Antigenotoxic Effects and Possible Mechanism of Red Yeast (Sporidiobolus pararoseus) on Aflatoxin B1-Induced Mutagenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Preparation of Red Yeast Extracts
2.3. Analysis of Chemical Constituent in Red Yeast
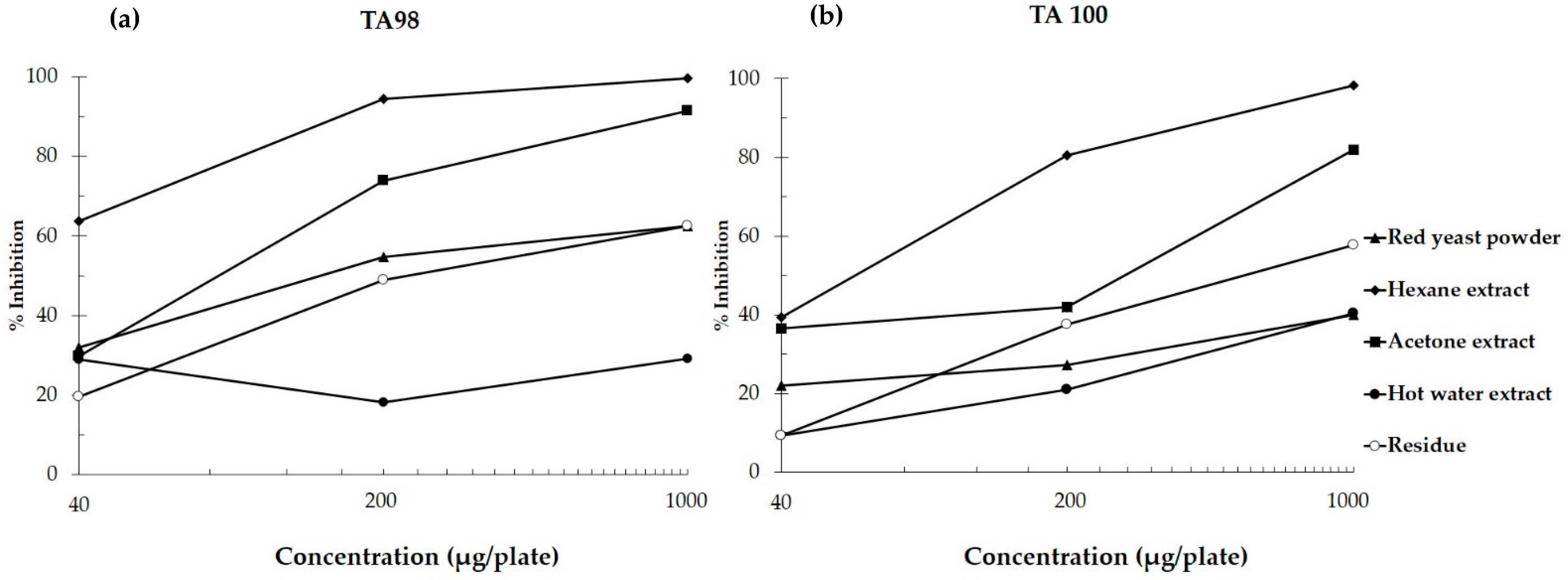
2.4. Mutagenicity and Antimutagenicity of Red Yeast Using Salmonella Mutation Assay
2.5. Animals
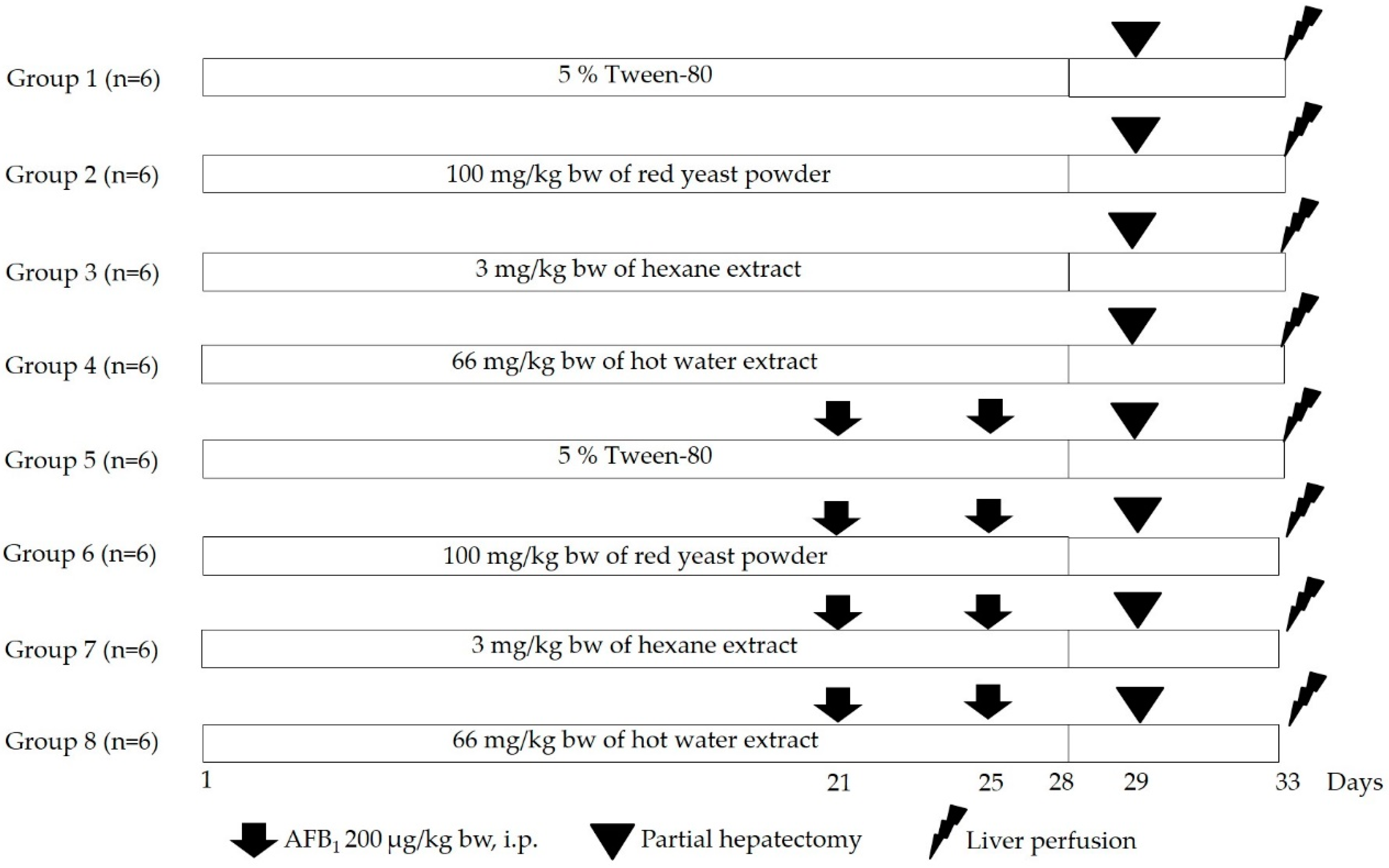
2.6. Clastogenicity and Anticlastogenicity of Red Yeast Using a Rat Liver Micronucleus Test
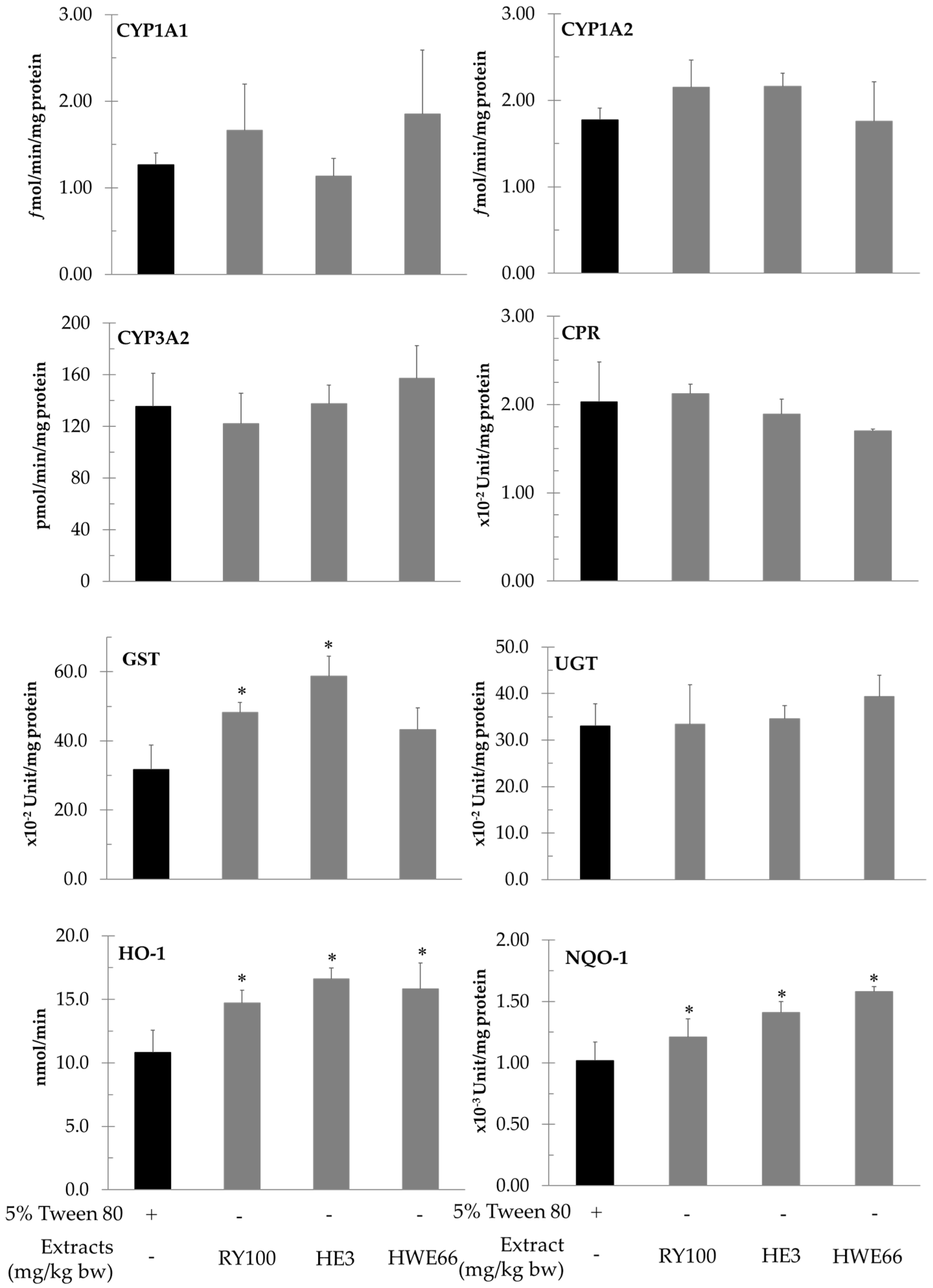
2.7. Determination of the Activities of Hepatic Phases I and II Xenobiotic Metabolizing Enzymes
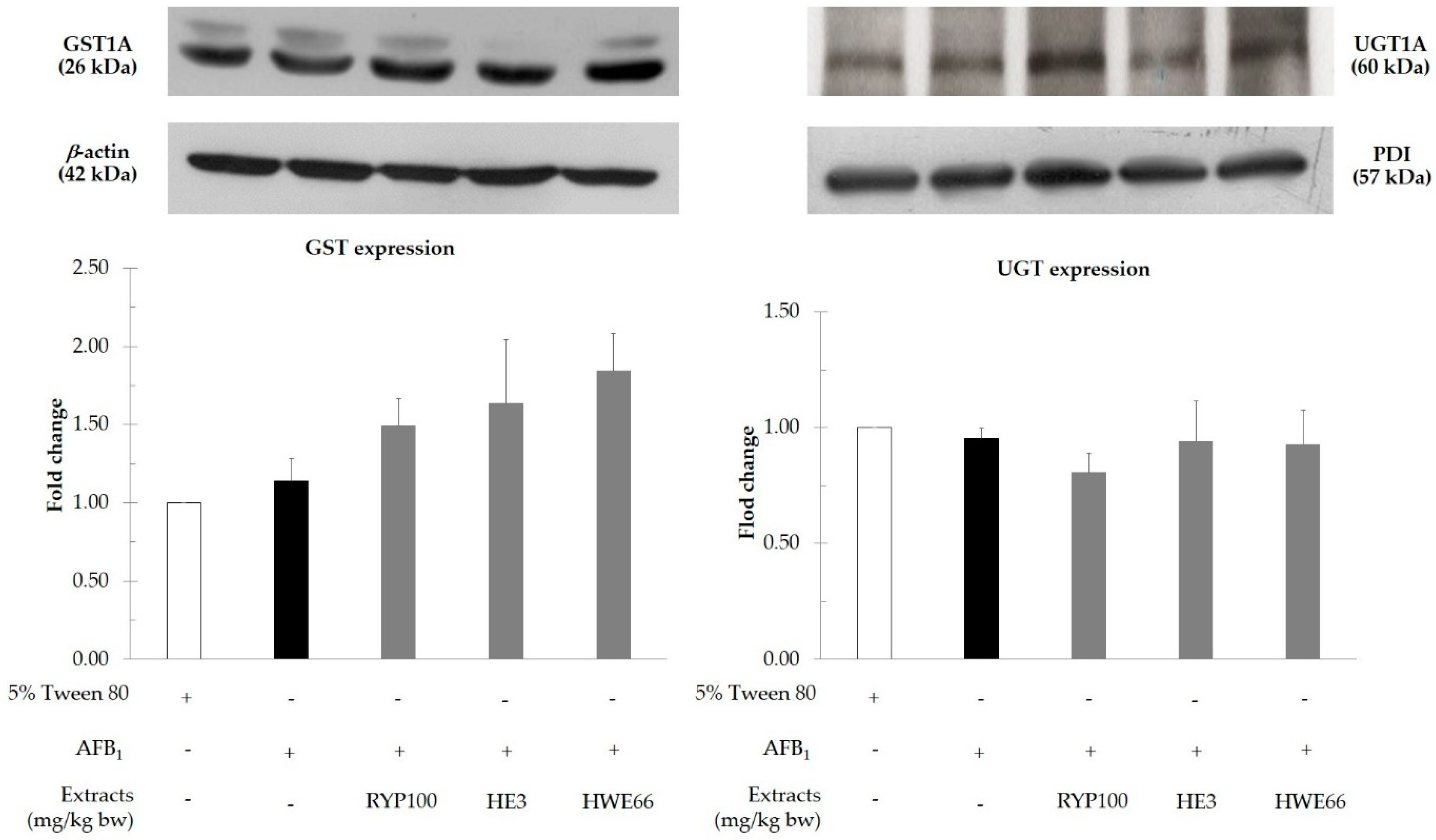
2.8. Determination of Protein Expression of Xenobiotic Metabolizing Enzymes
2.9. Statistical Analysis
3. Results
3.1. Chemical Ingredients in Red Yeast
3.2. Mutagenicity and Antimutagenicity of Red Yeast in Salmonella Mutation Assay
3.3. Clastogenicity and Anticlastogenicity of Red Yeast in Rats
3.4. Effect of Red Yeast on Xenobiotic Metabolizing Enzymes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hamid, A.S.; Tesfamariam, I.G.; Zhang, Y.; Zhang, Z.G. Aflatoxin B1-induced hepatocellular carcinoma in developing countries: Geographical distribution, mechanism of action and prevention. Oncol. Lett. 2013, 5, 1087–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawal, S.; Kim, J.E.; Coulombe, R., Jr. Aflatoxin B1 in poultry: Toxicology, metabolism and prevention. Vet. Sci. Res. J. 2010, 89, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Karlovsky, P.; Suman, M.; Berthiller, F.; De Meester, J.; Eisenbrand, G.; Perrin, I.; Oswald, I.P.; Speijers, G.; Chiodini, A.; Recker, T. Impact of food processing and detoxification treatments on mycotoxin contamination. Mycotoxin Res. 2016, 32, 179–205. [Google Scholar] [CrossRef] [PubMed]
- Firmin, S.; Morgavi, D.; Yiannikouris, A.; Boudra, H. Effectiveness of modified yeast cell wall extracts to reduce aflatoxin B1 absorption in dairy ewes. Int. J. Dairy Sci. 2011, 94, 5611–5619. [Google Scholar] [CrossRef] [PubMed]
- Yiannikouris, A.; André, G.; Buléon, A.; Jeminet, G.; Canet, I.; François, J.; Bertin, G.; Jouany, J.-P. Comprehensive conformational study of key interactions involved in zearalenone complexation with β-D-glucans. Biomacromolecules 2004, 5, 2176–2185. [Google Scholar] [CrossRef]
- Shetty, P.H.; Jespersen, L. Saccharomyces cerevisiae and lactic acid bacteria as potential mycotoxin decontaminating agents. Trends Food Sci. Tech. 2006, 17, 48–55. [Google Scholar] [CrossRef]
- Du, C.; Guo, Y.; Cheng, Y.; Han, M.; Zhang, W.; Qian, H. Anti-cancer effects of torulene, isolated from Sporidiobolus pararoseus, on human prostate cancer LNCaP and PC-3 cells via a mitochondrial signal pathway and the down-regulation of AR expression. RSC Adv. 2017, 7, 2466–2474. [Google Scholar] [CrossRef] [Green Version]
- Shi, Q.; Wang, H.; Du, C.; Zhang, W.; Qian, H. Tentative identification of torulene cis/trans geometrical isomers isolated from Sporidiobolus pararoseus by high-performance liquid chromatography-diode array detection-mass spectrometry and preparation by column chromatography. Anal. Sci. 2013, 29, 997–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawata, A.; Murakami, Y.; Suzuki, S.; Fujisawa, S. Anti-inflammatory activity of β-carotene, lycopene and tri-n-butylborane, a scavenger of reactive oxygen species. In Vivo 2018, 32, 255–264. [Google Scholar]
- Du, C.; Guo, Y.; Cheng, Y.; Han, M.; Zhang, W.; Qian, H. Torulene and torularhodin, protects human prostate stromal cells from hydrogen peroxide-induced oxidative stress damage through the regulation of Bcl-2/Bax mediated apoptosis. Free Radic. Res. 2017, 51, 113–123. [Google Scholar] [CrossRef]
- Novak, M.; Vetvicka, V. β-Glucans, history, and the present: Immunomodulatory aspects and mechanisms of action. J Immunotoxicol. 2008, 5, 47–57. [Google Scholar] [CrossRef]
- Tsai, P.-W.; Yang, C.-Y.; Chang, H.-T.; Lan, C.-Y. Human antimicrobial peptide LL-37 inhibits adhesion of Candida albicans by interacting with yeast cell-wall carbohydrates. PLoS ONE 2011, 6, e17755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, T.; Haldar, S.; Bedford, M.; Muthusami, N.; Samanta, I. Assessment of yeast cell wall as replacements for antibiotic growth promoters in broiler diets: Effects on performance, intestinal histo-morphology and humoral immune responses. J. Anim. Physiol. Anim. Nutr. 2012, 96, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Fruhauf, S.; Schwartz, H.; Ottner, F.; Krska, R.; Vekiru, E. Yeast cell based feed additives: Studies on aflatoxin B1 and zearalenone. Food Addit. Contam. Part A 2012, 29, 217–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapingkae, W.; Yindee, P.; Moonmanee, T. Effect of dietary red yeast (Sporidiobolus pararoseus) supplementation on small intestinal histomorphometry of laying hens. J. Anim. Plant Sci. 2016, 26, 909–915. [Google Scholar]
- Siddiqui, I.A.; Sanna, V.; Ahmad, N.; Sechi, M.; Mukhtar, H. Resveratrol nanoformulation for cancer prevention and therapy. Ann. N. Y. Acad. Sci. 2015, 1348, 20–31. [Google Scholar] [CrossRef]
- Maurici, D.; Aardema, M.; Corvi, R.; Kleber, M.; Krul, C.; Laurent, C.; Loprieno, N.; Pasanen, M.; Pfuhler, S.; Phillips, B.; et al. Genotoxicity and mutagenicity. Altern. Lab. Anim. 2005, 33 (Suppl. 1), 117–130. [Google Scholar] [CrossRef]
- Manowattana, A.; Techapun, C.; Watanabe, M.; Chaiyaso, T. Bioconversion of biodiesel-derived crude glycerol into lipids and carotenoids by an oleaginous red yeast Sporidiobolus pararoseus KM281507 in an airlift bioreactor. J. Biosci. Bioeng. 2018, 125, 59–66. [Google Scholar] [CrossRef]
- Sankam, P.; Punvittayagul, C.; Sringam, K.; Chaiyasut, C.; Wongpoomchai, R. Antimutagenicity and anticlastogenicity of glutinous purple rice hull using in vitro and in vivo testing systems. Mol. Cell. Toxicol. 2013, 9, 169–176. [Google Scholar] [CrossRef]
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.-I.; Lee, Y.C. Carbohydrate analysis by a phenol–sulfuric acid method in microplate format. Anal. Biochem. 2005, 339, 69–72. [Google Scholar] [CrossRef]
- Inboot, W.; Taya, S.; Chailungka, A.; Meepowpan, P.; Wongpoomchai, R. Genotoxicity and antigenotoxicity of the methanol extract of Cleistocalyx nervosum var. paniala seed using a Salmonella mutation assay and rat liver micronucleus tests. Mol. Cell. Toxicol. 2012, 8, 19–24. [Google Scholar] [CrossRef]
- Chariyakornkul, A.; Punvittayagul, C.; Taya, S.; Wongpoomchai, R. Inhibitory effect of purple rice husk extract on AFB1-induced micronucleus formation in rat liver through modulation of xenobiotic metabolizing enzymes. BMC Complement Altern. Med. 2019, 19, 237. [Google Scholar] [CrossRef] [Green Version]
- Suwannakul, N.; Punvittayagul, C.; Jarukamjorn, K.; Wongpoomchai, R. Purple rice bran extract attenuates the aflatoxin B1-induced initiation stage of hepatocarcinogenesis by alteration of xenobiotic metabolizing enzymes. Asian Pac. J. Cancer Prev. 2015, 16, 3371–3376. [Google Scholar] [CrossRef] [Green Version]
- Punvittayagul, C.; Wongpoomchai, R.; Taya, S.; Pompimon, W. Effect of pinocembrin isolated from Boesenbergia pandurata on xenobiotic-metabolizing enzymes in rat liver. Drug Metab. Lett. 2011, 5, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Rushing, B.R.; Selim, M.I. Aflatoxin B1: A review on metabolism, toxicity, occurrence in food, occupational exposure, and detoxification methods. Food Chem. Toxicol. 2019, 124, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Manowattana, A.; Seesuriyachan, P.; Techapun, C.; Chaiyaso, T. Optimization of carotenoids production by red yeast Sporobolomyces pararoseus TISTR5213 using waste glycerol as the sole carbon source. Asia Pac. J. Sci. Technol. 2012, 17, 607–621. [Google Scholar]
- He, Y.; Campbell, T.C. Effects of carotenoids on aflatoxin B1-induced mutagenesis in S. typhimurium TA 100 and TA 98. Nutr. Cancer 1990, 13, 243–253. [Google Scholar]
- Rauscher, R.; Edenharder, R.; Platt, K. In vitro antimutagenic and in vivo anticlastogenic effects of carotenoids and solvent extracts from fruits and vegetables rich in carotenoids. Mutat. Res. 1998, 413, 129–142. [Google Scholar] [CrossRef]
- Luzhna, L.; Kathiria, P.; Kovalchuk, O. Micronuclei in genotoxicity assessment: From genetics to epigenetics and beyond. Front. Genet. 2013, 4, 131. [Google Scholar] [CrossRef] [Green Version]
- Morita, T.; MacGregor, J.T.; Hayashi, M. Micronucleus assays in rodent tissues other than bone marrow. Mutagenesis 2011, 26, 223–230. [Google Scholar] [CrossRef] [Green Version]
- Ziglari, T.; Allameh, A. The significance of glutathione conjugation in aflatoxin metabolism. In Aflatoxins, Research Advances and Future Prospects; InTech: London, UK, 2013; pp. 267–286. [Google Scholar]
- Grancharov, K.; Naydenova, Z.; Lozeva, S.; Golovinsky, E. Natural and synthetic inhibitors of UDP-glucuronosyltransferase. Pharmacol. Ther. 2001, 89, 171–186. [Google Scholar] [CrossRef]
- Reddy, L.; Odhav, B.; Bhoola, K. Aflatoxin B1-induced toxicity in HepG2 cells inhibited by carotenoids: Morphology, apoptosis and DNA damage. Biol. Chem. 2006, 387, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Alija, A.; Bresgen, N.; Sommerburg, O.; Siems, W.; Eckl, P. Cytotoxic and genotoxic effects of β-carotene breakdown products on primary rat hepatocytes. Carcinogenesis 2004, 25, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Shnimizu, M.; Moriwaki, H. Cancer chemoprevention by carotenoids. Molecules 2012, 17, 3202–3242. [Google Scholar] [CrossRef] [PubMed]
- Pool-Zobel, B.; Veeriah, S.; Böhmer, F.-D. Modulation of xenobiotic metabolising enzymes by anticarcinogens—focus on glutathione S-transferases and their role as targets of dietary chemoprevention in colorectal carcinogenesis. Mutat. Res. 2005, 591, 74–92. [Google Scholar] [CrossRef]
- Karaca, A.; Yilmaz, S.; Kaya, E.; Altun, S. The effect of lycopene on hepatotoxicity of aflatoxin B1 in rats. Arch. Physiol. Biochem. 2019, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Zhang, J.; Cao, Z.; Ji, Q.; Han, Y.; Song, M.; Shao, B.; Li, Y. Lycopene attenuates AFB-1-induced renal injury with the activation of the Nrf2 antioxidant signaling pathway in mice. Food Funct. 2018, 9, 6427–6434. [Google Scholar] [CrossRef]
- Trejo-Solís, C.; Pedraza-Chaverrí, J.; Torres-Ramos, M.; Jiménez-Farfán, D.; Cruz Salgado, A.; Serrano-García, N.; Osorio-Rico, L.; Sotelo, J. Multiple molecular and cellular mechanisms of action of lycopene in cancer inhibition. Evid. Based Complement Alternat. Med. 2013, 2013, 705121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loboda, A.; Damulewicz, M.; Pyza, E.; Jozkowicz, A.; Dulak, J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: An evolutionarily conserved mechanism. Cell Mol. Life Sci. 2016, 73, 3221–3247. [Google Scholar] [CrossRef] [Green Version]
| Compounds | Contents (mg/g Extract) | ||||
|---|---|---|---|---|---|
| Total Carbohydrate a | Protein a | Phenolic Compounds a | Lycopene b | β-Carotene b | |
| Red yeast powder | 497.7 ± 12.6 | 62.3 ± 6.7 | 4.9 ± 0.4 | 0.024 ± 0.000 | 0.013 ± 0.001 |
| Hexane extract | 36.1 ± 0.0 | 12.0 ± 5.3 | 5.0 ± 1.1 | 0.449 ± 0.035 | 0.094 ± 0.009 |
| Acetone extract | 118.6 ± 14.3 | 8.8 ± 1.7 | 10.7 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Hot water extract | 835.7 ± 64.6 | 22.9 ± 0.6 | 1.5 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Residue | 192.0 ± 13.1 | 118.5 ± 2.1 | 11.2 ± 0.8 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Treatment | Concentration (per Plate) | Number of His+ Revertant Colonies (Mutagenic Index) | |||
|---|---|---|---|---|---|
| TA98 | TA100 | ||||
| +S9 | −S9 | +S9 | −S9 | ||
| DMSO | 50 µL | 29 ± 1 (1.00) | 24 ± 2 (1.00) | 136 ± 13 (1.00) | 92 ± 3 (1.00) |
| DW | 50 µL | 28 ± 4 (1.00) | 24 ± 1 (1.00) | 136 ± 23 (1.00) | 103 ± 5 (1.00) |
| 2AA | 0.05 µg | 871 ± 157 (29.29) | - | 815 ± 98 (6.07) | - |
| AF-2 | 0.10 µg | - | 264 ± 11 (11.01) | - | - |
| AF-2 | 0.01 µg | - | - | - | 917 ± 14 (10.00) |
| Red yeast powder | 40 µg | 26 ± 3 (0.80) | 22 ± 2 (0.90) | 125 ± 22 (0.90) | 84 ± 8 (0.93) |
| 200 µg | 23 ± 2 (0.77) | 22 ± 1 (0.89) | 103 ± 13 (0.75) | 85 ± 11 (0.95) | |
| 1000 µg | 28 ± 4 (0.95) | 19 ± 1 (0.77) | 113 ± 7 (0.84) | 99 ± 6 (1.09) | |
| 5000 µg | 27 ± 5 (0.90) | 22 ± 0 (0.91) | 79 ± 13 (0.81) | 103 ± 6 (1.15) | |
| Hexane extract | 40 µg | 27 ± 3 (0.90) | 22 ± 3 (0.89) | 125 ± 19 (0.91) | 74 ± 5 (1.81) |
| 200 µg | 25 ± 3 (0.84) | 24 ± 2 (0.97) | 106 ± 9 (0.78) | 83 ± 4 (0.91) | |
| 1000 µg | 23 ± 1 (0.77) | 23 ± 2 (0.94) | 88 ± 7 (0.66) | 77 ± 6 (0.85) | |
| 5000 µg | 19 ± 2 (0.66) K | 14 ± 0 (0.59) K | 78 ± 8 (0.57) K | 66 ± 1 (0.73) K | |
| Acetone extract | 40 µg | 28 ± 4 (0.95) | 25 ± 3 (1.02) | 128 ± 20 (0.93) | 84 ± 2 (0.84) |
| 200 µg | 28 ± 5 (0.94) | 25 ± 4 (1.03) | 131 ± 9 (0.96) | 90 ± 3 (0.99) | |
| 1000 µg | 29 ± 3 (0.98) | 23 ± 2 (0.95) | 95 ± 12 (0.70) | 88 ± 10 (0.98) | |
| 5000 µg | 19 ± 2 (0.64) K | 20 ± 1 (0.81) | 92 ± 7 (0.68) K | 86 ± 2 (0.95) | |
| Hot water extract | 40 µg | 29 ± 4 (1.06) | 23 ± 1 (0.98) | 137 ± 16 (1.03) | 98 ± 6 (0.96) |
| 200 µg | 26 ± 2 (0.93) | 24 ± 2 (1.03) | 150 ± 11 (1.15) | 107 ± 13 (1.07) | |
| 1000 µg | 27 ± 3 (0.97) | 23 ± 1 (0.98) | 143 ± 23 (1.06) | 93 ± 9 (0.92) | |
| 5000 µg | 26 ± 3 (0.95) | 21 ± 1 (0.89) | 161 ± 35 (1.18) | 92 ± 13(0.91) | |
| Residue | 40 µg | 28 ± 2 (0.95) | 24 ± 1 (0.97) | 122 ± 16 (0.89) | 86 ± 2 (0.95) |
| 200 µg | 21 ± 1 (0.73) | 22 ± 1 (0.92) | 121 ± 17 (0.89) | 86 ± 4 (0.95) | |
| 1000 µg | 28 ± 3 (0.96) | 23 ± 3 (0.95) | 116 ± 6 (0.86) | 88 ± 1 (0.97) | |
| 5000 µg | 30 ± 3 (1.03) | 24 ± 2 (0.99) | 110 ± 5 (0.80) | 79 ± 2 (0.87) | |
| Treatment | Concentration (per Plate) | TA98 | TA100 | ||
|---|---|---|---|---|---|
| Number of Revertant Colonies | %Inhibition | Number of Revertant Colonies | %Inhibition | ||
| DMSO | 50 µL | 30 ± 7 | - | 118 ± 6 | - |
| AFB1 | 1.25 ng | 1148 ± 30 * | - | 893 ± 42 * | - |
| β-carotene | 20 ng | 283 ± 27 ** | 77.4 | 293 ± 37 ** | 78.4 |
| Lycopene | 1000 ng | 526 ± 25 ** | 55.6 | 540 ± 16 ** | 48.4 |
| Treatment | Final Body Weight (g) | Number per 1000 Hepatocytes | Mitotic Index | %Inhibition | ||
|---|---|---|---|---|---|---|
| MNH | MN | BNH Cells | ||||
| Vehicle | 276 ± 18 | 3.9 ± 0.5 | 3.9 ± 0.5 | 1.52 ± 0.2 | 0.64 ± 0.1 | - |
| Vehicle + RYP 100 mg/kg bw | 262 ± 13 | 3.5 ± 0.7 | 3.6 ± 0.8 | 1.52 ± 0.3 | 0.73 ± 0.1 | - |
| Vehicle + HE 3 mg/kg bw | 277 ± 23 | 3.3 ± 0.5 | 3.6 ± 0.6 | 0.89 ± 0.2 * | 0.79 ± 0.1 | - |
| Vehicle + HWE 66 mg/kg bw | 276 ± 18 | 3.4 ± 0.8 | 3.5 ± 0.8 | 1.56 ± 0.6 | 0.57 ± 0.1 | - |
| AFB1 | 270 ± 15 | 13.3 ± 2.1 * | 13.6 ± 2.0 * | 2.28 ± 0.3 * | 1.29 ± 0.3 * | - |
| AFB1 + RYP 100 mg/kg bw | 288 ± 8 | 7.1 ± 1.6 ** | 7.3 ± 1.3 ** | 1.71 ± 0.4 ** | 1.28 ± 0.2 | 46.4 ± 12.0 |
| AFB1 + HE 3 mg/kg bw | 291 ± 16 | 6.0 ± 2.1 ** | 6.2 ± 2.0 ** | 1.70 ± 0.2 ** | 1.27 ± 0.3 | 54.6 ± 15.7 |
| AFB1 + HWE 66 mg/kg bw | 288 ± 10 | 9.2 ± 1.7 ** | 9.3 ± 1.7 ** | 1.93 ± 0.4 | 1.48 ± 0.1 | 26.5 ± 7.7 |
| Enzyme Activities (per mg Protein) | 5% Tween-80 | AFB1 | AFB1+ RYP 100 mg/kg bw | AFB1 + HE 3 mg/kg bw | AFB1 + HWE 66 mg/kg bw |
|---|---|---|---|---|---|
| Cytochrome P450 1A1 (fmol/min) | 1.30 ± 0.51 | 1.05 ± 0.25 | 1.19 ± 0.48 | 0.95 ± 0.14 | 0.92 ± 0.20 |
| Cytochrome P450 1A2 (fmol/min) | 0.55 ± 0.02 | 0.47 ± 0.11 | 0.61 ± 0.19 | 0.56 ± 0.04 | 0.60 ± 0.15 |
| Cytochrome P450 3A2 (pmol/min) | 113.56 ± 14.98 | 134.30 ± 15.86 | 108.98 ± 16.72 | 141.35 ± 32.38 | 122.49 ± 10.16 |
| Heme oxygenase (nmol/min) | 9.74 ± 0.55 | 10.42 ± 2.83 | 10.08 ± 1.03 | 9.42 ± 1.82 | 9.84 ± 1.78 |
| NADPH quinone reductase (×10−3 Unit) | 1.37 ± 0.22 | 1.65 ± 0.12 * | 1.71 ± 0.13 | 1.69 ± 0.13 | 1.46 ± 0.26 |
| NADPH-Cytochrome P450 reductase (×10−3 Unit) | 2.42 ± 0.13 | 2.58 ± 0.34 | 2.26 ± 0.41 | 2.29 ± 0.30 | 2.52 ± 0.35 |
| Glutathione-S-transferase (×10−2 Unit) | 34.17 ± 4.28 | 47.24 ± 2.93 * | 52.04 ± 1.66 ** | 59.84 ± 3.19 ** | 47.23 ± 4.58 |
| UDP-glucuronyltransferase (×10−2 Unit) | 34.40 ± 2.80 | 32.50 ± 5.10 | 31.10 ± 3.0 | 33.10 ± 2.50 | 36.20 ± 3.20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kittichaiworakul, R.; Taya, S.; Chariyakornkul, A.; Chaiyaso, T.; Wongpoomchai, R. Antigenotoxic Effects and Possible Mechanism of Red Yeast (Sporidiobolus pararoseus) on Aflatoxin B1-Induced Mutagenesis. Biomolecules 2021, 11, 734. https://doi.org/10.3390/biom11050734
Kittichaiworakul R, Taya S, Chariyakornkul A, Chaiyaso T, Wongpoomchai R. Antigenotoxic Effects and Possible Mechanism of Red Yeast (Sporidiobolus pararoseus) on Aflatoxin B1-Induced Mutagenesis. Biomolecules. 2021; 11(5):734. https://doi.org/10.3390/biom11050734
Chicago/Turabian StyleKittichaiworakul, Romteera, Sirinya Taya, Arpamas Chariyakornkul, Thanongsak Chaiyaso, and Rawiwan Wongpoomchai. 2021. "Antigenotoxic Effects and Possible Mechanism of Red Yeast (Sporidiobolus pararoseus) on Aflatoxin B1-Induced Mutagenesis" Biomolecules 11, no. 5: 734. https://doi.org/10.3390/biom11050734
APA StyleKittichaiworakul, R., Taya, S., Chariyakornkul, A., Chaiyaso, T., & Wongpoomchai, R. (2021). Antigenotoxic Effects and Possible Mechanism of Red Yeast (Sporidiobolus pararoseus) on Aflatoxin B1-Induced Mutagenesis. Biomolecules, 11(5), 734. https://doi.org/10.3390/biom11050734








