Unravelling the Molecular Mechanisms Underlying the Protective Effect of Lactate on the High-Pressure Resistance of Listeria monocytogenes
Abstract
:1. Introduction
2. Material and Methods
2.1. Cooked Ham Model Medium Formulation and Characterization
2.2. L. monocytogenes Strains and Pre-Culture Conditions
2.3. Preparation of the Samples and HPP
2.4. L. monocytogenes Enumeration and Data Analysis
2.5. Nucleic Acid Extraction and Sequencing
2.6. Bioinformatics and Data Analysis
2.7. Availability of Data and Material
2.8. Fatty Acid Profile of L. monocytogenes
3. Results and Discussion
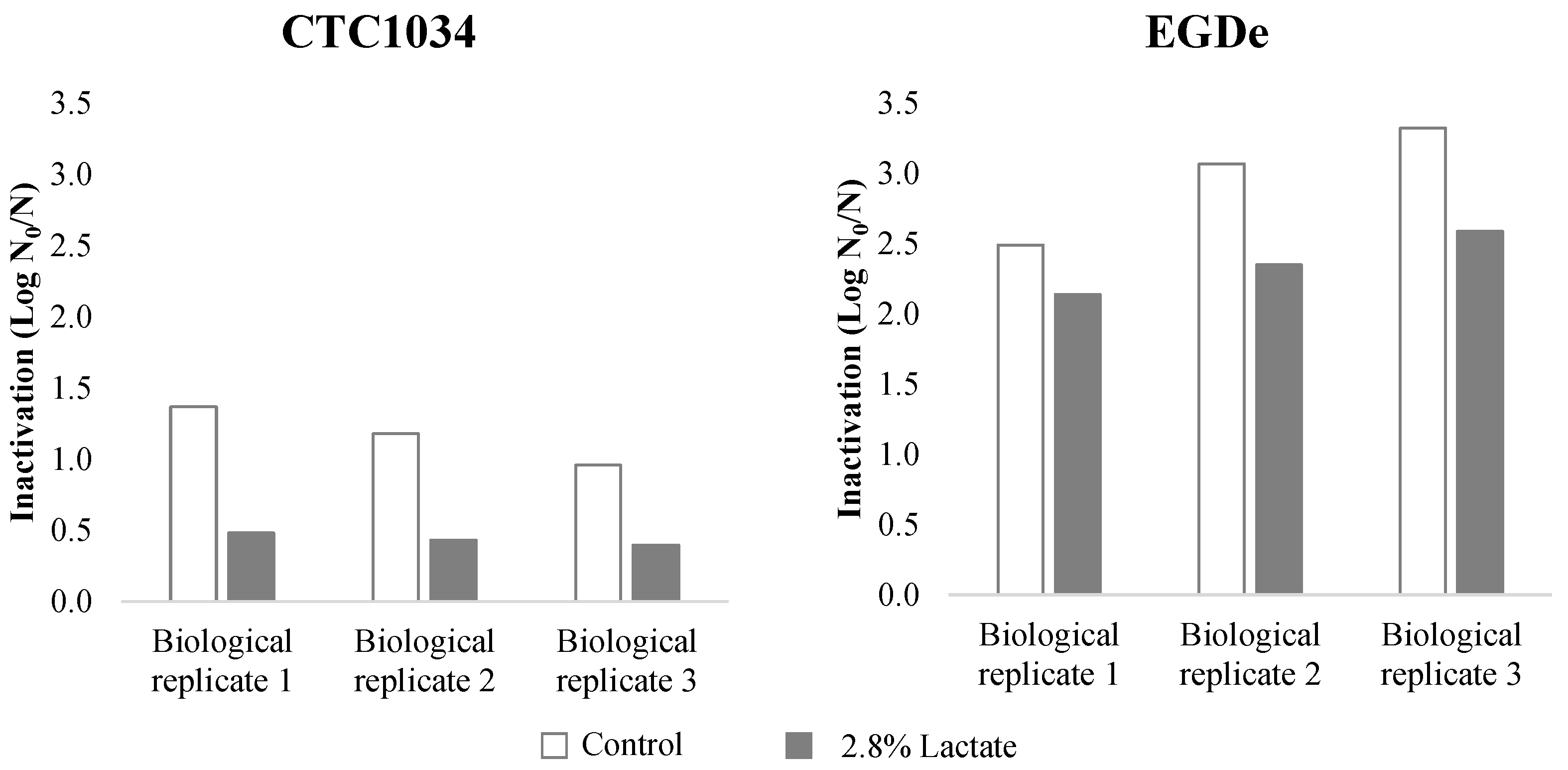
3.1. Inactivation of L. monocytogenes by HPP
3.2. Analysis of RNA-Seq Results. KEGG Annotation Classification and Pathway Enrichment Analysis of the DEGs
3.2.1. Comparison of L. monocytogenes CTC1034 and EGDe Genomes
3.2.2. Whole Transcriptome Analysis
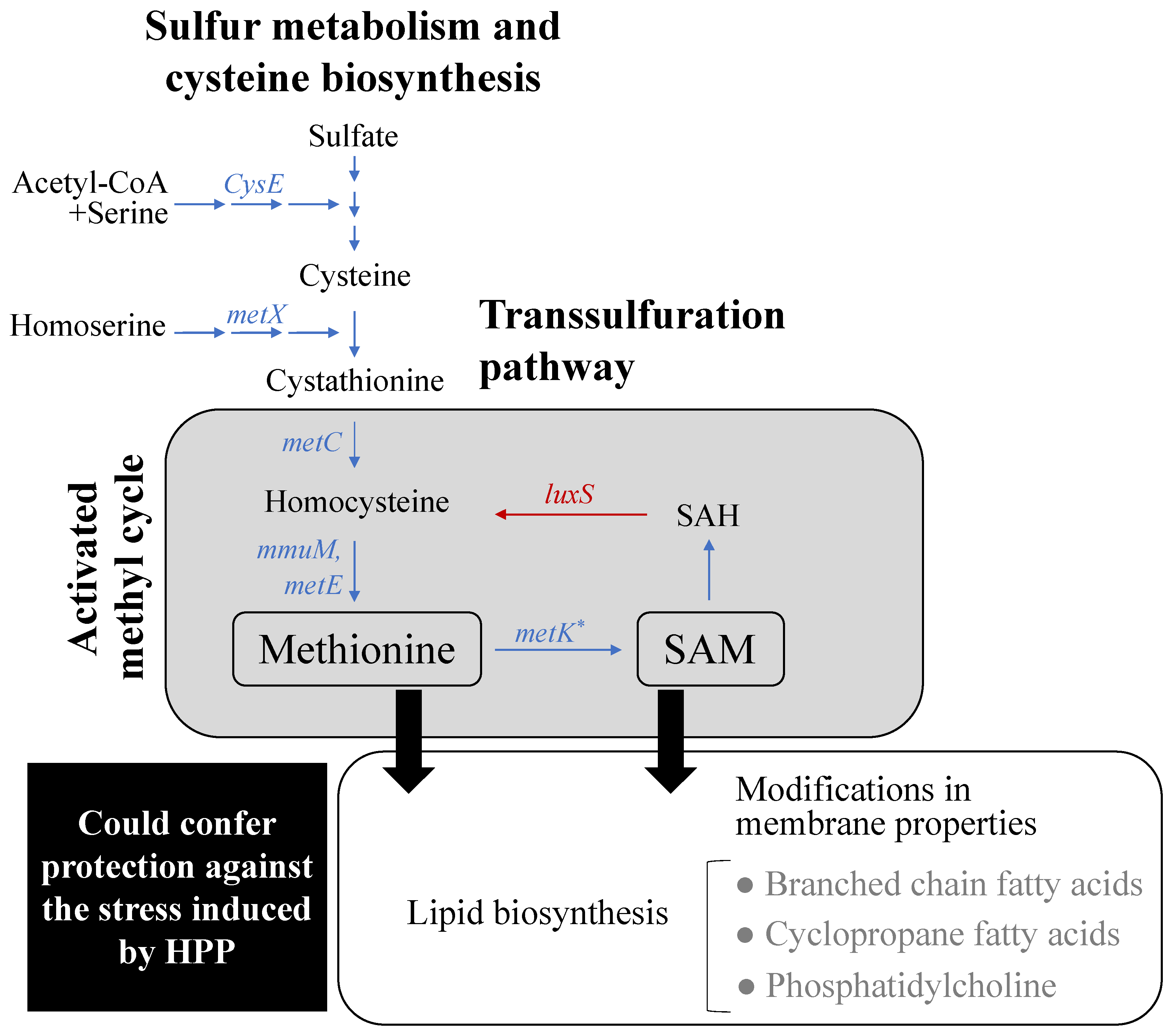
3.2.3. Effect of Lactate Exposure on L. monocytogenes
3.2.4. Effect of HPP on L. monocytogenes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FDA; USDA. Interagency Risk Assessment: Listeria Monocytogenes in Retail Delicatessens; Technical report; U.S. Department of Agriculture. Food Safety and Inspection Service: Washington, DC, USA, 2013. [Google Scholar]
- EFSA Commission Regulation (EC) No 2073/2005 of 15th November 2005 on microbiological criteria for foodstuffs. Off. J. Eur. Union 2005, L338, 1–26.
- FSIS 9 CFR Part 430: Control of Listeria monocytogenes in ready-to-eat meat and poultry products. Fed. Regist. 2003, 68, 34208–34254.
- Jofré, A.; Garriga, M.; Aymerich, T. Inhibition of Salmonella sp. Listeria monocytogenes and Staphylococcus aureus in cooked ham by combining antimicrobials, high hydrostatic pressure and refrigeration. Meat Sci. 2008, 78, 53–59. [Google Scholar] [CrossRef]
- Barmpalia, I.M.; Koutsoumanis, K.P.; Geornaras, I.; Belk, K.E.; Scanga, J.A.; Kendall, P.A.; Smith, G.C.; Sofos, J.N. Effect of antimicrobials as ingredients of pork bologna for Listeria monocytogenes control during storage at 4 or 10 °C. Food Microbiol. 2005, 22, 205–211. [Google Scholar] [CrossRef]
- Theron, M.M.; Lues, J.F.R. Organic Acids and Meat Preservation: A Review. Food Rev. Int. 2007, 23, 141–158. [Google Scholar] [CrossRef]
- Leistner, L. Basic aspects of food preservation by hurdle technology. Int. J. Food Microbiol. 2000, 55, 181–186. [Google Scholar] [CrossRef]
- Capozzi, V.; Fiocco, D.; Amodio, M.L.; Gallone, A.; Spano, G. Bacterial stressors in minimally processed food. Int. J. Mol. Sci. 2009, 10, 3076–3105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerasle, M.; Guillou, S.; Simonin, H.; Anthoine, V.; Chéret, R.; Federighi, M.; Membré, J.-M. Assessment of Salmonella and Listeria monocytogenes level in ready-to-cook poultry meat: Effect of various high pressure treatments and potassium lactate concentrations. Int. J. Food Microbiol. 2014, 186, 74–83. [Google Scholar] [CrossRef]
- Serra-Castelló, C.; Jofré, A.; Belletti, N.; Garriga, M.; Bover-Cid, S. Modelling the piezo-protection effect exerted by lactate on the high pressure resistance of Listeria monocytogenes in cooked ham. Food Res. Int. 2021, 140, 110003. [Google Scholar] [CrossRef]
- Bover-Cid, S.; Serra-Castelló, C.; Dalgaard, P.; Garriga, M.; Jofré, A. New insights on Listeria monocytogenes growth in pressurised cooked ham: A piezo-stimulation effect enhanced by organic acids during storage. Int. J. Food Microbiol. 2019, 290, 150–158. [Google Scholar] [CrossRef]
- Gorski, L.; Flaherty, D.; Mandrell, R.E. Competitive Fitness of Listeria monocytogenes Serotype 1/2a and 4b Strains in Mixed Cultures with and without Food in the U.S. Food and Drug Administration Enrichment Protocol. Appl. Environ. Microbiol. 2006, 72, 776–783. [Google Scholar] [CrossRef] [Green Version]
- Aymerich, T.; Jofré, A.; Garriga, M.; Hugas, M. Inhibition of Listeria monocytogenes and Salmonella by natural antimicrobials and high hydrostatic pressure in sliced cooked ham. J. Food Prot. 2005, 68, 173–177. [Google Scholar] [CrossRef]
- Hereu, A. Modeling the high pressure inactivation kinetics of Listeria monocytogenes on RTE cooked meat products. Innov. Food Sci. Emerg. Technol. 2012, 16, 305–315. [Google Scholar] [CrossRef]
- Hereu, A.; Bover-Cid, S.; Garriga, M.; Aymerich, T. High hydrostatic pressure and biopreservation of dry-cured ham to meet the Food Safety Objectives for Listeria monocytogenes. Int. J. Food Microbiol. 2012, 154, 107–112. [Google Scholar] [CrossRef]
- Morales, P.; Calzada, J.; Rodríguez, B.; de Paz, M.; Gaya, P.; Nuñez, M. Effect of cheese water activity and carbohydrate content on the barotolerance of Listeria monocytogenes scott A. J. Food Prot. 2006, 69, 1328–1333. [Google Scholar] [CrossRef]
- Jantzen, M.M.; Navas, J.; De Paz, M.; Rodríguez, B.; Da Silva, W.P.; Nuñez, M.; Martínez-Suárez, J.V. Evaluation of ALOA plating medium for its suitability to recover high pressure-injured Listeria monocytogenes from ground chicken meat. Lett. Appl. Microbiol. 2006, 43, 313–317. [Google Scholar] [CrossRef]
- Cocolin, L.; Manzano, M.; Aggio, D.; Cantoni, C.; Comi, G. A novel polymerase chain reaction (PCR)-denaturing gradient gel electrophoresis (DGGE) for the identification of Micrococcaceae strains involved in meat fermentations. Its application to naturally fermented Italian sausages. Meat Sci. 2001, 58, 59–64. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Stärk, M.; Huson, D.H. Analysis of 16S rRNA environmental sequences using MEGAN. BMC Genom. 2011, 12, S17. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core: Vienna, Austria, 2019. [Google Scholar]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009, 10, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro-Gómez, P.; Fontecha, J.; Rodríguez-Alcalá, L.M. A high-performance direct transmethylation method for total fatty acids assessment in biological and foodstuff samples. Talanta 2014, 128, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Calvo, M.V.; Martín-Hernández, M.C.; García-Serrano, A.; Castro-Gómez, M.P.; Alonso-Miravalles, L.; García-Martín, R.; Megino-Tello, J.; Alonso, L.; Fontecha, J. Comprehensive characterization of neutral and polar lipids of buttermilk from different sources and its milk fat globule membrane isolates. J. Food Compos. Anal. 2020, 86, 103386. [Google Scholar] [CrossRef]
- Jofré, A.; Garriga, M.; Aymerich, T. Inhibition of Listeria monocytogenes in cooked ham through active packaging with natural antimicrobials and high-pressure processing. J. Food Prot. 2007, 70, 2498–2502. [Google Scholar] [CrossRef] [PubMed]
- Stollewerk, K.; Jofré, A.; Comaposada, J.; Arnau, J.; Garriga, M. NaCl-free processing, acidification, smoking and high pressure: Effects on growth of Listeria monocytogenes and Salmonella enterica in QDS processed® dry-cured ham. Food Control 2014, 35, 56–64. [Google Scholar] [CrossRef]
- Jofré, A.; Champomier-Vergès, M.; Anglade, P.; Baraige, F.; Martín, B.; Garriga, M.; Zagorec, M.; Aymerich, T. Protein synthesis in lactic acid and pathogenic bacteria during recovery from a high pressure treatment. Res. Microbiol. 2007, 158, 512–520. [Google Scholar] [CrossRef]
- Stasiewicz, M.J.; Wiedmann, M.; Bergholz, T.M. The transcriptional response of Listeria monocytogenes during adaptation to growth on lactate and diacetate includes synergistic changes that increase fermentative acetoin production. Appl. Environ. Microbiol. 2011, 77, 5294–5306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpenter, C.E.; Broadbent, J.R. External concentration of organic acid anions and pH: Key independent variables for studying how organic acids inhibit growth of bacteria in mildly acidic foods. J. Food Sci. 2009, 74, R12-5. [Google Scholar] [CrossRef]
- Mellin, J.R.; Tiensuu, T.; Bécavin, C.; Gouin, E.; Johansson, J.; Cossart, P. A riboswitch-regulated antisense RNA in Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 2013, 110, 13132–13137. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.; Murrieta, C.M.; Rule, D.C.; Miller, K.W. Exogenous or L-Rhamnose-derived 1,2-Propanediol is metabolized via a pduD-dependent pathway in Listeria innocua. Appl. Environ. Microbiol. 2008, 74, 7073–7079. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Orsi, R.H.; den Bakker, H.C.; Wiedmann, M.; Boor, K.J.; Bergholz, T.M. Transcriptomic analysis of the adaptation of Listeria monocytogenes to growth on vacuum-packed cold smoked salmon. Appl. Environ. Microbiol. 2015, 81, 6812–6824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anast, J.M.; Schmitz-Esser, S. The transcriptome of Listeria monocytogenes during co-cultivation with cheese rind bacteria suggests adaptation by induction of ethanolamine and 1,2-propanediol catabolism pathway genes. PLoS ONE 2020, 15, e0233945. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Smid, E.J.; Boeren, S.; Notebaart, R.A.; Abee, T. Bacterial microcompartment-dependent 1,2-Propanediol utilization stimulates anaerobic growth of Listeria monocytogenes EGDe. Front. Microbiol. 2019, 10, 2660. [Google Scholar] [CrossRef]
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to stress conditions encountered in food and food processing environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roe, A.J.; McLaggan, D.; Davidson, I.; O’Byrne, C.; Booth, I.R. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol. 1998, 180, 767–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feehily, C.; O’Byrne, C.P.; Karatzas, K.A.G. Functional γ-aminobutyrate shunt in Listeria monocytogenes: Role in acid tolerance and succinate biosynthesis. Appl. Environ. Microbiol. 2013, 79, 74–80. [Google Scholar] [CrossRef] [Green Version]
- Gabel, C.V.; Berg, H.C. The speed of the flagellar rotary motor of Escherichia coli varies linearly with protonmotive force. Proc. Natl. Acad. Sci. USA 2003, 100, 8748–8751. [Google Scholar] [CrossRef] [Green Version]
- Porter, S.L.; Wadhams, G.H.; Armitage, J.P. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 2011, 9, 153–165. [Google Scholar] [CrossRef]
- Lado, B.H.; Yousef, A.E. Characteristics of Listeria monocytogenes important to food processors. In Listeria, Listeriosis, and Food Safety; Ryser, E., Marth, E., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 157–213. [Google Scholar]
- Brul, S.; Coote, P. Preservative agents in foods: Mode of action and microbial resistance mechanisms. Int. J. Food Microbiol. 1999, 50, 1–17. [Google Scholar] [CrossRef]
- Doores, S. Organic acids. In Antimicrobials in Food; Davidson, P., Sofos, J., Branen, A., Eds.; CRC Press: Boca Raton, FL, USA, 2005; pp. 91–141. [Google Scholar]
- Ten Brink, B.; Otto, R.; Hansen, U.P.; Konings, W.N. Energy recycling by lactate efflux in growing and nongrowing cells of Streptococcus cremoris. J. Bacteriol. 1985, 162, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Suo, Y.; Gao, S.; Baranzoni, G.M.; Xie, Y.; Liu, Y. Comparative transcriptome RNA-Seq analysis of Listeria monocytogenes with sodium lactate adaptation. Food Control 2018, 91, 193–201. [Google Scholar] [CrossRef]
- Roe, A.J.; O’Byrne, C.; McLaggan, D.; Booth, I.R. Inhibition of Escherichia coli growth by acetic acid: A problem with methionine biosynthesis and homocysteine toxicity. Microbiology 2002, 148, 2215–2222. [Google Scholar] [CrossRef] [Green Version]
- Pinhal, S.; Ropers, D.; Geiselmann, J.; de Jong, H. Acetate metabolism and the inhibition of bacterial growth by acetate. J. Bacteriol. 2019, 201, e00147-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferla, M.P.; Patrick, W.M. Bacterial methionine biosynthesis. Microbiology 2014, 160, 1571–1584. [Google Scholar] [CrossRef] [Green Version]
- Walvekar, A.S.; Laxman, S. Methionine at the heart of anabolism and signaling: Perspectives from Budding Yeast. Front. Microbiol. 2019, 10, 2624. [Google Scholar] [CrossRef]
- Aktas, M.; Gleichenhagen, J.; Stoll, R.; Narberhaus, F. S-adenosylmethionine-binding properties of a bacterial phospholipid N-methyltransferase. J. Bacteriol. 2011, 193, 3473–3481. [Google Scholar] [CrossRef] [Green Version]
- Loi, V.V.; Rossius, M.; Antelmann, H. Redox regulation by reversible protein S-thiolation in bacteria. Front. Microbiol. 2015, 6, 187. [Google Scholar] [CrossRef] [Green Version]
- Gründling, A.; Burrack, L.S.; Bouwer, H.G.A.; Higgins, D.E. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl. Acad. Sci. USA 2004, 101, 12318–12323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hingston, P.A.; Piercey, M.J.; Truelstrup Hansen, L. Genes associated with desiccation and osmotic stress in Listeria monocytogenes as revealed by insertional mutagenesis. Appl. Environ. Microbiol. 2015, 81, 5350–5362. [Google Scholar] [CrossRef] [Green Version]
- O’Neil, H.S.; Marquis, H. Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect. Immun. 2006, 74, 6675–6681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, J.P.; Bittencourt, C.R.; Ross, T. Differential gene expression of Listeria monocytogenes during high hydrostatic pressure processing. Microbiology 2008, 154, 462–475. [Google Scholar] [CrossRef] [Green Version]
- Parveen, N.; Cornell, K.A. Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol. Microbiol. 2011, 79, 7–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravim, F.; da Silva, L.F.; Souza, D.T.; Lippman, S.I.; Broach, J.R.; Fernandes, A.A.R.; Fernandes, P.M.B. High hydrostatic pressure activates transcription factors involved in Saccharomyces cerevisiae stress tolerance. Curr. Pharm. Biotechnol. 2012, 13, 2712–2720. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; Levine, R.L. Methionine in proteins defends against oxidative stress. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2009, 23, 464–472. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, W.M. Involvement of methionine in bacterial lipid synthesis. J. Bacteriol. 1959, 78, 709–713. [Google Scholar] [CrossRef] [Green Version]
- Cheftel, J.C. Review: High-pressure, microbial inactivation and food preservation. Food Sci. Technol. Int. 1995, 1, 75–90. [Google Scholar] [CrossRef]
- Macdonald, A.G. The effects of pressure on the molecular structure and physiological functions of cell membranes. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 1984, 304, 47–68. [Google Scholar] [CrossRef]
- Brooks, N.J. Pressure effects on lipids and bio-membrane assemblies. IUCrJ 2014, 1, 470–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; O’Riordan, M.X.D. Branched-chain fatty acids promote Listeria monocytogenes intracellular infection and virulence. Infect. Immun. 2010, 78, 4667–4673. [Google Scholar] [CrossRef] [Green Version]
- Geiger, O.; López-Lara, I.M.; Sohlenkamp, C. Phosphatidylcholine biosynthesis and function in bacteria. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2013, 1831, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Grogan, D.W.; Cronan, J.E.J. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 1997, 61, 429–441. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Gänzle, M.G. Influence of cyclopropane fatty acids on heat, high pressure, acid and oxidative resistance in Escherichia coli. Int. J. Food Microbiol. 2016, 222, 16–22. [Google Scholar] [CrossRef]
- Gänzle, M.; Liu, Y. Mechanisms of pressure-mediated cell death and injury in Escherichia coli: From fundamentals to food applications. Front. Microbiol. 2015, 6, 599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jochim, A.; Shi, T.; Belikova, D.; Schwarz, S.; Peschel, A.; Heilbronner, S. Methionine limitation impairs pathogen expansion and biofilm formation capacity. Appl. Environ. Microbiol. 2019, 85, e00177-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Fatty Acid | Condition | |||
|---|---|---|---|---|
| Control | Lactate | HPP | Lactate + HPP | |
| C10:0 | 0.02 ± 0.03 | 0.09 ± 0.12 | 0.03 ± 0.04 | 0.08 ± 0.01 |
| C12:0 | 1.03 ± 0.32 | 0.87 ± 0.07 | 0.73 ± 0.08 | 0.77 ± 0.03 |
| C13 iso | 0.12 ± 0.01 | 0.12 ± 0.10 | 0.06 ± 0.00 | 0.06 ± 0.03 |
| C13 anteiso | 0.23 ± 0.08 | 0.27 ± 0.06 | 0.26 ± 0.07 | 0.26 ± 0.06 |
| C14 iso | 1.26 ± 0.01 | 1.22 ± 0.05 | 1.22 ± 0.15 | 1.21 ± 0.16 |
| C14 | 4.59 ± 0.40 | 3.99 ± 0.61 | 4.04 ± 0.77 | 3.99 ± 0.89 |
| C15 iso | 14.05 ± 0.83 | 15.45 ± 0.09 | 14.31 ± 0.28 | 14.59 ± 0.18 |
| C15 anteiso | 39.72 ± 3.06 | 41.78 ± 1.28 | 41.24 ± 0.31 | 41.14 ± 0.45 |
| C15 | 0.42 ± 0.18 | 0.49 ± 0.06 | 0.51 ± 0.05 | 0.51 ± 0.12 |
| C16 iso | 3.13 ± 0.03 | 3.26 ± 0.35 | 3.60 ± 0.58 | 3.40 ± 0.21 |
| C16 | 5.90 ± 2.51 | 4.17 ± 0.38 | 4.29 ± 0.60 | 4.03 ± 0.15 |
| C16:1 | 2.62 ± 0.00 | 2.44 ± 1.19 | 2.79 ± 0.86 | 2.63 ± 0.58 |
| C17 iso | 4.63 ± 0.35 | 4.82 ± 0.05 | 5.23 ± 0.07 | 5.06 ± 0.20 |
| C17 anteiso | 16.75 ± 1.39 | 17.28 ± 0.60 | 17.91 ± 0.73 | 18.10 ± 0.53 |
| C18 | 1.72 ± 0.71 | 1.11 ± 0.11 | 1.12 ± 0.13 | 1.38 ± 0.05 |
| C18:1 cis9 | 3.31 ± 1.28 | 2.33 ± 0.10 | 2.40 ± 0.21 | 2.39 ± 0.17 |
| C18:1 cis11 | 0.02 ± 0.03 | 0.04 ± 0.05 | 0.01 ± 0.02 | 0.00 ± 0.00 |
| C19:0 | 0.11 ± 0.04 | 0.08 ± 0.01 | 0.09 ± 0.01 | 0.26 ± 0.10 |
| C18:2 | 0.36 ± 0.17 | 0.20 ± 0.09 | 0.17 ± 0.14 | 0.16 ± 0.10 |
| BCFA a | 79.89 ± 5.67 | 84.20 ± 1.57 | 83.82 ± 0.59 | 83.82 ± 0.24 |
| iso BCFA | 23.19 ± 1.15 | 24.88 ± 0.36 | 24.41 ± 0.52 | 24.31 ± 0.37 |
| anteiso BCFA | 56.70 ± 4.53 | 59.32 ± 1.93 | 59.41 ± 1.12 | 59.50 ± 0.13 |
| iso/anteiso | 0.41 ± 0.02 | 0.42 ± 0.02 | 0.41 ± 0.02 | 0.41 ± 0.01 |
| C13 BCFA | 0.35 ± 0.07 | 0.39 ± 0.04 | 0.32 ± 0.08 | 0.32 ± 0.02 |
| C15 BCFA | 53.77 ± 3.89 | 57.23 ± 1.37 | 55.55 ± 0.59 | 55.73 ± 0.63 |
| C17 BCFA | 21.38 ± 1.74 | 22.10 ± 0.65 | 23.14 ± 0.66 | 23.16 ± 0.73 |
| C15 BCFA/C17 BCFA | 2.51 ± 0.02 | 2.59 ± 0.01 | 2.40 ± 0.05 | 2.41 ± 0.10 |
| C15 anteiso/C17 anteiso | 2.37 ± 0.01 | 2.42 ± 0.01 | 2.31 ± 0.08 | 2.27 ± 0.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serra-Castelló, C.; Ferrocino, I.; Jofré, A.; Cocolin, L.; Bover-Cid, S.; Rantsiou, K. Unravelling the Molecular Mechanisms Underlying the Protective Effect of Lactate on the High-Pressure Resistance of Listeria monocytogenes. Biomolecules 2021, 11, 677. https://doi.org/10.3390/biom11050677
Serra-Castelló C, Ferrocino I, Jofré A, Cocolin L, Bover-Cid S, Rantsiou K. Unravelling the Molecular Mechanisms Underlying the Protective Effect of Lactate on the High-Pressure Resistance of Listeria monocytogenes. Biomolecules. 2021; 11(5):677. https://doi.org/10.3390/biom11050677
Chicago/Turabian StyleSerra-Castelló, Cristina, Ilario Ferrocino, Anna Jofré, Luca Cocolin, Sara Bover-Cid, and Kalliopi Rantsiou. 2021. "Unravelling the Molecular Mechanisms Underlying the Protective Effect of Lactate on the High-Pressure Resistance of Listeria monocytogenes" Biomolecules 11, no. 5: 677. https://doi.org/10.3390/biom11050677






