Molecular Simulation-Based Investigation of Highly Potent Natural Products to Abrogate Formation of the nsp10–nsp16 Complex of SARS-CoV-2
Abstract
1. Introduction
2. Methods
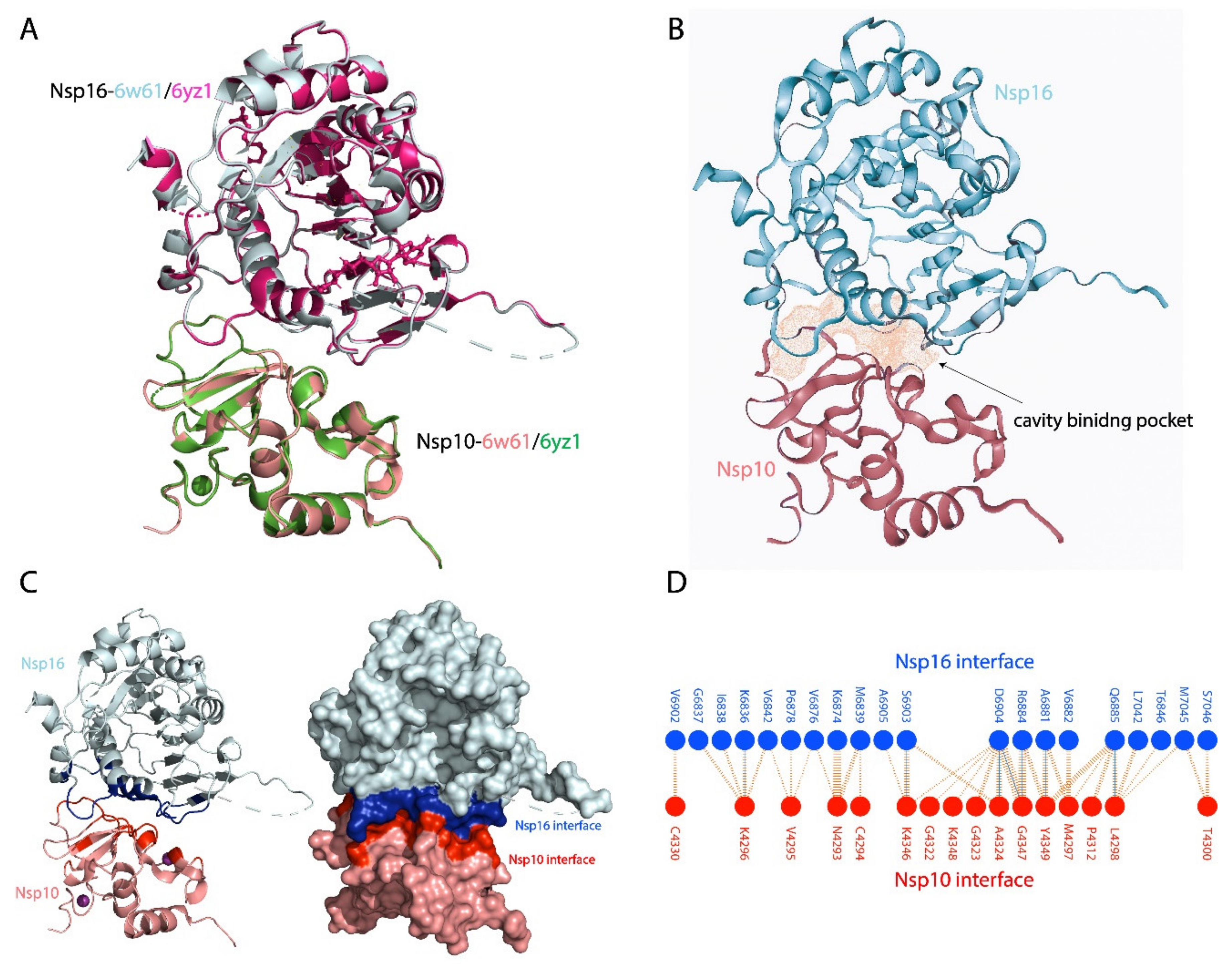
2.1. nsp10–nsp16 Structure
2.2. Virtual Screening of Compounds Interacting with nsp10
2.3. Molecular Dynamics Simulation of nsp10 Compound Complexes
2.4. Binding Free Energy Calculation
3. Results and Discussion
3.1. nsp10–nsp16 Structure and Interface and Cavity
3.2. Virtual Drug Screening and Molecular Docking
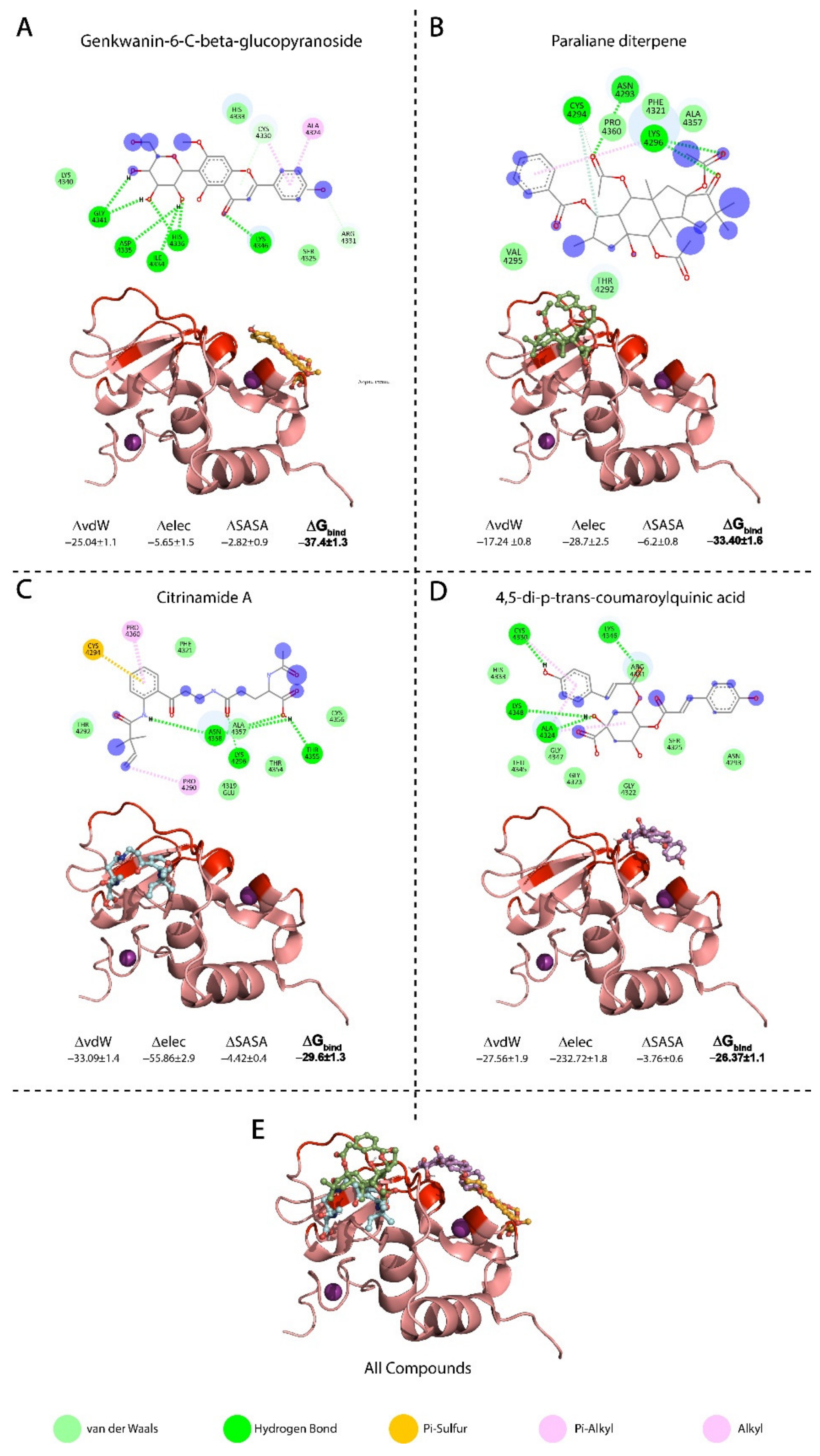
3.3. Binding Models of the Top Compounds with nsp10 Interface Residues
3.3.1. Binding Mode of Genkwanin-6-C-beta-glucopyranos
3.3.2. Binding Mode of Paraliane Diterpene
3.3.3. Binding Mode of Citrinamide A
3.3.4. Binding Mode of 4,5-di-p-trans-coumaroylquinic Acid
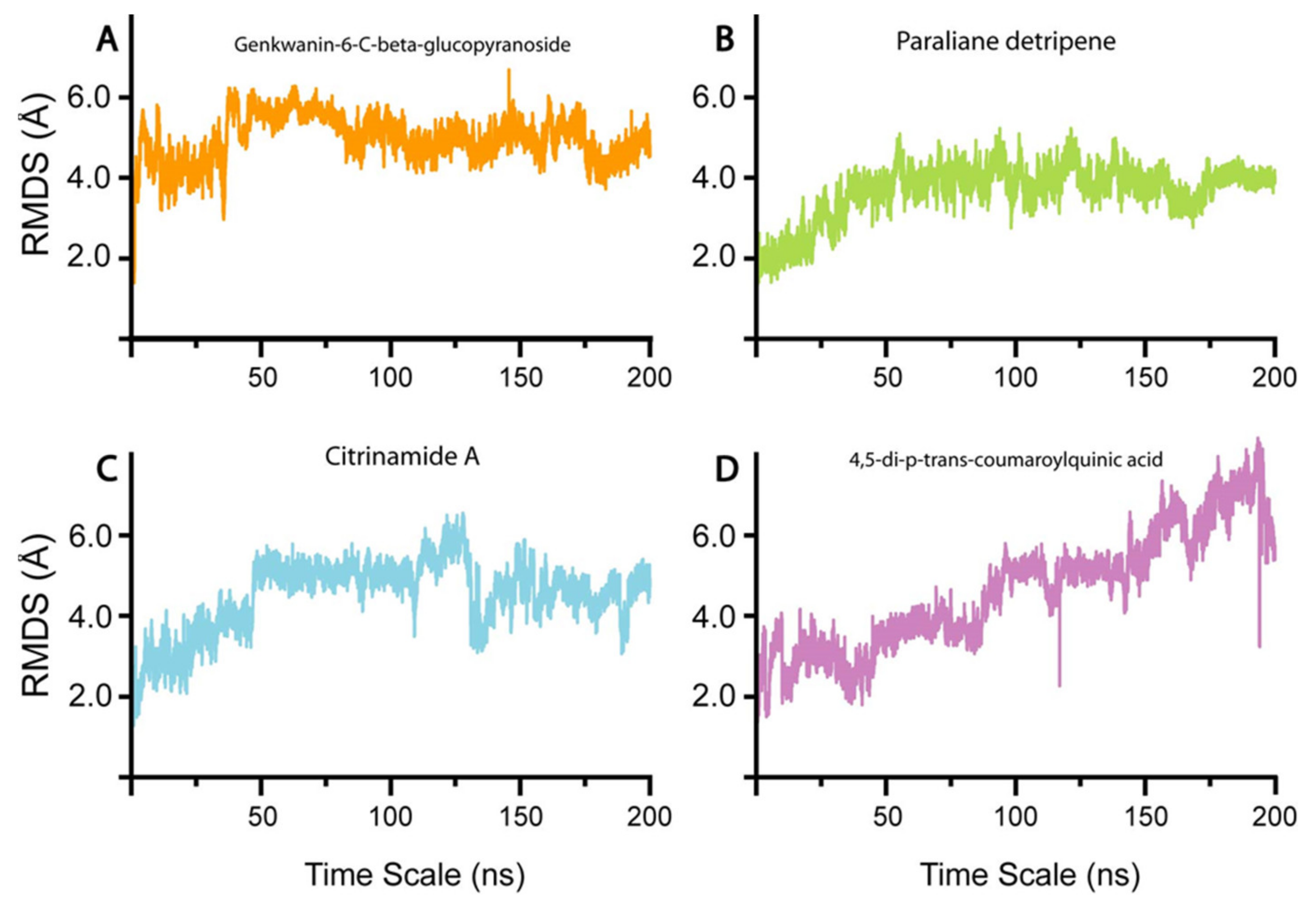
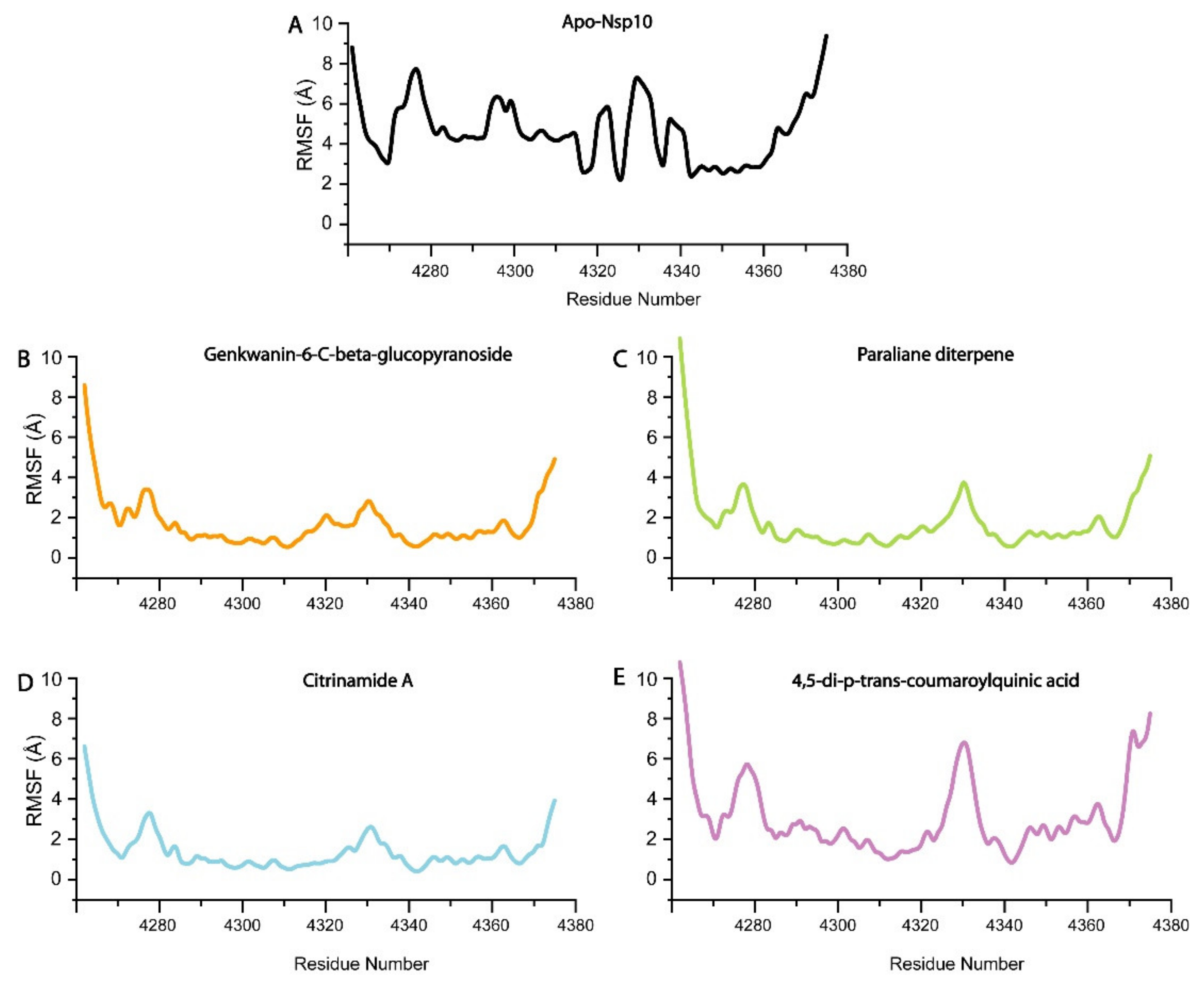
3.4. Conformational Dynamics and Binding Energies of nsp10 in Complex with the Selected Compounds
3.4.1. Genkwanin-6-C-beta-Glucopyronoside–nsp10 Complex
3.4.2. Paraliane Diterpene–nsp10 Complex
3.4.3. Citrinamide A–nsp10 Complex
3.4.4. 4,5-di-p-trans-coumaroylquinic acid–nsp10 Complex
4. Summary
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Phan, T. Novel coronavirus: From discovery to clinical diagnostics. Infect. Genet. Evol. 2020, 79, 104211. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, L.; Du, H.; Zhang, J.; Li, Y.Y.; Qu, J.; Zhang, W.; Wang, Y.; Bao, S.; Li, Y.; et al. SARS-CoV-2 Infection in Children. N. Engl. J. Med. 2020, 382, 1663–1665. [Google Scholar] [CrossRef]
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar] [CrossRef]
- Masters, P.S. Coronavirus genomic RNA packaging. Virology 2019, 537, 198–207. [Google Scholar] [CrossRef]
- Harcourt, B.H.; Jukneliene, D.; Kanjanahaluethai, A.; Bechill, J.; Severson, K.M.; Smith, C.M.; Rota, P.A.; Baker, S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004, 78, 13600–13612. [Google Scholar] [CrossRef]
- Malik, Y.A. Properties of Coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020, 42, 3–11. [Google Scholar]
- Prentice, E.; McAuliffe, J.; Lu, X.; Subbarao, K.; Denison, M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004, 78, 9977–9986. [Google Scholar] [CrossRef]
- Decroly, E.; Debarnot, C.; Ferron, F.; Bouvet, M.; Coutard, B.; Imbert, I.; Gluais, L.; Papageorgiou, N.; Sharff, A.; Bricogne, G.; et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011, 7, e1002059. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.S.; Saikatendu, K.S.; Subramanian, V.; Neuman, B.W.; Brooun, A.; Griffith, M.; Moy, K.; Yadav, M.K.; Velasquez, J.; Buchmeier, M.J.; et al. Crystal Structure of Nonstructural Protein 10 from the Severe Acute Respiratory Syndrome Coronavirus Reveals a Novel Fold with Two Zinc-Binding Motifs. J. Virol. 2006, 80, 7894–7901. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wu, L.; Shaw, N.; Gao, Y.; Wang, J.; Sun, Y.; Lou, Z.; Yan, L.; Zhang, R.; Rao, Z. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc. Natl. Acad. Sci. USA 2015, 112, 9436–9441. [Google Scholar] [CrossRef]
- Rogstam, A.; Nyblom, M.; Christensen, S.; Sele, C.; Talibov, V.O.; Lindvall, T.; Rasmussen, A.A.; André, I.; Fisher, Z.; Knecht, W.; et al. Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2. Int. J. Mol. Sci. 2020, 21, 7375. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Lemus, M.; Minasov, G.; Shuvalova, L.; Inniss, N.L.; Kiryukhina, O.; Brunzelle, J.; Satchell, K.J.F. High-resolution structures of the SARS-CoV-2 2’-O-methyltransferase reveal strategies for structure-based inhibitor design. Sci. Signal. 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, C.; Majer, O.; Frohner, I.E.; Lesiak-Markowicz, I.; Hildering, K.S.; Glaser, W.; Stockinger, S.; Decker, T.; Akira, S.; Müller, M.; et al. Conventional dendritic cells mount a type I IFN response against Candida spp. requiring novel phagosomal TLR7-mediated IFN-β signaling. J. Immunol. 2011, 186, 3104–3112. [Google Scholar] [CrossRef]
- Roth-Cross, J.K.; Bender, S.J.; Weiss, S.R. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 2008, 82, 9829–9838. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, S.-I.; Abe, F.; Kanno, A.; Tanimura, N.; Mori Saitoh, Y.; Fukui, R.; Shibata, T.; Sato, K.; Ichinohe, T.; Hayashi, M.; et al. TLR7 mediated viral recognition results in focal type I interferon secretion by dendritic cells. Nat. Commun. 2017, 8, 1592. [Google Scholar] [CrossRef]
- Daffis, S.; Szretter, K.J.; Schriewer, J.; Li, J.; Youn, S.; Errett, J.; Lin, T.-Y.; Schneller, S.; Zust, R.; Dong, H.; et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 2010, 468, 452–456. [Google Scholar] [CrossRef]
- Viswanathan, T.; Arya, S.; Chan, S.-H.; Qi, S.; Dai, N.; Misra, A.; Park, J.-G.; Oladunni, F.; Kovalskyy, D.; Hromas, R.A.; et al. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020, 11, 3718. [Google Scholar] [CrossRef]
- Züst, R.; Cervantes-Barragan, L.; Habjan, M.; Maier, R.; Neuman, B.W.; Ziebuhr, J.; Szretter, K.J.; Baker, S.C.; Barchet, W.; Diamond, M.S.; et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011, 12, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.N.; Phuong, V.N.; Chen, H.C.; Chou, C.H.; Cheng, H.C.; Wu, C.H. Detection of the Severe Acute Respiratory Syndrome-Related Coronavirus and Alphacoronavirus in the Bat Population of Taiwan. Zoonoses Public Health 2016, 63, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Lima, C.D. Processing the message: Structural insights into capping and decapping mRNA. Curr. Opin. Struct. Biol. 2005, 15, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Lemus, M.; Minasov, G.; Shuvalova, L.; Inniss, N.L.; Kiryukhina, O.; Wiersum, G.; Kim, Y.; Jedrzejczak, R.; Maltseva, N.I.; Endres, M.; et al. The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. bioRxiv 2020. [Google Scholar] [CrossRef]
- Almazán, F.; DeDiego, M.L.; Galán, C.; Escors, D.; Álvarez, E.; Ortego, J.; Sola, I.; Zuñiga, S.; Alonso, S.; Moreno, J.L.; et al. Construction of a Severe Acute Respiratory Syndrome Coronavirus Infectious cDNA Clone and a Replicon To Study Coronavirus RNA Synthesis. J. Virol. 2006, 80, 10900–10906. [Google Scholar] [CrossRef]
- Ke, M.; Chen, Y.; Wu, A.; Sun, Y.; Su, C.; Wu, H.; Jin, X.; Tao, J.; Wang, Y.; Ma, X.; et al. Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein nsp10 can suppress the 2′-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res. 2012, 167, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Y.; Wu, A.; Xu, S.; Pan, R.; Zeng, C.; Jin, X.; Ge, X.; Shi, Z.; Ahola, T.; et al. Coronavirus nsp10/nsp16 Methyltransferase Can Be Targeted by nsp10-Derived Peptide In Vitro and In Vivo To Reduce Replication and Pathogenesis. J. Virol. 2015, 89, 8416–8427. [Google Scholar] [CrossRef]
- Maurya, S.K.; Maurya, A.K.; Mishra, N.; Siddique, H.R. Virtual screening, ADME/T, and binding free energy analysis of anti-viral, anti-protease, and anti-infectious compounds against NSP10/NSP16 methyltransferase and main protease of SARS CoV-2. J. Recept. Signal Transduct. 2020, 40, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liu, L.; Manning, M.; Bonahoom, M.; Lotvola, A.; Yang, Z.; Yang, Z.-Q. Structural analysis, virtual screening and molecular simulation to identify potential inhibitors targeting 2’-O-ribose methyltransferase of SARS-CoV-2 coronavirus. J. Biomol. Struct. Dyn. 2020, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Chen, H.; Ye, F.; Chen, Z.; Yang, F.; Zheng, Y.; Cao, Y.; Qiao, J.; Yang, S.; Lu, G. Crystal structure of SARS-CoV-2 nsp10/nsp16 2’-O-methylase and its implication on antiviral drug design. Signal Transduct. Target. Ther. 2020, 5, 131. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Heng, W.; Wang, Y.; Qiu, J.; Wei, X.; Peng, S.; Saleem, S.; Khan, M.; Ali, S.S.; Wei, D.Q. In silico and in vitro evaluation of kaempferol as a potential inhibitor of the SARS-CoV-2 main protease (3CLpro). Phytother. Res. PTR 2021. [Google Scholar] [CrossRef]
- Ntie-Kang, F.; Telukunta, K.K.; Döring, K.; Simoben, C.V.A.; Moumbock, A.F.; Malange, Y.I.; Njume, L.E.; Yong, J.N.; Sippl, W.; Günther, S. NANPDB: A Resource for Natural Products from Northern African Sources. J. Nat. Prod. 2017, 80, 2067–2076. [Google Scholar] [CrossRef]
- Simoben, C.V.; Qaseem, A.; Moumbock, A.F.A.; Telukunta, K.K.; Günther, S.; Sippl, W.; Ntie-Kang, F. Pharmacoinformatic Investigation of Medicinal Plants from East Africa. Mol. Inform. 2020, 39, e2000163. [Google Scholar] [CrossRef] [PubMed]
- Krafcikova, P.; Silhan, J.; Nencka, R.; Boura, E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 2020, 11, 3717. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newslett. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Schöning-Stierand, K.; Diedrich, K.; Fährrolfes, R.; Flachsenberg, F.; Meyder, A.; Nittinger, E.; Steinegger, R.; Rarey, M. ProteinsPlus: Interactive analysis of protein–ligand binding interfaces. Nucleic Acids Res. 2020, 48, W48–W53. [Google Scholar] [CrossRef]
- Suzuki, M. PyMOL Tutorial: Interaction Interface. Available online: http://www.protein.osaka-u.ac.jp/rcsfp/supracryst/suzuki/jpxtal/Katsutani/en/interface.php (accessed on 7 January 2021).
- Laskowski, R.A. PDBsum new things. Nucleic Acids Res. 2009, 37, D355–D359. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. Publ. Protein Soc. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Grob, S. Molinspiration Cheminformatics Free Web Services. Available online: https://www.molinspiration.com (accessed on 20 December 2020).
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. WIREs Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Price, D.J., III; Brooks, C.L. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004, 121, 10096–10103. [Google Scholar] [CrossRef]
- Meza, J.C. Steepest descent. WIREs Comput. Stat. 2010, 2, 719–722. [Google Scholar] [CrossRef]
- Watowich, S.J.; Meyer, E.S.; Hagstrom, R.; Josephs, R. A stable, rapidly converging conjugate gradient method for energy minimization. J. Comput. Chem. 1988, 9, 650–661. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Kräutler, V.; van Gunsteren, W.F.; Hünenberger, P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Tahir Khan, M.; Saleem, S.; Junaid, M.; Ali, A.; Shujait Ali, S.; Khan, M.; Wei, D.Q. Structural insights into the mechanism of RNA recognition by the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein. Comput. Struct. Biotechnol. J. 2020, 18, 2174–2184. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Sk, M.F.; Jonniya, N.A.; Roy, R.; Poddar, S.; Kar, P. Computational Investigation of Structural Dynamics of SARS-CoV-2 Methyltransferase-Stimulatory Factor Heterodimer nsp16/nsp10 Bound to the Cofactor SAM. Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef]
- Kassem, M.E.; Shoela, S.; Marzouk, M.M.; Sleem, A.A. A sulphated flavone glycoside from Livistona australis and its antioxidant and cytotoxic activity. Nat. Prod. Res. 2012, 26, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Abdelgaleil, S.A.; el-Aswad, A.F.; Nakatani, M. Molluscicidal and anti-feedant activities of diterpenes from Euphorbia paralias L. Pest Manag. Sci. 2002, 58, 479–482. [Google Scholar] [CrossRef]
- Fattahian, M.; Ghanadian, M.; Ali, Z.; Khan, I.A. Jatrophane and rearranged jatrophane-type diterpenes: Biogenesis, structure, isolation, biological activity and SARs (1984–2019). Phytochem. Rev. 2020, 1–72. [Google Scholar] [CrossRef]
- Barile, E.; Fattorusso, E.; Ialenti, A.; Ianaro, A.; Lanzotti, V. Paraliane and pepluane diterpenes as anti-inflammatory agents: First insights in structure-activity relationships. Bioorg. Med. Chem. Lett. 2007, 17, 4196–4200. [Google Scholar] [CrossRef]
- Fukuda, T.; Hasegawa, Y.; Sakabe, Y.; Tomoda, H.; Ōmura, S. Citrinamides, New Potentiators of Antifungal Miconazole Activity, Produced by Penicillium sp. FKI-1938. J. Antibiot. 2008, 61, 550–555. [Google Scholar] [CrossRef]
- Hammoda, H.M.; Ghazy, N.M.; Harraz, F.M.; Radwan, M.M.; ElSohly, M.A.; Abdallah, I.I. Chemical constituents from Tribulus terrestris and screening of their antioxidant activity. Phytochemistry 2013, 92, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Vettoretti, G.; Moroni, E.; Sattin, S.; Tao, J.; Agard, D.A.; Bernardi, A.; Colombo, G. Molecular Dynamics Simulations Reveal the Mechanisms of Allosteric Activation of Hsp90 by Designed Ligands. Sci. Rep. 2016, 6, 23830. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Kumar, S.; MaqsoodAli, M.; Khan, S. Virtual Screening of Molecular Properties and Bioactivity Score of Boswellic Acid Derivatives in Search of Potent Anti-Inflammatory Lead Molecule. Int. J. Interdiscip. Multidiscip. Stud. 2013, 1, 8–12. [Google Scholar]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
| Compound Name | Vina Score |
|---|---|
| Delta-tocopherol | −8.1 |
| 1-O-linoleoyl-3-O-beta-D-galactopyranosyl-syn-glycerol | −8.2 |
| N1,N10-di-dihydrocaffeoylspermidine | −8.0 |
| Citrinamide A | −7.4 |
| (9Z,12Z)-octadecadienoic acid glucoside | −7.1 |
| Paraliane deterpine | −7.1 |
| 4,5-di-p-trans-coumaroylquinic acid | −7.2 |
| 2,3-dihydro-5,7-dihydroxy-3-((3Z,6Z,9Z,12Z,15Z)-octadeca-3,6,9,12,15-pentaenyl) chromen-4-one | −6.8 |
| 10-epi-cubebol-(alpha-xylopyranoside-triacetate) | −7.3 |
| Genkwanin-6-C-beta-glucopyranoside | −7.2 |
| Compound Name | MW. | Source | Molecule Class | Biological Activity | Lipinski Violation | AMES * Toxicity | Rat Oral LD50 (mol/kg) | Max. Tolerated Dose (Human) (log Mg/Kg/day) | IC50 In Vitro (µM) | T.Pyriformis Toxicity ** (log µg/L) | HBD | HBA | Rotatable Bonds No. | TPSA | Bioactivity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| genkwanin-6-C-β-glucopyranoside | 446 | Livistona australis | Flaonoid | Antioxidant | 1 | - | 2.77 | 0.405 | 0.029–0.035 | 0.284 | 6 | 10 | 4 | 170.05 Å2 | 0.41 |
| Paraliane deterpine | 598.68 | Euphorbia paralias | Terpenoid | Molluscicidal | 1 | - | 3.43 | −0.308 | 0.1–10 | 0.284 | 1 | 10 | 9 | 142.50 Å2 | 0.09 |
| Citrinamide A | 431.48 | Penicillium citrinum | Alkaloid | Miconazole | 0 | - | 1.96 | 0.883 | 0.005–0.009 | 0.284 | 4 | 6 | 15 | 141.67 Å2 | 0.18 |
| 4,5-di-p-trans-coumaroylquinic acid | 484.45 | Tribulus terrstris | Phenolic | Antioxidant | 0 | - | 2.32 | 0.169 | 0.039 | 0.284 | 5 | 10 | 9 | 170.82 Å2 | 0.43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammad, A.; Alshawaf, E.; Marafie, S.K.; Abu-Farha, M.; Al-Mulla, F.; Abubaker, J. Molecular Simulation-Based Investigation of Highly Potent Natural Products to Abrogate Formation of the nsp10–nsp16 Complex of SARS-CoV-2. Biomolecules 2021, 11, 573. https://doi.org/10.3390/biom11040573
Mohammad A, Alshawaf E, Marafie SK, Abu-Farha M, Al-Mulla F, Abubaker J. Molecular Simulation-Based Investigation of Highly Potent Natural Products to Abrogate Formation of the nsp10–nsp16 Complex of SARS-CoV-2. Biomolecules. 2021; 11(4):573. https://doi.org/10.3390/biom11040573
Chicago/Turabian StyleMohammad, Anwar, Eman Alshawaf, Sulaiman K. Marafie, Mohamed Abu-Farha, Fahd Al-Mulla, and Jehad Abubaker. 2021. "Molecular Simulation-Based Investigation of Highly Potent Natural Products to Abrogate Formation of the nsp10–nsp16 Complex of SARS-CoV-2" Biomolecules 11, no. 4: 573. https://doi.org/10.3390/biom11040573
APA StyleMohammad, A., Alshawaf, E., Marafie, S. K., Abu-Farha, M., Al-Mulla, F., & Abubaker, J. (2021). Molecular Simulation-Based Investigation of Highly Potent Natural Products to Abrogate Formation of the nsp10–nsp16 Complex of SARS-CoV-2. Biomolecules, 11(4), 573. https://doi.org/10.3390/biom11040573








