, J.; Bayliss, R. Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: Insights into selectivity and drug design. Biochem. J. 2010, 427, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Meyerowitz, J.G.; Weiss, W.A.; Gustafson, W.C. A new “angle” on kinase inhibitor design: Prioritizing amphosteric activity above kinase inhibition. Mol. Cell. Oncol. 2015, 2, e975641. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Nowakowski, J.; Cronin, C.N.; McRee, D.E.; Knuth, M.W.; Nelson, C.G.; Pavletich, N.P.; Rogers, J.; Sang, B.-C.; Scheibe, D.N.; Swanson, R.V.; et al. Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography. Structure 2002, 10, 1659–1667. [Google Scholar] [CrossRef]
- Bayliss, R.; Sardon, T.; Vernos, I.; Conti, E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell 2003, 12, 851–862. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. Ccp4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Abraham, M.J.; van der Spoel, D.; Lindahl, E.; Hess, B.; the GROMACS Development Team. GROMACS User Manual Version. 2018. Available online: www.gromacs.org (accessed on 21 March 2018).
- Gutiérrez, I.S.; Lin, F.Y.; Vanommeslaeghe, K.; Lemkul, J.A.; Armacost, K.A.; Brooks, C.L., III; MacKerell, A.D., Jr. Parametrization of halogen bonds in the CHARMM general force field: Improved treatment of ligand-protein interactions. Bioorganic Med. Chem. 2016, 24, 4812–4825. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Berendsen, H.J.; Postma, J.V.; van Gunsteren, W.F.; DiNola, A.R.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463. [Google Scholar] [CrossRef]
- Amadei, A.; Linssen, A.B.; Berendsen, H.J. Essential dynamics of proteins. Proteins: Struct. Funct. Bioinform. 1993, 17, 412–425. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Satinover, D.L.; Leach, C.A.; Stukenberg, P.T.; Brautigan, D.L. Activation of Aurora-A kinase by protein phosphatase inhibitor-2, a bifunctional signaling protein. Proc. Natl. Acad. Sci. USA 2004, 101, 8625–8630. [Google Scholar] [CrossRef]
- Burgess, S.G.; Bayliss, R. The structure of C290A: C393A Aurora A provides structural insights into kinase regulation. Acta Crystallogr. Sect. F: Struct. Biol. Commun. 2015, 71, 315–319. [Google Scholar] [CrossRef]
- Rowan, F.C.; Richards, M.; Bibby, R.A.; Thompson, A.; Bayliss, R.; Blagg, J. Insights into Aurora-A kinase activation using unnatural amino acids incorporated by chemical modification. Acs Chem. Biol. 2013, 8, 2184–2191. [Google Scholar] [CrossRef]
- Willems, E.; Dedobbeleer, M.; Digregorio, M.; Lombard, A.; Lumapat, P.N.; Rogister, B. The functional diversity of Aurora kinases: A comprehensive review. Cell Div. 2018, 13, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Nolen, B.; Taylor, S.; Ghosh, G. Regulation of protein kinases: Controlling activity through activation segment conformation. Mol. Cell 2004, 15, 661–675. [Google Scholar] [CrossRef] [PubMed]
- Roeschert, I.; Poon, E.; Henssen, A.G.; Garcia, H.D.; Gatti, M.; Giansanti, C.; Eilers, M. Combined inhibition of Aurora-A and ATR kinases results in regression of MYCN-amplified neuroblastoma. Nat. Cancer 2021, 2, 312–326. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musavizadeh, Z.; Grottesi, A.; Guarguaglini, G.; Paiardini, A. Phosphorylation, Mg-ADP, and Inhibitors Differentially Shape the Conformational Dynamics of the A-Loop of Aurora-A. Biomolecules 2021, 11, 567. https://doi.org/10.3390/biom11040567
Musavizadeh Z, Grottesi A, Guarguaglini G, Paiardini A. Phosphorylation, Mg-ADP, and Inhibitors Differentially Shape the Conformational Dynamics of the A-Loop of Aurora-A. Biomolecules. 2021; 11(4):567. https://doi.org/10.3390/biom11040567
Chicago/Turabian StyleMusavizadeh, Zahra, Alessandro Grottesi, Giulia Guarguaglini, and Alessandro Paiardini. 2021. "Phosphorylation, Mg-ADP, and Inhibitors Differentially Shape the Conformational Dynamics of the A-Loop of Aurora-A" Biomolecules 11, no. 4: 567. https://doi.org/10.3390/biom11040567
APA StyleMusavizadeh, Z., Grottesi, A., Guarguaglini, G., & Paiardini, A. (2021). Phosphorylation, Mg-ADP, and Inhibitors Differentially Shape the Conformational Dynamics of the A-Loop of Aurora-A. Biomolecules, 11(4), 567. https://doi.org/10.3390/biom11040567






