Five Tetramic Acid Derivatives Isolated from the Iranian Fungus Colpoma quercinum CCTU A372
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Experiment
2.2. Fungal Material and Morphological Analysis
2.3. Molecular Analysis, Sequencing, and Phylogenetic Analysis
2.4. Scale-up Production, Extraction, and Isolation
2.5. Antimicrobial Activity Assay
2.6. Cytotoxicity Assay
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hyde, K.D.; Xu, J.; Rapior, S.; Jeewon, R.; Lumyong, S.; Niego, A.G.T.; Abeywickrama, P.D.; Aluthmuhandiram, J.V.S.; Brahamanage, R.S.; Brooks, S.; et al. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef] [Green Version]
- Bills, G.F.; Gloer, J.B. Biologically active secondary metabolites from the fungi. Microbiol. Spectr. 2016, 4, 6. [Google Scholar] [CrossRef]
- Karwehl, S.; Stadler, M. Exploitation of fungal biodiversity for discovery of novel antibiotics. Curr. Top. Microbiol. Immunol. 2016, 398, 303–338. [Google Scholar]
- Narmani, A.; Teponno, R.B.; Helaly, S.E.; Arzanlou, M.; Stadler, M. Cytotoxic, anti-biofilm and antimicrobial polyketides from the plant associated fungus Chaetosphaeronema achilleae. Fitoterapia 2019, 139, 104390. [Google Scholar] [CrossRef] [PubMed]
- Macabeo, A.P.G.; Cruz, A.J.C.; Narmani, A.; Arzanlou, M.; Babai-Ahari, A.; Pilapil, L.A.E.; Garcia, K.Y.M.; Huch, V.; Stadler, M. Tetrasubstituted α-pyrone derivatives from the endophytic fungus, Neurospora udagawae. Phytochem. Lett. 2020, 35, 147–151. [Google Scholar] [CrossRef]
- Lambert, C.; Pourmoghaddam, M.J.; Cedeño-Sanchez, M.; Surup, F.; Khodaparast, S.A.; Krisai-Greilhuber, I.; Voglmayr, H.; Stradal, T.E.B.; Stadler, M. Resolution of the Hypoxylon fuscum complex (Hypoxylaceae, Xylariales) and discovery and biological characterization of two of its prominent secondary metabolites. J. Fungi 2021, 7, 131. [Google Scholar] [CrossRef]
- Shao, L.; Marin-Felix, Y.; Surup, F.; Stchigel, A.M.; Stadler, M. Seven new cytotoxic and Antimicrobial Xanthoquinodins from Jugulospora vestita. J. Fungi 2020, 6, 188. [Google Scholar] [CrossRef]
- Mapook, A.; Macabeo, A.P.G.; Thongbai, B.; Hyde, K.D.; Stadler, M. Polyketide-derived secondary metabolites from a Dothideomycetes fungus, Pseudopalawania siamensis gen. et sp. nov., (Muyocopronales) with antimicrobial and cytotoxic activities. Biomolecules 2020, 10, 569. [Google Scholar] [CrossRef] [Green Version]
- Noumeur, S.R.; Teponno, R.B.; Helaly, S.E.; Wang, X.-W.; Harzallah, D.; Houbraken, J.; Crous, P.W.; Stadler, M. Diketopiperazines from Batnamyces globulariicola, gen. & sp. nov. (Chaetomiaceae), a fungus associated with roots of the medicinal plant Globularia alypum in Algeria. Mycol. Prog. 2020, 19, 589–603. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Dai, D.Q.; Tang, L.Z.; Aptroot, A.; Castañeda-Ruiz, R.F.; Druzhinina, I.S.; Cai, F.; Ekanayaka, A.H.; Erdoğdu, M.; et al. A dynamic portal for a community-driven, continuously updated classification of fungi and fungus-like organisms: Outlineoffungi.org. Mycosphere 2020, 11, 1514–1526. [Google Scholar] [CrossRef]
- Narmani, A.; Teponno, R.B.; Arzanlou, M.; Surup, F.; Helaly, S.E.; Wittstein, K.; Praditya, D.F.; Babai-Ahari, A.; Steinmann, E.; Stadler, M. Cytotoxic, antimicrobial and antiviral secondary metabolites produced by the plant pathogenic fungus Cytospora sp. CCTU A309. Fitoterapia 2019, 134, 314–322. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylo-genetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. ISBN 978-0-12-372180-8. [Google Scholar]
- Narmani, A.; Pichai, S.; Palani, P.; Arzanlou, M.; Surup, F.; Stadler, M. Daldinia sacchari (Hypoxylaceae) from India produces the new cytochalasins Saccalasins A and B and belongs to the D. eschscholtzii species complex. Mycol. Prog. 2019, 18, 175–185. [Google Scholar] [CrossRef]
- Becker, K.; Wessel, A.-C.; Luangsa-Ard, J.J.; Stadler, M. Viridistratins A−C, antimicrobial and cytotoxic benzo[j]fluoranthenes from stromata of Annulohypoxylon viridistratum (Hypoxylaceae, Ascomycota). Biomolecules 2020, 10, 805. [Google Scholar] [CrossRef]
- Brel, O.; Touré, S.; Levasseur, M.; Lechat, C.; Pellissier, L.; Wolfender, J.-L.; Van-Elslande, E.; Litaudon, M.; Dusfour, I.; Stien, D.; et al. Paecilosetin derivatives as potent qntimicrobial qgents from Isaria farinosa. J. Nat. Prod. 2020, 83, 2915–2922. [Google Scholar] [CrossRef] [PubMed]
- Halecker, S.; Surup, F.; Kuhnert, E.; Mohr, K.I.; Brock, N.L.; Dickschat, J.S.; Junker, C.; Schulz, B.; Stadler, M. Hymenosetin, a 3-decalinoyltetramic acid antibiotic from cultures of the ash dieback pathogen, Hymenoscyphus pseudoalbidus. Phytochemistry 2014, 100, 86–91. [Google Scholar] [CrossRef]
- Matsumori, N.; Kaneno, D.; Murata, M.; Nakamura, H.; Tachibana, K. Stereochemical determination of acyclic structures based on carbon-proton spin-coupling constants. A method of configuration analysis for natural products. J. Org. Chem. 1999, 64, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Li, Q.; Ju, J. Naturally occurring tetramic acid products: Isolation, structure elucidation and biological activity. RSC Adv. 2014, 4, 50566–50593. [Google Scholar] [CrossRef]
- Vesonder, R.F.; Tjarks, L.W.; Rohwedder, W.K.; Burmeister, H.R.; Laugal, J.A. Equisetin, an antibiotic from Fusarium equiseti NRRL 5537, identified as a derivative of N-methyl-2,4-pyrollidone. J. Antibiot. 1979, 32, 759–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellwig, V.; Grothe, T.; Mayer-Bartschmid, A.; Endermann, R.; Geschke, F.U.; Henkel, T.; Stadler, M. Altersetin, a new anti-biotic from cultures of endophytic Alternaria spp. taxonomy, fermentation, isolation, structure elucidation and biological activities. J. Antibiot. (Tokyo) 2002, 55, 881–892. [Google Scholar] [CrossRef] [Green Version]
- Putri, S.P.; Kinoshita, H.; Ihara, F.; Igarashi, Y.; Nihira, T. Ophiosetin, a new tetramic acid derivative from the mycopathogenic fungus Elaphocordyceps ophioglossoides. J. Antibiot. 2010, 63, 195–198. [Google Scholar] [CrossRef] [Green Version]
- Anoumedem, E.G.M.; Mountessou, B.Y.G.; Kouam, S.F.; Narmani, A.; Surup, F. Simplicilones A and B isolated from the endophytic fungus Simplicillium subtropicum SPC3. Antibiotics 2020, 9, 753. [Google Scholar] [CrossRef]
- Sato, M.; Dander, J.E.; Sato, C.; Hung, Y.S.; Gao, S.S.; Tang, M.C.; Hang, L.; Winter, J.M.; Garg, N.K.; Watanabe, K.; et al. Col-laborative biosynthesis of maleimide- and succinimide-containing natural products by fungal polyketide megasynthases. J. Am. Chem. Soc. 2017, 139, 5317–5320. [Google Scholar] [CrossRef] [Green Version]
- Schobert, R.; Schlenk, A. Tetramic and tetronic acids: An update on new derivatives and biological aspects. Bioorganic Med. Chem. 2008, 16, 4203–4221. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, P.; Tang, M.C.; Zou, Y.; Gao, S.S.; Hung, Y.S.; Zhao, M.; Watanabe, K.; Houk, K.N.; Tang, Y. Biochemical character-ization of a eukaryotic decalin-forming Diels-Alderase. J. Am. Chem. Soc. 2016, 138, 15837–15840. [Google Scholar] [CrossRef] [Green Version]
- Ondeyka, J.G.; Smith, S.K.; Zink, D.L.; Vicente, F.; Basilio, A.; Bills, G.F.; Polishook, J.D.; Garlisi, C.; McGuinness, D.; Smith, E.; et al. Isolation, structure elucidation and antibacterial activity of a new tetramic acid, ascosetin. J. Antibiot. 2014, 67, 527–531. [Google Scholar] [CrossRef]
- Wu, B.; Wiese, J.; Labes, A.; Kramer, A.; Schmaljohann, R.; Imhoff, J.F. Lindgomycin, an unusual antibiotic polyketide from a marine fungus of the Lindgomycetaceae. Mar. Drugs 2015, 13, 4617–4632. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Tamayo-Castillo, G.; Pang, C.; Clardy, J.; Cao, S.; Kim, K.H. Diketopiperazines from Costa Rican endolichenic fungus Colpoma sp. CR1465A. Bioorganic Med. Chem. Lett. 2016, 26, 2438–2441. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Chen, S.; Li, J.; Liu, L. The biological and chemical diversity of tetramic acid compounds from marine-derived microorganisms. Mar. Drugs 2020, 18, 114. [Google Scholar] [CrossRef] [Green Version]
- Larson, E.C.; Lim, A.L.; Pond, C.D.; Craft, M.; Čavužić, M.; Waldrop, G.L.; Schmidt, E.W.; Barrows, L.R. Pyrrolocin C and equisetin inhibit bacterial acetyl-CoA carboxylase. PLoS ONE 2020, 15, e0233485. [Google Scholar] [CrossRef] [PubMed]
- Graupner, P.R.; Carr, A.; Clancy, E.; Gilbert, J.; Bailey, K.L.; Derby, J.A.; Gerwick, B.C. The macrocidins: Novel cyclic tetramic acids with herbicidal activity produced by Phoma macrostoma. J. Nat. Prod. 2003, 66, 1558–1561. [Google Scholar] [CrossRef] [PubMed]
- Matio Kemkuignou, B.; Treiber, L.; Zeng, H.; Schrey, H.; Schobert, R.; Stadler, M. Macrooxazoles A–D, new 2,5-disubstituted oxazole-4-carboxylic acid derivatives from the plant pathogenic fungus Phoma macrostoma. Molecules 2020, 25, 5497. [Google Scholar] [CrossRef] [PubMed]
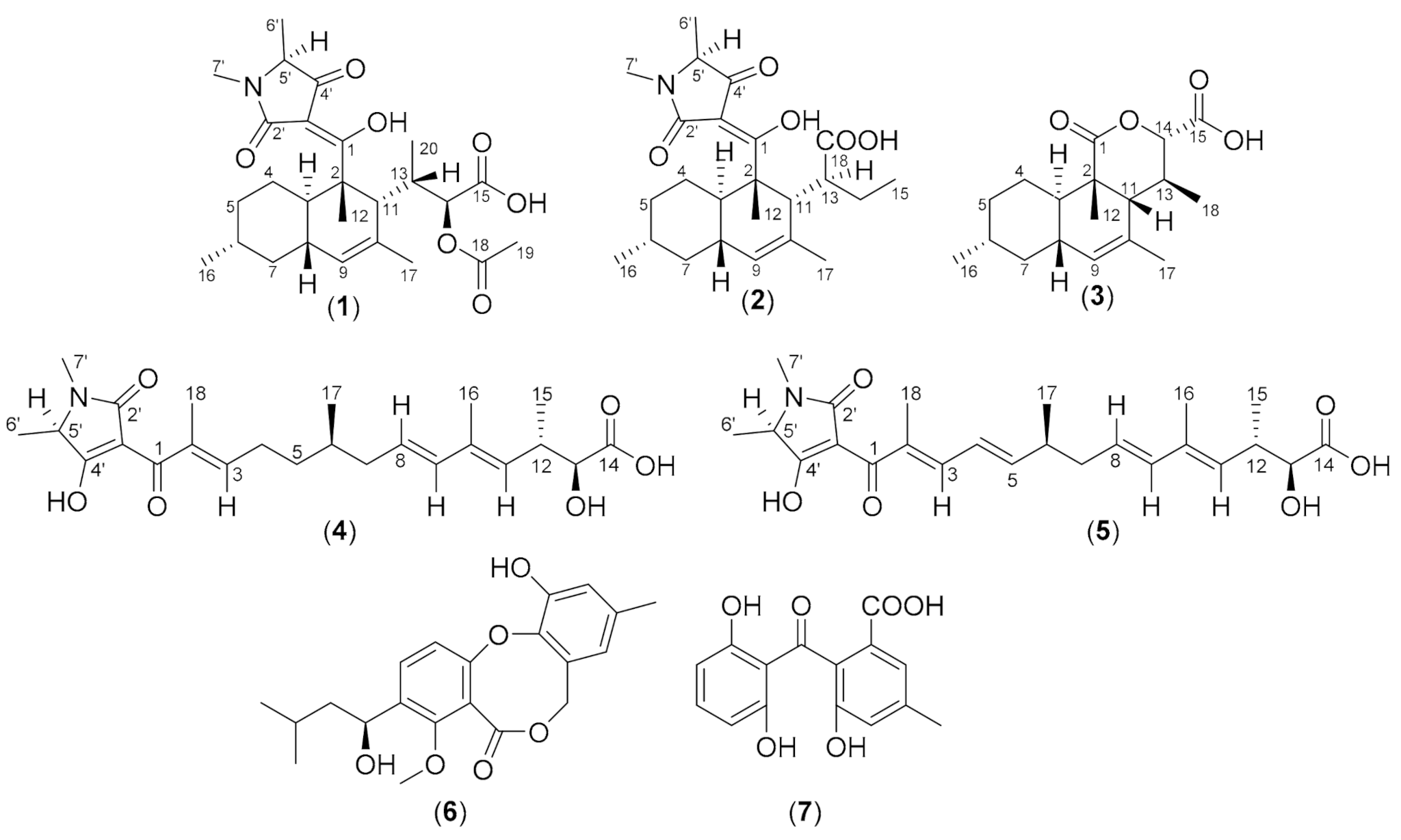
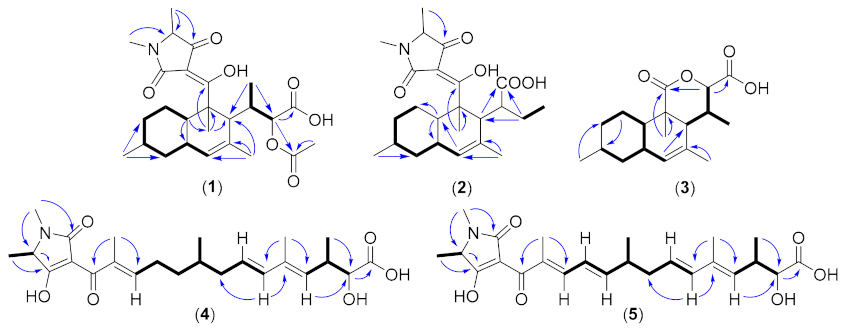

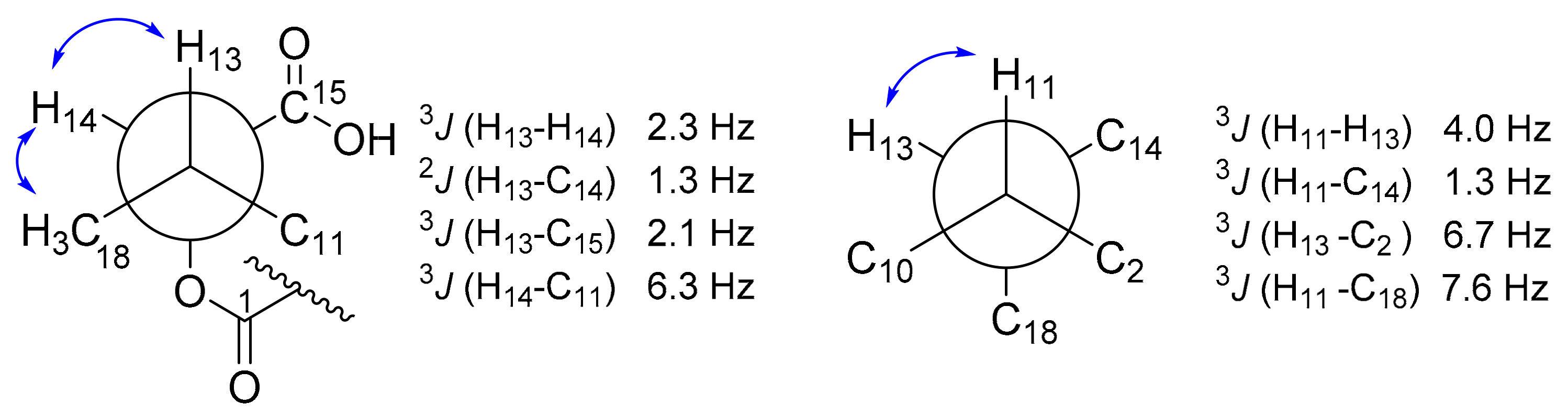

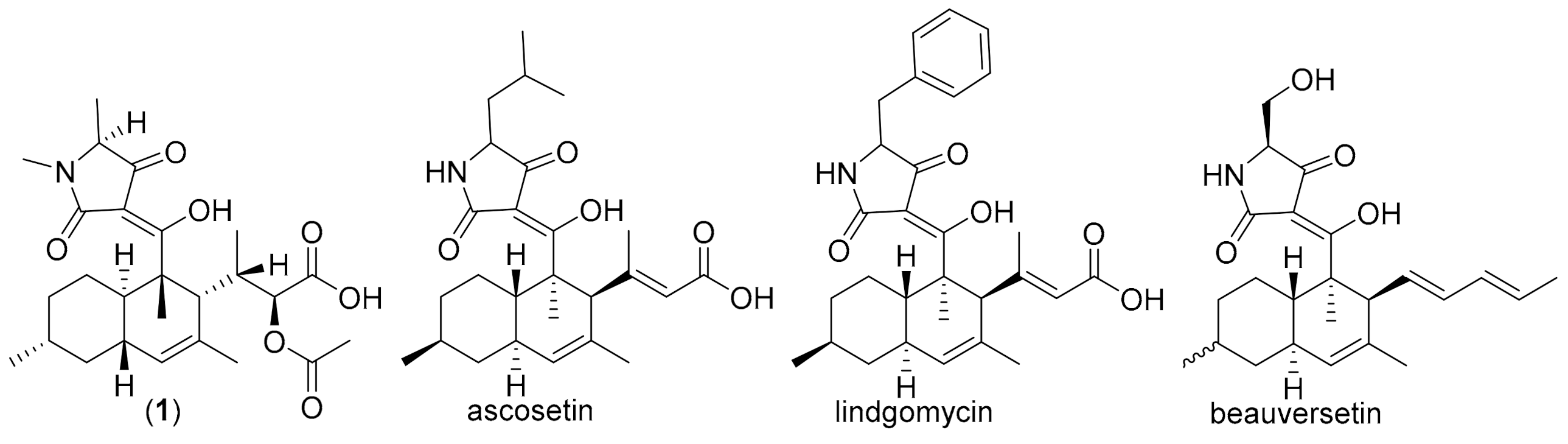
| Pos. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δH, m (J in Hz) | δC, Type | δH, m (J in Hz) | δC, Type | δH, m (J in Hz) | δC, Type | |
| 1 | - | 200.7, C | - | 200.1, C | - | 177.3, C |
| 2 | - | 51.7, C | - | 49.8, C | - | 46.5, C |
| 3 | 1.91, m | 41.3, CH | 2.17, m | 40.1, CH | 1.35, m | 44.2, CH |
| 4 | 1.96; 0.98, m | 30.0, CH2 | 2.11; 1.02, m | 30.0, CH2 | 1.68; 1.21, m | 28.5, CH2 |
| 5 | 1.80; 1.08, m | 37.6, CH2 | 1.15, m | 37.3, CH2 | 1.79; 0.86, m | 37.2, CH2 |
| 6 | 1.54, m | 35.1, CH | 1.52, m | 35.3, CH | 1.45, m | 34.0, CH |
| 7 | 1.83; 0.81, m | 43.4, CH2 | 1.72; 0.96, m | 42.6, CH2 | 1.83; 0.74, m | 43.9, CH2 |
| 8 | 1.85, m | 41.5, CH | 1.80, m | 42.4, CH | 1.86, m | 38.8, CH |
| 9 | 5.37, s | 130.0, CH | 5.20, s | 128.8, CH | 5.39, s | 130.5, CH |
| 10 | - | 132.7, C | - | 132.8, C | - | 131.5, C |
| 11 | 3.02, s (br) | 49.8, CH | 2.24, s (br) | 53.0, CH | 2.18, d (4.04) | 48.4, CH |
| 12 | 1.41, s | 16.0, CH3 | 1.38, s | 14.7, CH3 | 1.11, s | 19.5, CH3 |
| 13 | 2.02, t (7.6) | 37.8, CH | 2.92, m | 50.2, CH | 2.53, m | 32.2, CH |
| 14 | 4.65, d (8.6) | 77.1, CH | 1.86, 1.17, m | 29.5, CH2 | 4.89, d (1.3) | 83.0, CH |
| 15 | - | 174.3, C | 0.72, t (7.2) | 13.7, CH3 | - | 174.4, C |
| 16 | 0.93, d (7.0) | 23.1, CH3 | 0.92, d (7.0) | 23.1, CH3 | 0.91, d (7.0) | 22.8, CH3 |
| 17 | 1.78, s | 24.4, CH3 | 1.77, s | 15.5, CH3 | 1.66, s | 21.8, CH3 |
| 18 | - | 172.5, C | - | 177.5, C | 1.18, d (7.0) | 15.5, CH3 |
| 19 | 2.08, s | 21.3, CH3 | - | - | - | - |
| 20 | 1.01, d (7.7) | 17.4, CH3 | - | - | - | - |
| 1′ | - | - | - | - | - | - |
| 2′ | - | n.d | - | n.d | - | - |
| 3′ | - | n.d | - | n.d | - | - |
| 4′ | - | 194.9, C | - | 194.5, C | - | - |
| 5′ | 3.73, s (br) | 62.8, CH | 3.74, m | 62.9, CH | - | - |
| 6′ | 1.33, d (7.0) | 15.4, CH3 | 1.34, br | 15.5, CH3 | - | - |
| 7′ | 2.99, s | 27.2, CH3 | 3.01, s | 27.1, CH3 | - | - |
| Pos. | 4 | 5 | ||
|---|---|---|---|---|
| δH, m (J in Hz) | δC, Type | δH, m (J in Hz) | δC, Type | |
| 1 | - | 186.0, C | - | 184.4, C |
| 2 | - | 130.4. C | - | 128.6, C |
| 3 | 6.98, t (7.2) | 146.9, CH | 7.64, d (9.2) | 143.7, CH |
| 4 | 2.32, m | 27.7, CH2 | 6.53, dd, (15.0, 11.0) | 125.9, CH |
| 5 | 1.54; 1.40, m | 36.3, CH2 | 6.15, dd (15.0, 7.7) | 151.9, CH |
| 6 | 1.63, m | 34.4, CH | 2.45, dt (14.0, 7.0 (2x)) | 39.5, CH |
| 7 | 2.15; 1.99, m | 41.4, CH2 | 2.15; 2.33, m | 41.2, CH2 |
| 8 | 5.60, dt (15.0, 10.0) | 127.6, C | 5.58, m | 126.8, CH |
| 9 | 6.05, d (15.0) | 137.8, CH | 6.07, d (15.0) | 138.2, CH |
| 10 | - | 135.6, C | - | 135.6, C |
| 11 | 5.37, d (10.0) | 131.2, CH | 5.39, d (10.0) | 131.7, CH |
| 12 | 3.05, ddd (10.0, 7.0, 4.0) | 37.9, CH | 3.03, m | 37.9, CH |
| 13 | 4.06, d (4.0) | 75.7, CH | 4.06, d (4.0) | 75.7, CH |
| 14 | - | 177.1, C | - | 177.1, C |
| 15 | 1.09, d (7.0) | 18.1, CH3 | 1.09, d (7.0)ov | 18.1, CH3 |
| 16 | 1.74, d (1.1) | 13.1, CH3 | 1.74, d (1.0) | 13.1, CH3 |
| 17 | 0.93, d (7.0) | 20.0, CH3 | 1.08, d (7.0)ov | 20.1, CH3 |
| 18 | 1.90, s | 12.6, CH3 | 1.99, s | 12.8, CH3 |
| 1′ | - | - | - | - |
| 2′ | - | 174.9, C | - | 175.2, C |
| 3′ | - | n.d | - | n.d |
| 4′ | - | 195.8, C | - | 196.1, C |
| 5′ | 3.80, q (6.9) | 63,1, CH | 3.80, q (6.9) | 63.3, CH |
| 6′ | 2.97, s | 27.0, CH3 | 2.98, s | 27.0, CH3 |
| 7′ | 1.34, d (7.0) | 15.5, CH3 | 1.35, d (7.0) | 15.5, CH3 |
| Microorganism | 1 | 2 | 3 | 4 | 5 | Ref |
|---|---|---|---|---|---|---|
| MIC (µg/mL) | (µg/mL) | |||||
| Bacillus subtilis DSM 10 | n.i | 67 | n.i | n.i | n.i | 8.3 a |
| Staphylococcus aureus DSM 346 | n.i | n.1 | n.i | n.i | n.i | 1.7 a |
| Mucor hiemalis DSM 2656 | n.i | 67 | n.i | n.i | n.i | 4.2 b |
| Pichia anomala DSM 6766 | n.i | n.i | n.i | n.i | n.i | 4.2 b |
| Rhodotorula glutinis DSM 10134 | n.i | n.i | n.i | n.i | n.i | 1.0 b |
| Acinetobacter baumanii DSM 30008 | n.i | n.i | n.i | n.i | n.i | 0.26 c |
| Escherichia coli | n.i | n.i | n.i | n.i | n.i | 1.7 d |
| Cell lines | IC50 (µM) | Ref e (µM) | ||||
| mouse fibroblast L-929 | n.d | 57.5 | n.d | n.d | n.d | 8.3 × 10−4 |
| endocervical adenocarcinoma KB-3.1 | n.d | 5.7 | n.d | n.d | n.d | 5.3 × 10−5 |
| human breast adenocarcinoma MCF-7 | n.t | 7.9 | n.t | n.t | n.t | 7.1 × 10−5 |
| human lung carcinoma A-549 | n.t | 16.5 | n.t | n.t | n.t | 5.5 × 10−5 |
| human prostate cancer PC-3 | n.t | 17.9 | n.t | n.t | n.t | 2.7 × 10−4 |
| ovarian carcinoma SK-OV-3 | n.t | 8.1 | n.t | n.t | n.t | 2.4 × 10−4 |
| squamous cell carcinoma A-431 | n.t | 6.2 | n.t | n.t | n.t | 8.3 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Primahana, G.; Narmani, A.; Surup, F.; Teponno, R.B.; Arzanlou, M.; Stadler, M. Five Tetramic Acid Derivatives Isolated from the Iranian Fungus Colpoma quercinum CCTU A372. Biomolecules 2021, 11, 783. https://doi.org/10.3390/biom11060783
Primahana G, Narmani A, Surup F, Teponno RB, Arzanlou M, Stadler M. Five Tetramic Acid Derivatives Isolated from the Iranian Fungus Colpoma quercinum CCTU A372. Biomolecules. 2021; 11(6):783. https://doi.org/10.3390/biom11060783
Chicago/Turabian StylePrimahana, Gian, Abolfazl Narmani, Frank Surup, Rémy Bertrand Teponno, Mahdi Arzanlou, and Marc Stadler. 2021. "Five Tetramic Acid Derivatives Isolated from the Iranian Fungus Colpoma quercinum CCTU A372" Biomolecules 11, no. 6: 783. https://doi.org/10.3390/biom11060783







