Validation of Recombinant Chicken Liver Bile Acid Binding Protein as a Tool for Cholic Acid Hosting
Abstract
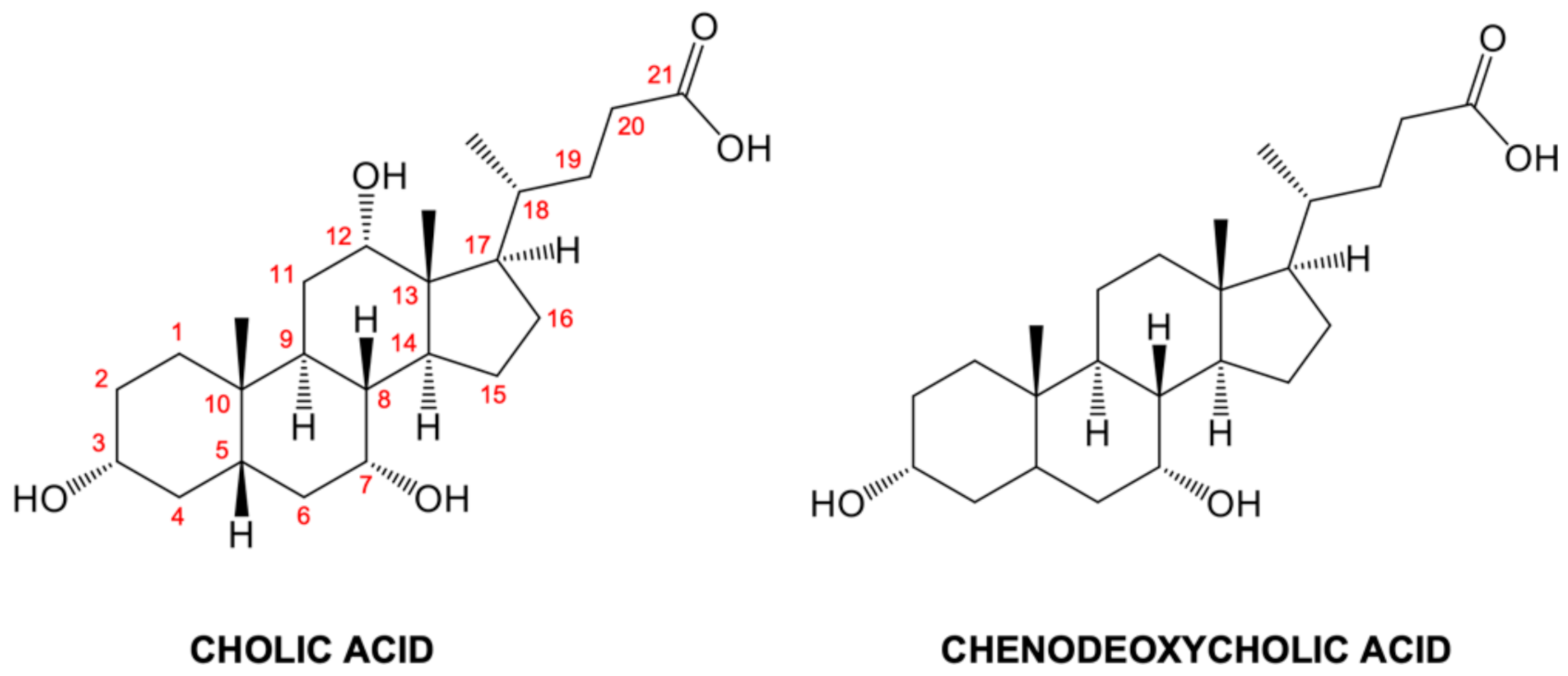
1. Introduction
2. Results and Discussion
2.1. An Efficient Protocol for Expression and Purification of Recombinant cL-BABP in E. coli
2.2. Crystallization of cL-BABP and Its Complex with CA
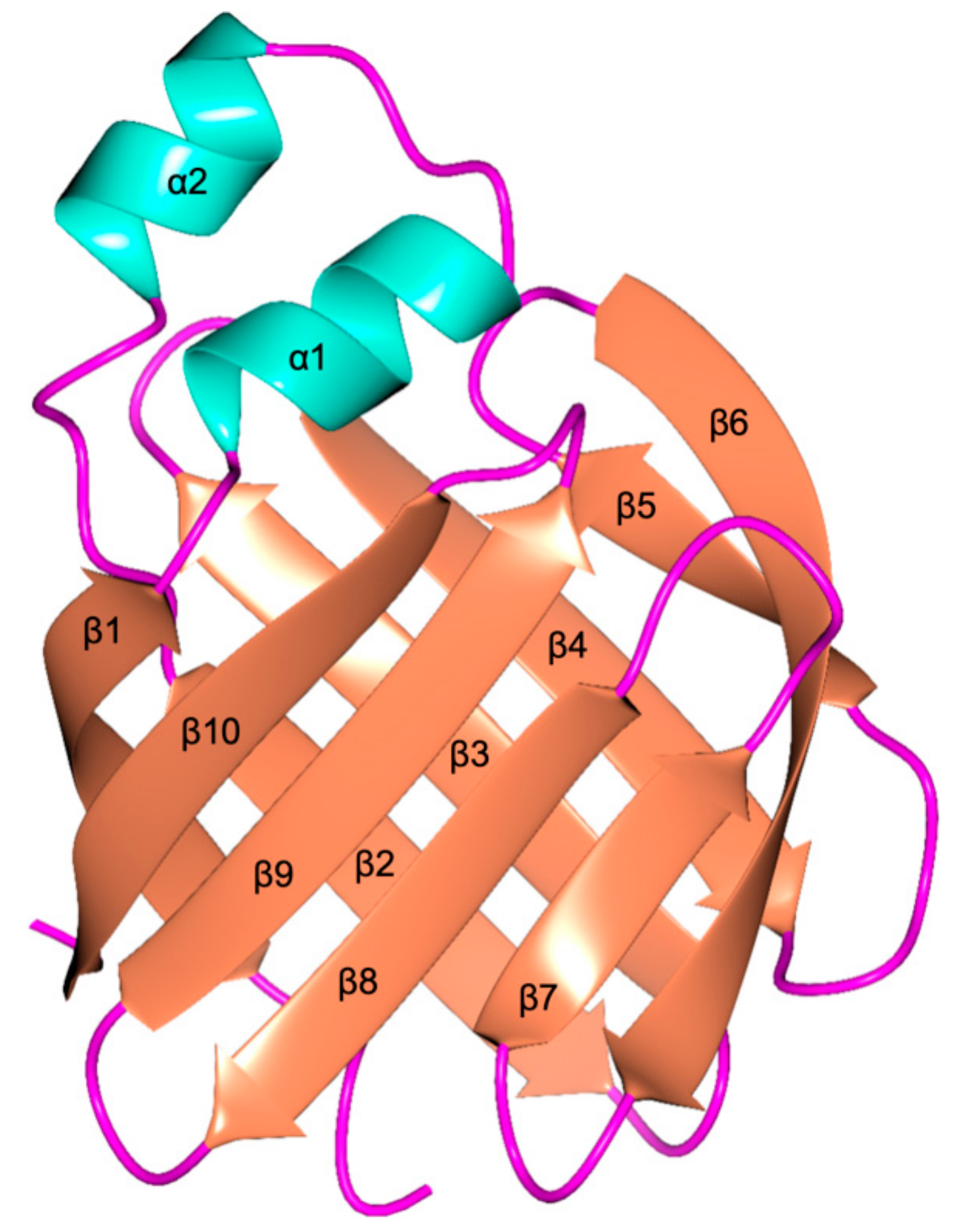
2.3. Structural Analysis of the Recombinant cL-BABP
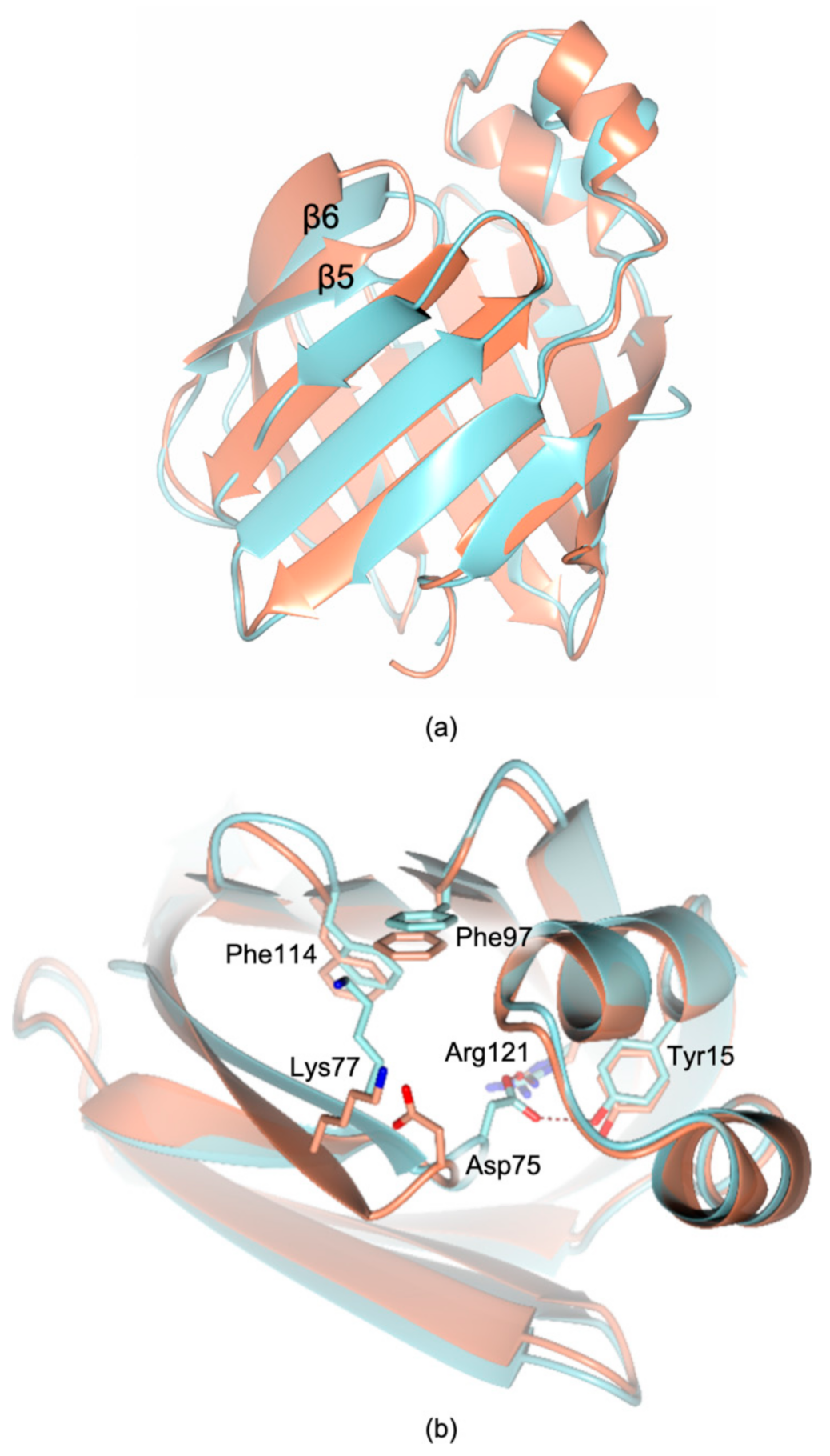
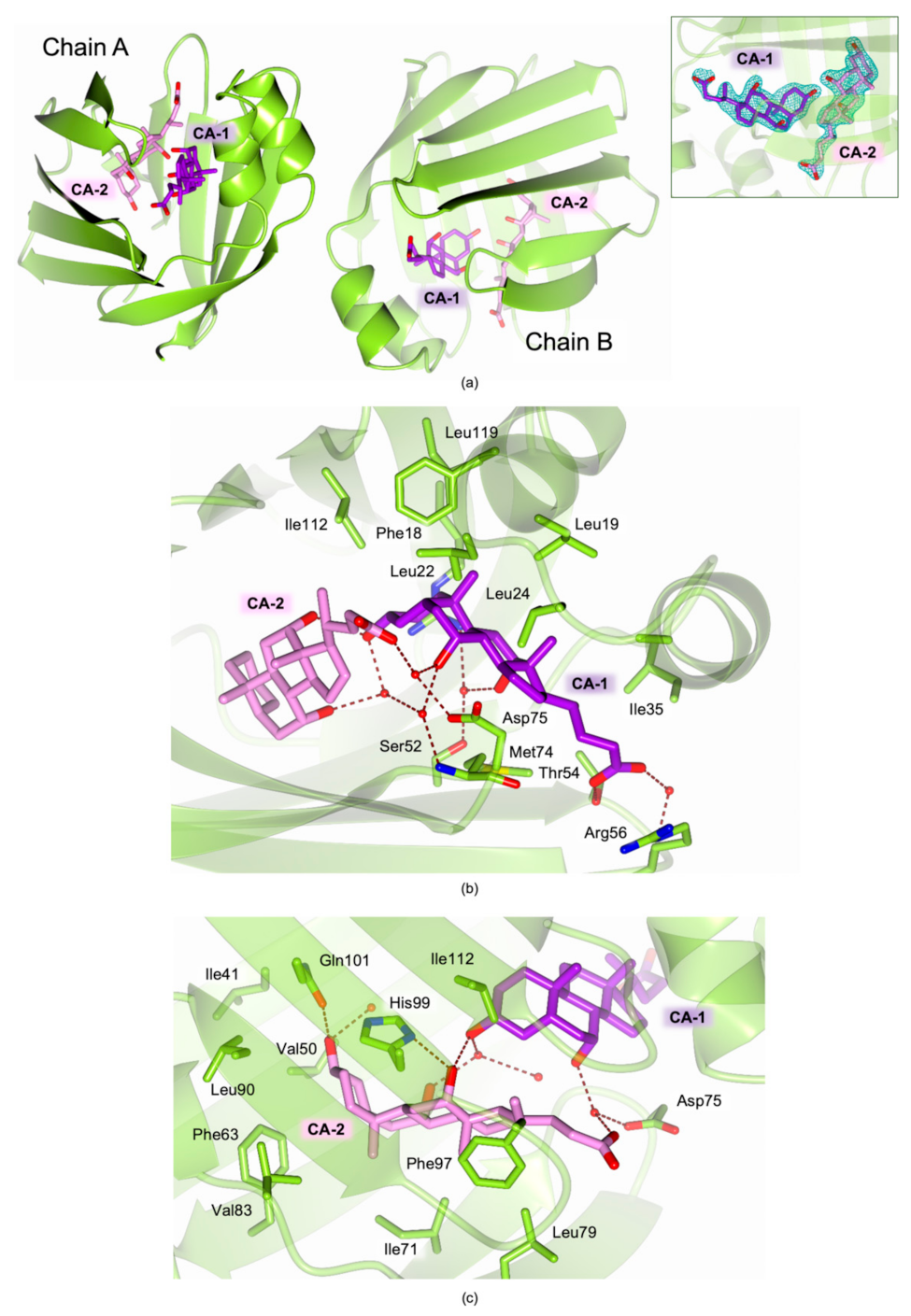
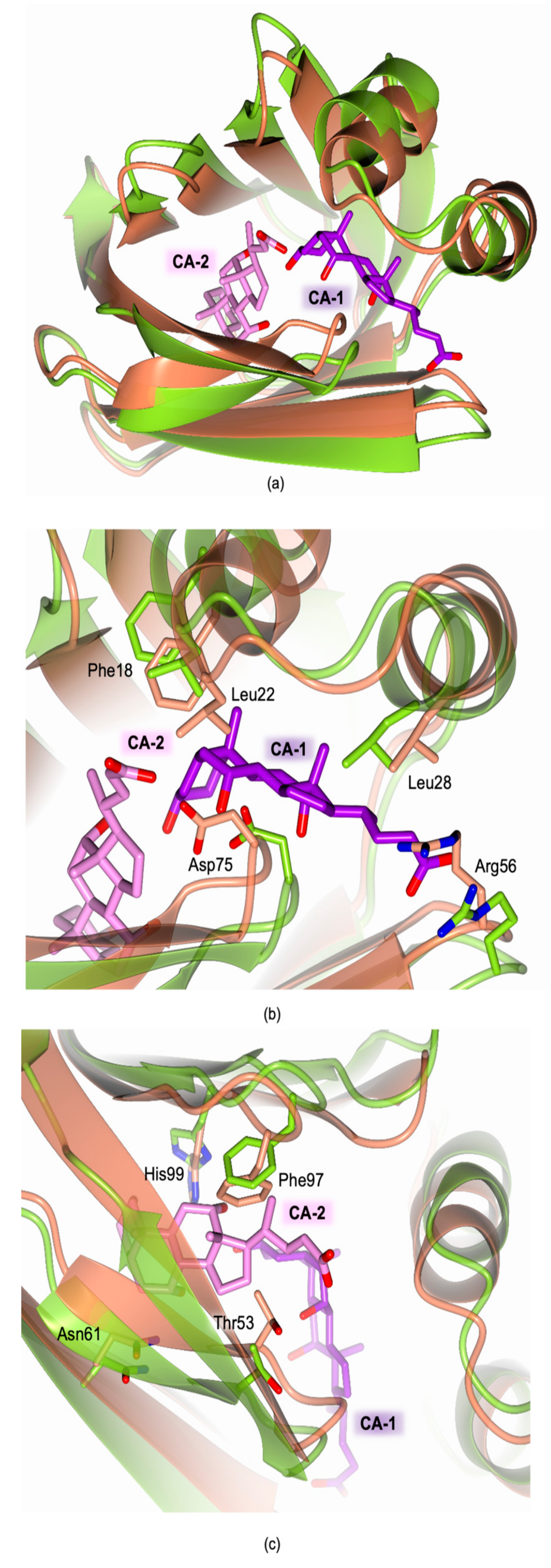
2.4. Structural Characterization of Recombinant cL-BABP in Complex with CA
3. Materials and Methods
3.1. Cloning
3.2. cL-BABP Expression and Purification
3.3. Crystallization
3.4. Data Collection, Structure Solution, and Refinement
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chiang, J.Y.L. Bile Acid Metabolism and Signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.W. The Enzymes, Regulation, and Genetics of Bile Acid Synthesis. Annu. Rev. Biochem. 2003, 72, 137–174. [Google Scholar] [CrossRef] [PubMed]
- Carey, M.C.; Small, D.M. Micelle Formation by Bile Salts. Physical-Chemical and Thermodynamic Considerations. Arch. Intern. Med. 1972, 130, 506–527. [Google Scholar] [CrossRef] [PubMed]
- Houten, S.M.; Watanabe, M.; Auwerx, J. Endocrine Functions of Bile Acids. EMBO J. 2006, 25, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Weisiger, R.A. When Is a Carrier Not a Membrane Carrier? The Cytoplasmic Transport of Amphipathic Molecules. Hepatology 1996, 24, 1288–1295. [Google Scholar] [CrossRef]
- Bernlohr, D.A.; Simpson, M.A.; Hertzel, A.V.; Banaszak, L.J. Intracellular Lipid-Binding Proteins and Their Genes. Annu. Rev. Nutr. 1997, 17, 277–303. [Google Scholar] [CrossRef]
- Marcelino, A.M.C.; Smock, R.G.; Gierasch, L.M. Evolutionary Coupling of Structural and Functional Sequence Information in the Intracellular Lipid-Binding Protein Family. Proteins 2006, 63, 373–384. [Google Scholar] [CrossRef]
- Ockner, R.K.; Manning, J.A.; Poppenhausen, R.B.; Ho, W.K. A Binding Protein for Fatty Acids in Cytosol of Intestinal Mucosa, Liver, Myocardium, and Other Tissues. Science 1972, 177, 56–58. [Google Scholar] [CrossRef]
- Banaszak, L.; Winter, N.; Xu, Z.; Bernlohr, D.A.; Cowan, S.; Jones, T.A. Lipid-Binding Proteins: A Family of Fatty Acid and Retinoid Transport Proteins. Adv. Protein Chem. 1994, 45, 89–151. [Google Scholar] [CrossRef]
- Veerkamp, J.H.; Maatman, R.G. Cytoplasmic Fatty Acid-Binding Proteins: Their Structure and Genes. Prog. Lipid Res. 1995, 34, 17–52. [Google Scholar] [CrossRef]
- Glatz, J.F.; van der Vusse, G.J. Cellular Fatty Acid-Binding Proteins: Their Function and Physiological Significance. Prog. Lipid Res. 1996, 35, 243–282. [Google Scholar] [CrossRef]
- Storch, J.; Thumser, A.E. The Fatty Acid Transport Function of Fatty Acid-Binding Proteins. Biochim. Biophys Acta 2000, 1486, 28–44. [Google Scholar] [CrossRef]
- Schaap, F.G.; van der Vusse, G.J.; Glatz, J.F.C. Evolution of the Family of Intracellular Lipid Binding Proteins in Vertebrates. Mol. Cell Biochem. 2002, 239, 69–77. [Google Scholar] [CrossRef]
- Haunerland, N.H.; Spener, F. Fatty Acid-Binding Proteins--Insights from Genetic Manipulations. Prog. Lipid Res. 2004, 43, 328–349. [Google Scholar] [CrossRef]
- Zanotti, G.; Berni, R. Plasma Retinol-Binding Protein: Structure and Interactions with Retinol, Retinoids, and Transthyretin. Vitam. Horm. 2004, 69, 271–295. [Google Scholar] [CrossRef]
- Menozzi, I.; Vallese, F.; Polverini, E.; Folli, C.; Berni, R.; Zanotti, G. Structural and Molecular Determinants Affecting the Interaction of Retinol with Human CRBP1. J. Struct. Biol. 2017, 197, 330–339. [Google Scholar] [CrossRef]
- Capaldi, S.; Guariento, M.; Perduca, M.; Pietro, S.M.D.; Santomé, J.A.; Monaco, H.L. Crystal Structure of Axolotl (Ambystoma Mexicanum) Liver Bile Acid-Binding Protein Bound to Cholic and Oleic Acid. Proteins Struct. Funct. Bioinform. 2006, 64, 79–88. [Google Scholar] [CrossRef]
- Nichesola, D.; Perduca, M.; Capaldi, S.; Carrizo, M.E.; Righetti, P.G.; Monaco, H.L. Crystal Structure of Chicken Liver Basic Fatty Acid-Binding Protein Complexed with Cholic Acid. Biochemistry 2004, 43, 14072–14079. [Google Scholar] [CrossRef]
- Monaco, H.L. Review: The Liver Bile Acid-Binding Proteins. Biopolymers 2009, 91, 1196–1202. [Google Scholar] [CrossRef]
- Di Pietro, S.M.; Córsico, B.; Perduca, M.; Monaco, H.L.; Santomé, J.A. Structural and Biochemical Characterization of Toad Liver Fatty Acid-Binding Protein. Biochemistry 2003, 42, 8192–8203. [Google Scholar] [CrossRef]
- Tarter, M.; Capaldi, S.; Carrizo, M.E.; Ambrosi, E.; Perduca, M.; Monaco, H.L. Crystal Structure of Human Cellular Retinol-Binding Protein II to 1.2 A Resolution. Proteins 2008, 70, 1626–1630. [Google Scholar] [CrossRef] [PubMed]
- Hertzel, A.V.; Bernlohr, D.A. The Mammalian Fatty Acid-Binding Protein Multigene Family: Molecular and Genetic Insights into Function. Trends Endocrinol. Metab. 2000, 11, 175–180. [Google Scholar] [CrossRef]
- Thompson, J.; Reese-Wagoner, A.; Banaszak, L. Liver Fatty Acid Binding Protein: Species Variation and the Accommodation of Different Ligands. Biochim. Biophys. Acta 1999, 1441, 117–130. [Google Scholar] [CrossRef]
- Capaldi, S.; Saccomani, G.; Fessas, D.; Signorelli, M.; Perduca, M.; Monaco, H.L. The X-Ray Structure of Zebrafish (Danio Rerio) Ileal Bile Acid-Binding Protein Reveals the Presence of Binding Sites on the Surface of the Protein Molecule. J. Mol. Biol. 2009, 385, 99–116. [Google Scholar] [CrossRef]
- Winter, N.S.; Gordon, J.I.; Banaszak, L.J. Characterization of Crystalline Rat Liver Fatty Acid Binding Protein Produced in Escherichia Coli. J. Biol. Chem. 1990, 265, 10955–10958. [Google Scholar] [CrossRef]
- Thompson, J.; Winter, N.; Terwey, D.; Bratt, J.; Banaszak, L. The Crystal Structure of the Liver Fatty Acid-Binding Protein. A Complex with Two Bound Oleates. J. Biol. Chem. 1997, 272, 7140–7150. [Google Scholar] [CrossRef] [PubMed]
- Scapin, G.; Spadon, P.; Pengo, L.; Mammi, M.; Zanotti, G.; Monaco, H.L. Chicken Liver Basic Fatty Acid-Binding Protein (PI = 9.0). Purification, Crystallization and Preliminary X-Ray Data. FEBS Lett. 1988, 240. [Google Scholar] [CrossRef]
- Wagner, M.; Zollner, G.; Trauner, M. New Molecular Insights into the Mechanisms of Cholestasis. J. Hepatol. 2009, 51, 565–580. [Google Scholar] [CrossRef]
- Chuang, S.; Velkov, T.; Horne, J.; Porter, C.J.H.; Scanlon, M.J. Characterization of the Drug Binding Specificity of Rat Liver Fatty Acid Binding Protein. J. Med. Chem. 2008, 51, 3755–3764. [Google Scholar] [CrossRef]
- Terreno, E.; Dastrù, W.; Delli Castelli, D.; Gianolio, E.; Geninatti Crich, S.; Longo, D.; Aime, S. Advances in Metal-Based Probes for MR Molecular Imaging Applications. Curr. Med. Chem. 2010, 17, 3684–3700. [Google Scholar] [CrossRef]
- Tomaselli, S.; Pagano, K.; Boulton, S.; Zanzoni, S.; Melacini, G.; Molinari, H.; Ragona, L. Lipid Binding Protein Response to a Bile Acid Library: A Combined NMR and Statistical Approach. FEBS J. 2015, 282, 4094–4113. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pagano, K.; Tomaselli, S.; Zanzoni, S.; Assfalg, M.; Molinari, H.; Ragona, L. Bile Acid Binding Protein: A Versatile Host of Small Hydrophobic Ligands for Applications in the Fields of MRI Contrast Agents and Bio-Nanomaterials. Comput. Struct Biotechnol. J. 2013, 6. [Google Scholar] [CrossRef] [PubMed]
- Ragona, L.; Catalano, M.; Luppi, M.; Cicero, D.; Eliseo, T.; Foote, J.; Fogolari, F.; Zetta, L.; Molinari, H. NMR Dynamic Studies Suggest That Allosteric Activation Regulates Ligand Binding in Chicken Liver Bile Acid-Binding Protein. J. Biol. Chem. 2006, 281, 9697–9709. [Google Scholar] [CrossRef] [PubMed]
- Eberini, I.; Rocco, A.G.; Ientile, A.R.; Baptista, A.M.; Gianazza, E.; Tomaselli, S.; Molinari, H.; Ragona, L. Conformational and Dynamics Changes Induced by Bile Acids Binding to Chicken Liver Bile Acid Binding Protein. Proteins Struct. Funct. Bioinform. 2008, 71, 1889–1898. [Google Scholar] [CrossRef]
- Benvenuti, M.; Mangani, S. Crystallization of Soluble Proteins in Vapor Diffusion for X-Ray Crystallography. Nat. Protoc. 2007, 2, 1633–1651. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Cryst. D Biol. Cryst. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Evans, P.R. An Introduction to Data Reduction: Space-Group Determination, Scaling and Intensity Statistics. Acta Cryst. D Biol. Cryst. 2011, 67, 282–292. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 Suite and Current Developments. Acta Cryst. D Biol. Cryst. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Vagin, A.; Teplyakov, A. Molecular Replacement with MOLREP. Acta Cryst. Sect. D Biol. Cryst. 2010, 66, 22–25. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the Refinement of Macromolecular Crystal Structures. Acta Cryst. Sect. D Biol. Cryst. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Cryst. D Biol. Cryst. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Winn, M.D.; Isupov, M.N.; Murshudov, G.N. Use of TLS Parameters to Model Anisotropic Displacements in Macromolecular Refinement. Acta Cryst. D Biol. Cryst. 2001, 57, 122–133. [Google Scholar] [CrossRef]
- Painter, J.; Merritt, E.A. Optimal Description of a Protein Structure in Terms of Multiple Groups Undergoing TLS Motion. Acta Cryst. Sect. D Biol. Cryst. 2006, 62, 439–450. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Potterton, L.; McNicholas, S.; Krissinel, E.; Gruber, J.; Cowtan, K.; Emsley, P.; Murshudov, G.N.; Cohen, S.; Perrakis, A.; Noble, M. Developments in the CCP4 Molecular-Graphics Project. Acta Cryst. D Biol. Cryst. 2004, 60, 2288–2294. [Google Scholar] [CrossRef]

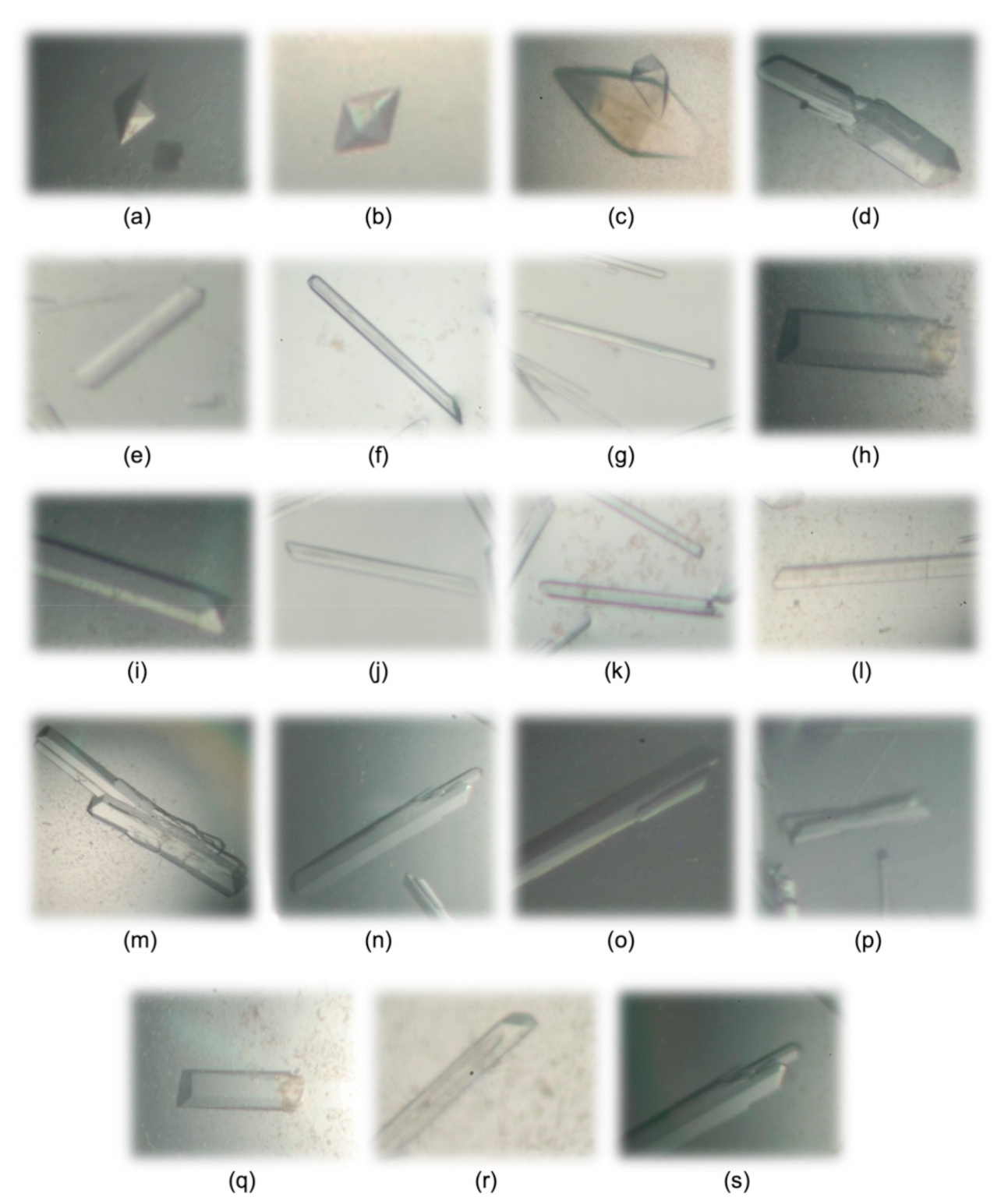
| Composition of Precipitant Solutions (Solution Number in Crystallization Kits from Hampton Research and Jena Bioscience) * | Temperature | Crystal Picture | Crystal Habitus | Resolution in X-Ray Diffraction Screening (Å) |
|---|---|---|---|---|
| 2.1 M DL-malic acid pH 7.0 (23 Index–HR) | R.T. | Figure 3a | Octahedral | 2.14 |
| 2.4 M sodium malonate pH 7.0 (27 Index–HR) | 8 °C | Figure 3b | Octahedral | 2.32 |
| 0.1 M BIS-TRIS pH 5.5, 25% w/v PEG 3350 (42 Index–HR) | R.T. | Figure 3c | Rhombohedral | 2.10 |
| 0.1 M Tris pH 8.5, 25% w/v PEG 3350 (45 Index–HR) | 8 °C | Figure 3d | Parallelepipedal | >2.50 |
| 0.2 M sodium chloride, 0.1 M HEPES pH 7.5, 25% w/v PEG 3350 (72 Index–HR) | 8 °C | Figure 3e | Parallelepipedal | 2.13 |
| 0.2 M lithium sulfate, 0.1 M BIS-TRIS pH 6.5, 25% w/v PEG 3350 (75 Index–HR) | 8 °C | Figure 3f | Needle -shaped | 1.47 |
| 0.2 M ammonium acetate, 0.1 M BIS-TRIS pH 6.5, 25% w/v PEG 3350 (79 Index–HR) | 8 °C | Figure 3g | Needle -shaped | 2.07 |
| 0.2 M ammonium acetate, 0.1 M HEPES pH 7.5, 25% w/v PEG 3350 (80 Index–HR) | 8 °C | Figure 3h | Parallelepipedal | 2.13 |
| 0.15 M potassium bromide, 30% w/v PEGme 2000 (96 Index–HR) | 8 °C | Figure 3i | Parallelepipedal | >2.50 |
| 0.2 M potassium chloride, 20% w/v PEG 3350 (8 PEG-Ion–HR) | 8 °C | Figure 3j | Needle -shaped | 1.88 |
| 0.2 M sodium thiocyanate, 20% w/v PEG 3350 (13 PEG-Ion–HR) | 8 °C | Figure 3k | Needle -shaped | 1.94 |
| 0.2 M ammonium formate, 20% w/v PEG 3350 (23 PEG-Ion–HR) | 8 °C | Figure 3l | Needle -shaped | 1.92 |
| 0.2 M potassium acetate, 20% w/v PEG 3350 (29 PEG-Ion–HR) | 8 °C | Figure 3m | Parallelepipedal | >2.50 |
| 0.1 M HEPES pH 7.5, 1.4 M sodium citrate (38 CS–HR) | 8 °C | Figure 3n | Parallelepipedal | 2.00 |
| 20% w/v PEG 4000, 20% v/v 2-propanol, 0.1 M tri-sodium citrate pH 5.6 (C2 JBSB-2–JB) | 8 °C | Figure 3o | Parallelepipedal | 1.40 |
| 20% w/v PEG 4000, 10% v/v 2-propanol 0.1 M HEPES pH 7.5 (C3 JBSB-2–JB) | 8 °C | Figure 3p | Parallelepipedal | 1.50 |
| 30% w/v PEG 4000, 0.1 M sodium acetate pH 4.6, 0.2 M ammonium acetate (C6 JBSB-2–JB) | 8 °C | Figure 3q | Parallelepipedal | 1.61 |
| 20% w/v PEG 10,000, 0.1 M HEPES pH 7.5 (E2 JBSB-3–JB) | 8 °C | Figure 3r | Parallelepipedal | >2.50 |
| cL-BABP | cL-BABP-CA | |
|---|---|---|
| PDB ID codes | 7O0J | 7O0K |
| DATA COLLECTION STATISTICS | ||
| Diffraction source | I04 (DLS) | I04 (DLS) |
| Wavelength (Å) | 0.8920 | 0.9795 |
| Temperature (K) | 100 | 100 |
| Detector | Eiger2 XE 16M | Eiger2 XE 16M |
| Crystal-detector distance (mm) | 177.2 | 240.5 |
| Exposure time per image (s) | 0.2 | 0.2 |
| Space group | P212121 | P212121 |
| No. of subunit in ASU | 1 | 2 |
| a, b, c (Å) | 60.68, 62.92, 77.57 | 39.01, 58.93, 71.29 |
| Resolution range (Å) | 71.29–1.40 (1.48–1.40) | 62.92–1.83 (1.93–1.83) |
| Total no. of reflections | 306,679 (42,484) | 198,330 (28,944) |
| No. of unique reflections | 32,871 (4682) | 26,839 (3830) |
| Completeness (%) | 99.3 (98.5) | 99.8 (99.6) |
| Multiplicity | 9.3 (9.1) | 7.4 (7.6) |
| (I/σ(I)) | 24.3 (3.1) | 13.6 (2.4) |
| Rmeas | 0.040 (0.688) | 0.061 (0.856) |
| Overall B factor from Wilson plot (Å2) | 18.9 | 32.2 |
| REFINEMENTS STATISTICS | ||
| Resolution range (Å) | 32.55–1.40 (1.44–1.40) | 48.87–1.83 (1.88–1.83) |
| Completeness (%) | 99.2 (98.1) | 99.7 (99.2) |
| No. of reflections, working set | 31,165 (2238) | 25,436 (1834) |
| No. of reflections, test set | 1657 (111) | 1326 (86) |
| Final Rcryst | 0.1579 (0.247) | 0.2291 (0.337) |
| Final Rfree | 0.2065 (0.247) | 0.2838 (0.374) |
| No. of non-H atoms | ||
| Protein | 1048 | 1921 |
| CA | - | 116 |
| Water | 213 | 112 |
| Total | 1261 | 2149 |
| R.m.s. deviations Bonds (Å) | 0.020 | 0.012 |
| Angles (°) | 2.400 | 2.140 |
| Average B factors (Å2) | 29.9 | 40.5 |
| Estimate error on coordinates based on R value (Å) | 0.054 | 0.153 |
| Ramachandran plot | ||
| Most favoured (%) | 99.2 | 98.8 |
| Allowed (%) | 0.8 | 1.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tassone, G.; Orlandini, M.; Olivucci, M.; Pozzi, C. Validation of Recombinant Chicken Liver Bile Acid Binding Protein as a Tool for Cholic Acid Hosting. Biomolecules 2021, 11, 645. https://doi.org/10.3390/biom11050645
Tassone G, Orlandini M, Olivucci M, Pozzi C. Validation of Recombinant Chicken Liver Bile Acid Binding Protein as a Tool for Cholic Acid Hosting. Biomolecules. 2021; 11(5):645. https://doi.org/10.3390/biom11050645
Chicago/Turabian StyleTassone, Giusy, Maurizio Orlandini, Massimo Olivucci, and Cecilia Pozzi. 2021. "Validation of Recombinant Chicken Liver Bile Acid Binding Protein as a Tool for Cholic Acid Hosting" Biomolecules 11, no. 5: 645. https://doi.org/10.3390/biom11050645
APA StyleTassone, G., Orlandini, M., Olivucci, M., & Pozzi, C. (2021). Validation of Recombinant Chicken Liver Bile Acid Binding Protein as a Tool for Cholic Acid Hosting. Biomolecules, 11(5), 645. https://doi.org/10.3390/biom11050645








