Translational Applications of Artificial Intelligence and Machine Learning for Diagnostic Pathology in Lymphoid Neoplasms: A Comprehensive and Evolutive Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample
2.2. Performance Analysis
2.3. Science Mapping Analysis (SMA)
2.3.1. SMA for Cognitive Framework
- (a)
- Motor themes (MT): Present a high density and a strong centrality signifying the most developed themes for the research area studied (upper-right quadrant);
- (b)
- Basic and transversal themes (BT): Represent themes shared for several disciplines; thus, their foundations are well-established (lower-right quadrant);
- (c)
- Emerging or declining themes (ED): Have a weak density and a low centrality and, thus, represent marginal areas of knowledge (lower-left quadrant);
- (d)
- Highly developed or isolated themes (HDI): Show a high density, meaning a significant internal development. However, they are less connected with other themes in the research field because of their low centrality values (upper-left quadrant).
2.3.2. SMA for Social Framework
3. Results
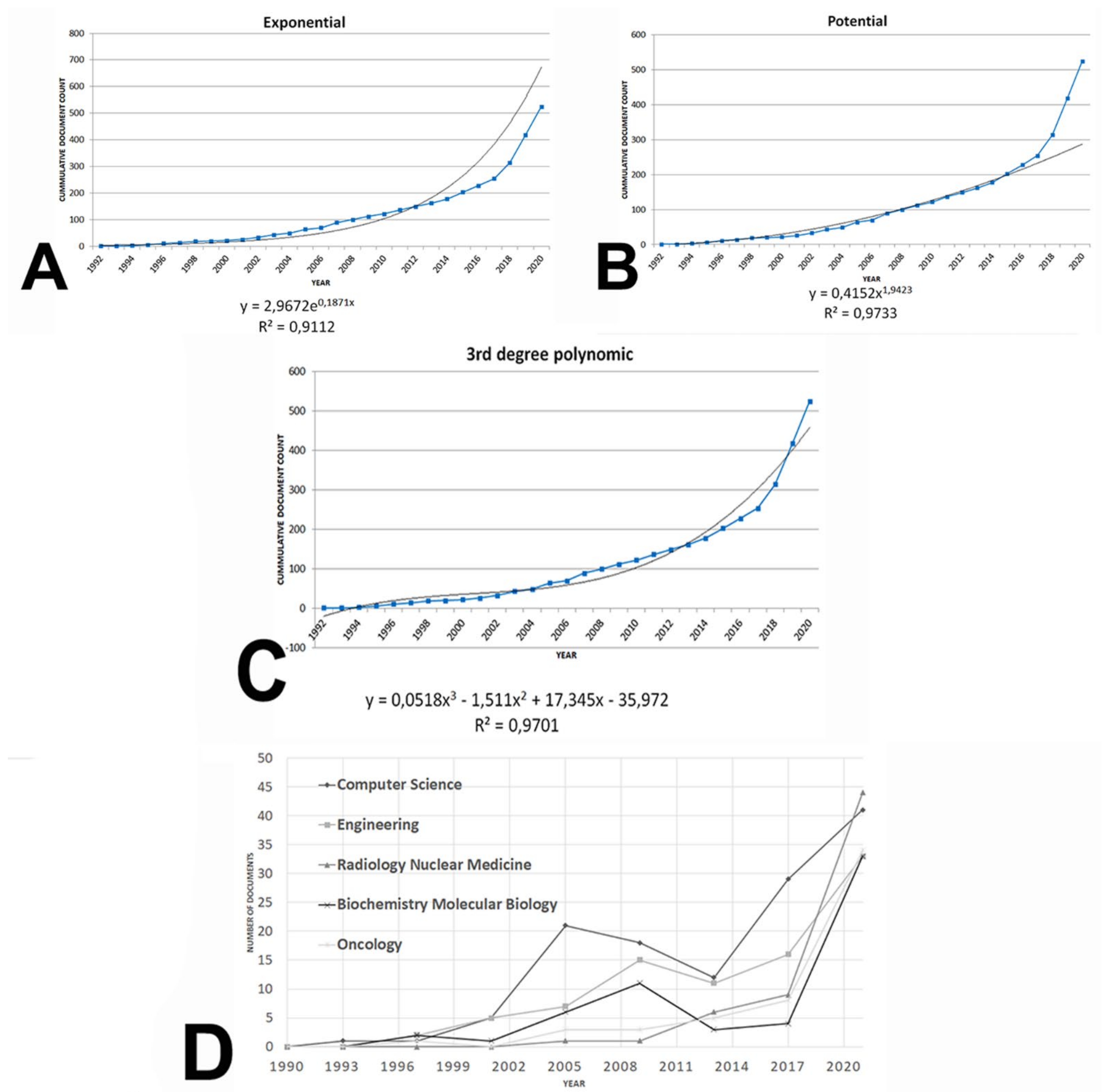
3.1. Performance Analysis
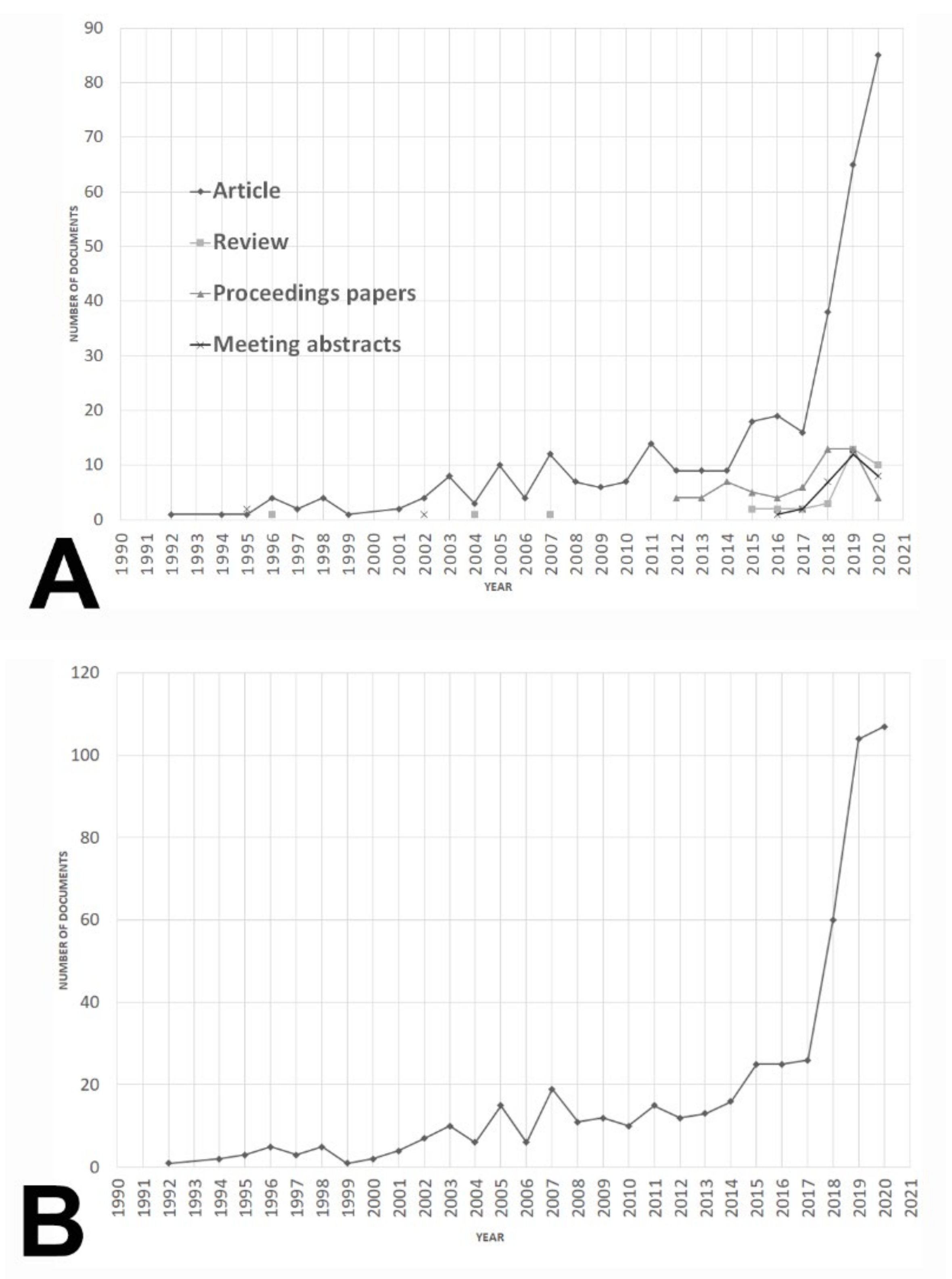
3.1.1. Document Type
3.1.2. Research Areas
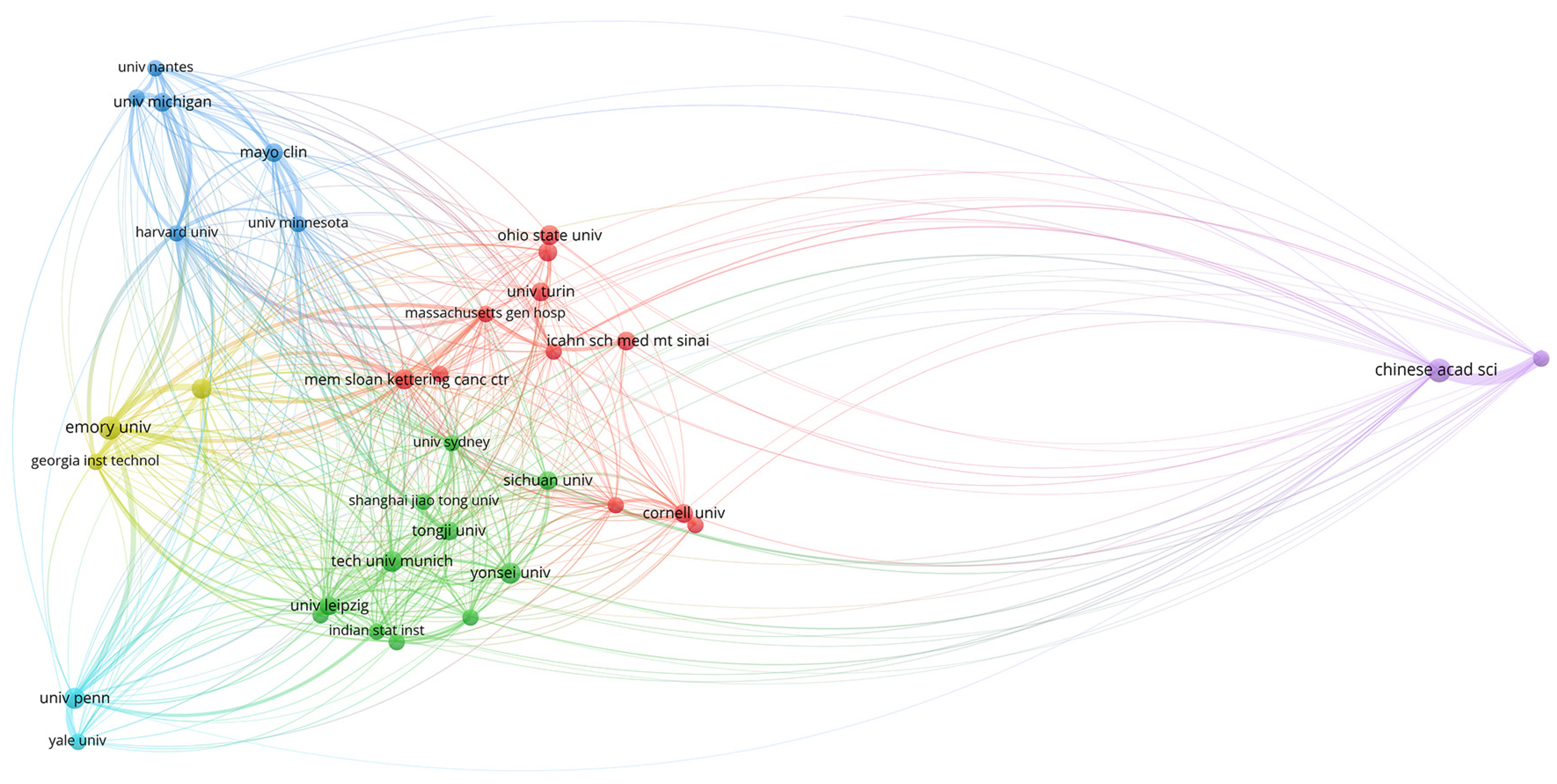
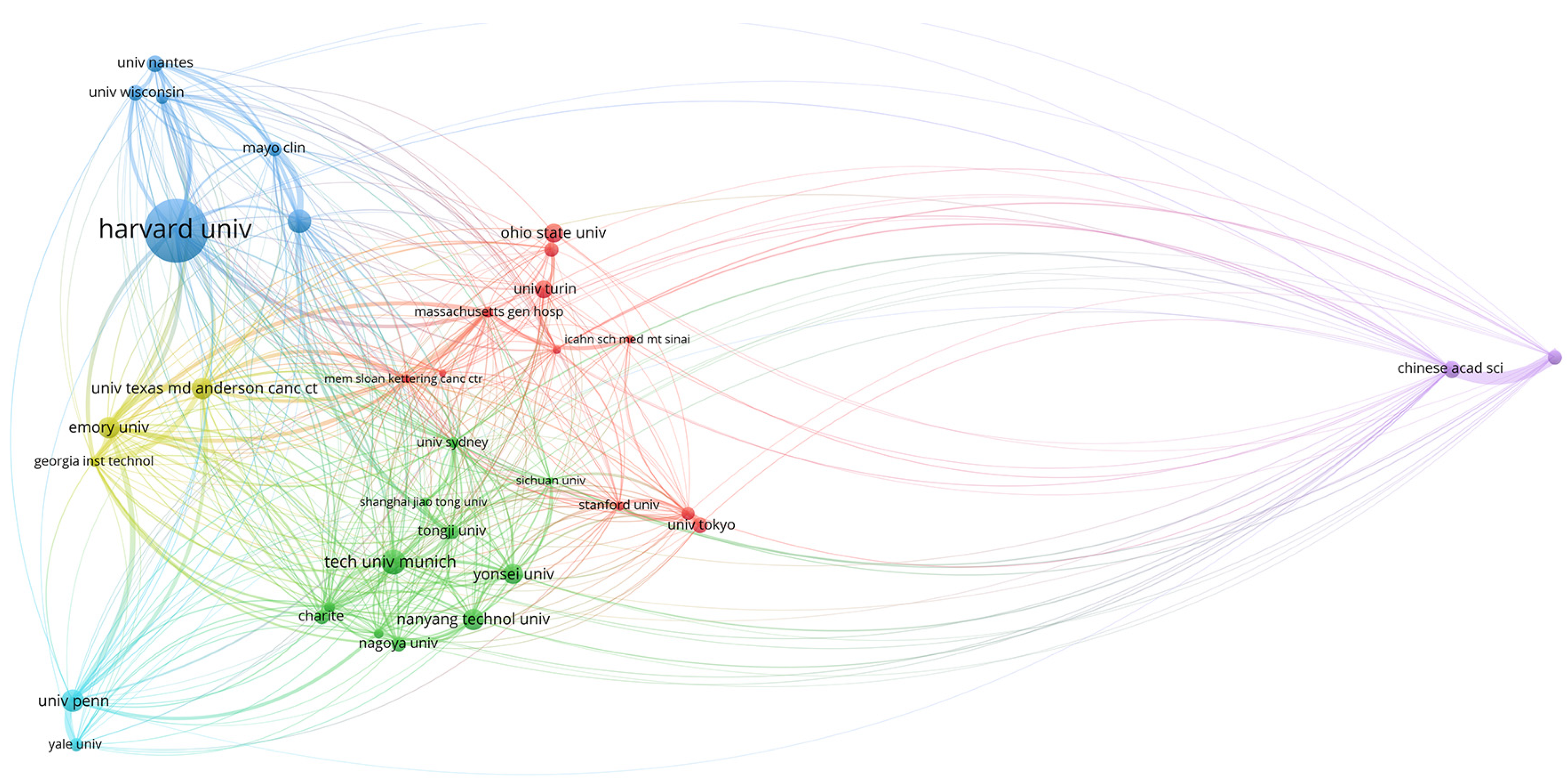
3.1.3. Organizations and Research Centers
3.1.4. Source Titles
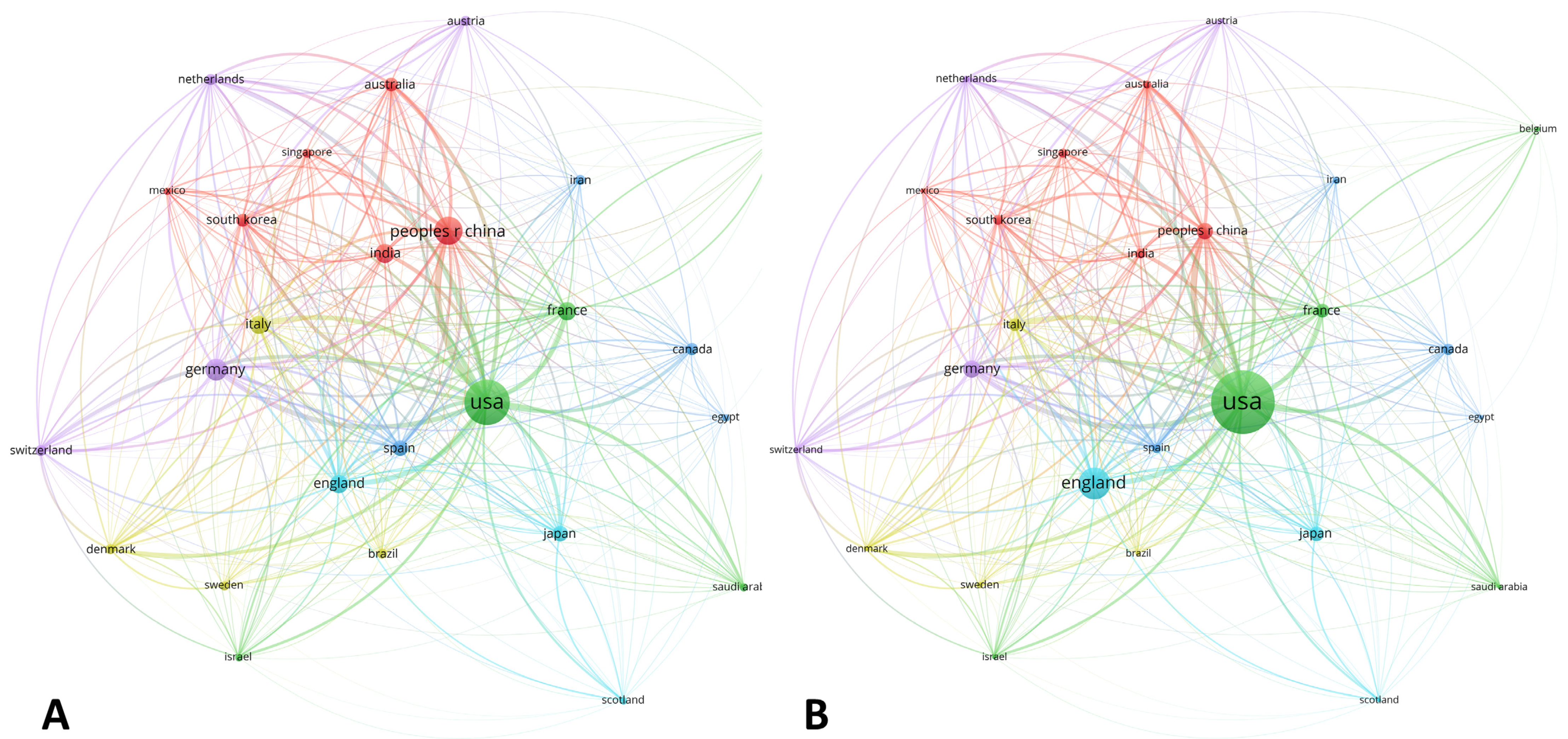
3.1.5. Country Distribution
3.2. SMA
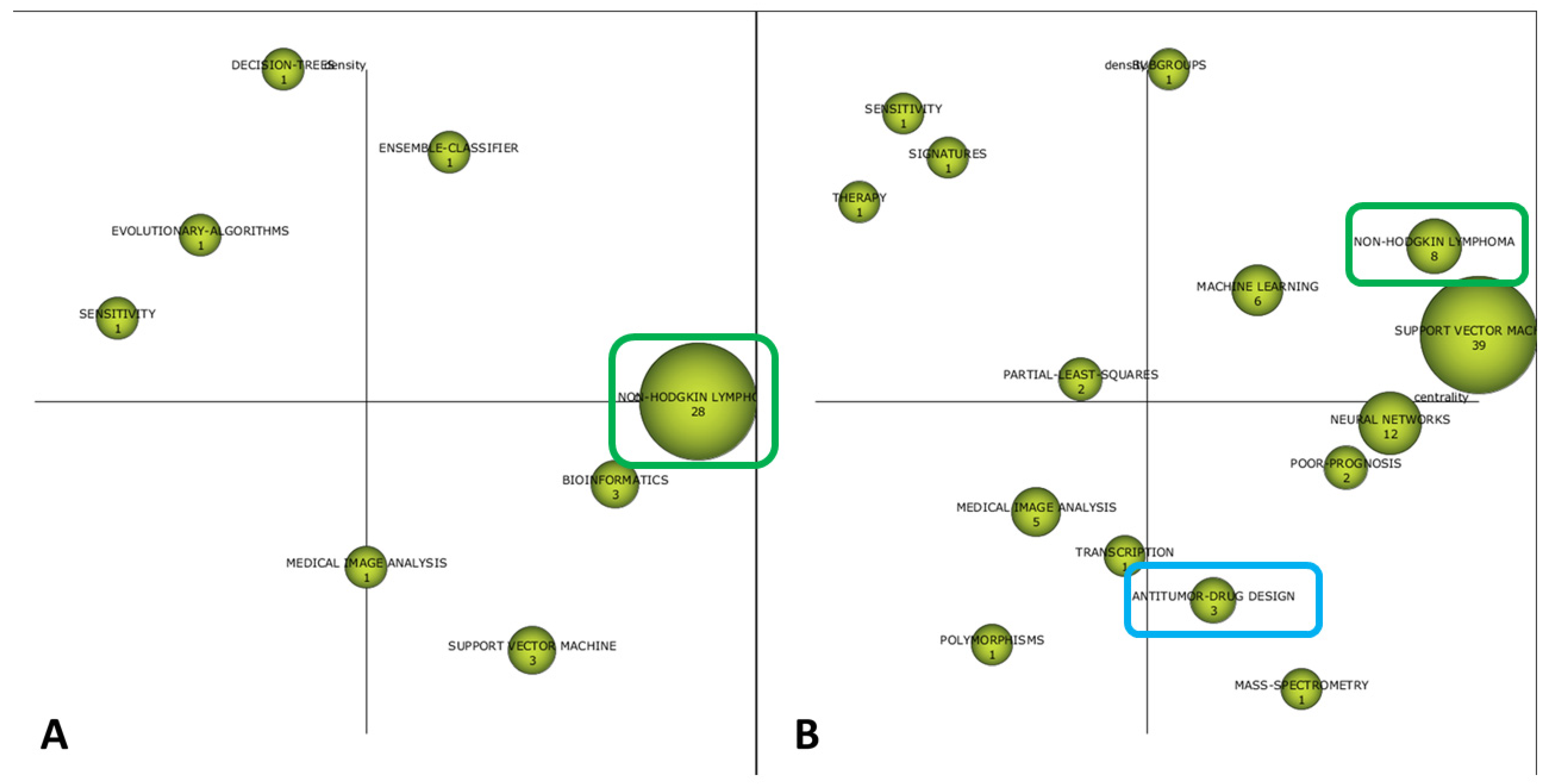
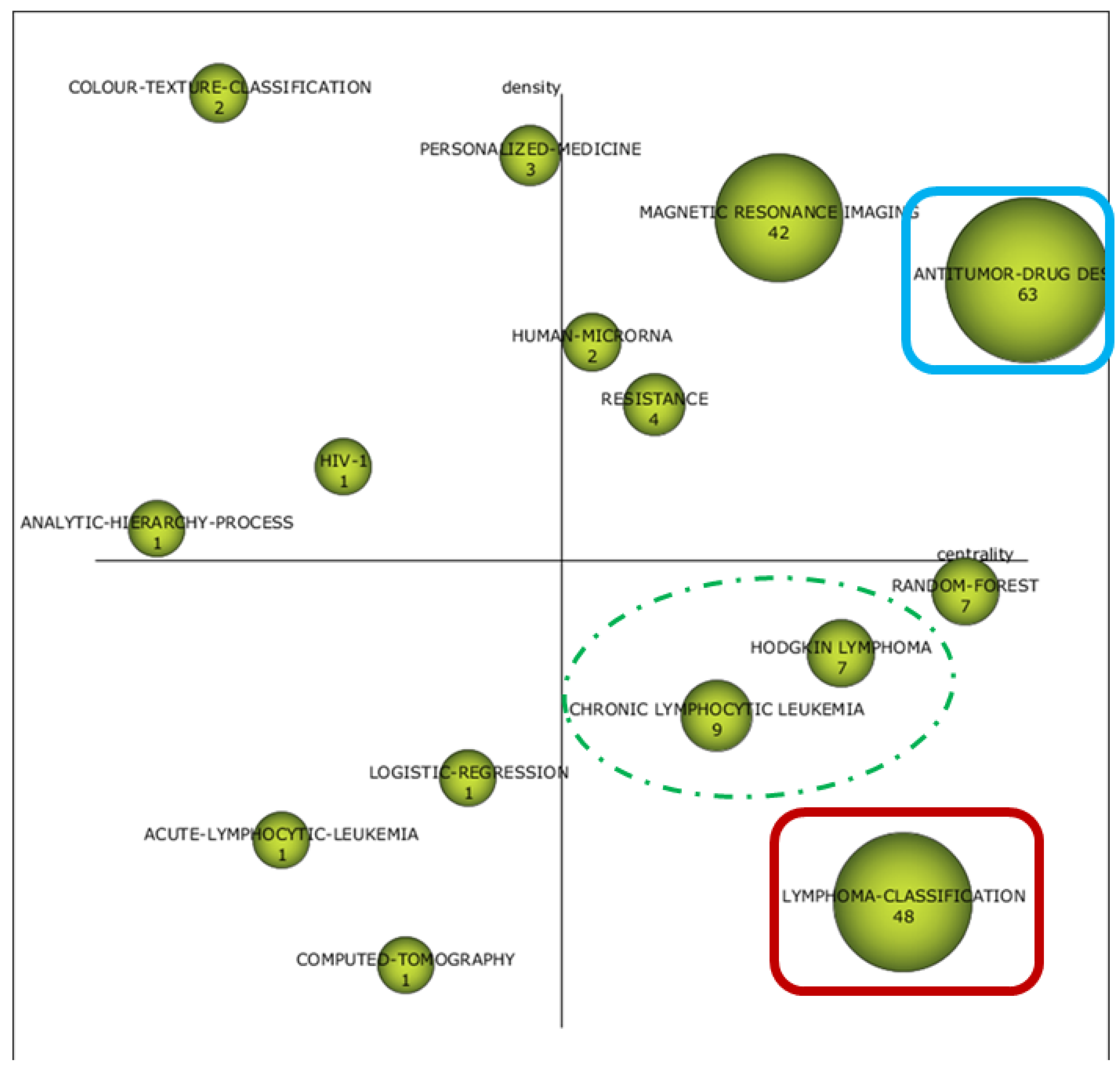
3.2.1. SMA for Cognitive Framework
3.2.2. SMA for Social Framework
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Flores, M.; Glusman, G.; Brogaard, K.; Price, N.D.; Hood, L. P4 medicine: How systems medicine will transform the healthcare sector and society. Pers. Med. 2013, 10, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Beam, A.L.; Kohane, I.S. Big Data and Machine Learning in Health Care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef] [PubMed]
- Die Bundesregierung. Eckpunkte der Bundesregierung für eine Strategie Künstliche Intelligenz; Die Bundesregierung: Berlin, Germany, 2018. [Google Scholar]
- Villani, C. For a Meaningful Artificial Intelligence towards a French and European Strategy; Conseil National du Numérique: Paris, France, 2018. [Google Scholar]
- Chinese State Council. Notice of the State Council on Issuing the Development Plan for the New Generation of Artificial Intelligence; Chinese State Council: Beijing, China, 2017. [Google Scholar]
- European Commission. Coordinated Plan on Artificial Intelligence; European Commission: Brussels, Belgium, 2018. [Google Scholar]
- Executive Office of the President of the United States. Artificial Intelligence, Automation, and the Economy; Executive Office of the President of the United States: Washington, DC, USA, 2016. [Google Scholar]
- Executive Office of the President of the United States. The National Artificial Intelligence Research And Development Strategic Plan; Executive Office of the President of the United States: Washington, DC, USA, 2016. [Google Scholar]
- Executive Office of the President of the United States. Preparing For The Future Of Artificial Intelligence; Executive Office of the President of the United States: Washington, DC, USA, 2016. [Google Scholar]
- Clark, G.; Hancock, M.; Hall, D.W.; Pesenti, J. AI Sector Deal; Department for Digital, Culture, Media & Sport: London, UK; Department for Business, Energy & Industrial Strategy: London, UK, 2019. [Google Scholar]
- Hall, D.W.; Pesenti, J. Growing the Artificial Intelligence Industry in the UK; Department for Digital, Culture, Media & Sport: London, UK; Department for Business, Energy & Industrial Strategy: London, UK, 2017. [Google Scholar]
- Gonon-Demoulian, R.; Goldman, J.M.; Nicolini, F.E. History of chronic myeloid leukemia: A paradigm in the treatment of cancer. Bull. Cancer 2014, 101, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Thomas, X. Acute Promyelocytic Leukemia: A History over 60 Years-From the Most Malignant to the most Curable Form of Acute Leukemia. Oncol. Ther. 2019, 7, 33–65. [Google Scholar] [CrossRef]
- Radakovich, N.; Nagy, M.; Nazha, A. Artificial Intelligence in Hematology: Current Challenges and Opportunities. Curr. Hematol. Malig. Rep. 2020, 15, 203–210. [Google Scholar] [CrossRef]
- Shouval, R.; Fein, J.A.; Savani, B.; Mohty, M.; Nagler, A. Machine learning and artificial intelligence in haematology. Br. J. Haematol. 2021, 192, 239–250. [Google Scholar] [CrossRef]
- Bobee, V.; Drieux, F.; Marchand, V.; Sater, V.; Veresezan, L.; Picquenot, J.M.; Viailly, P.J.; Lanic, M.D.; Viennot, M.; Bohers, E.; et al. Combining gene expression profiling and machine learning to diagnose B-cell non-Hodgkin lymphoma. Blood Cancer J. 2020, 10, 59. [Google Scholar] [CrossRef]
- Zhao, M.; Mallesh, N.; Hollein, A.; Schabath, R.; Haferlach, C.; Haferlach, T.; Elsner, F.; Luling, H.; Krawitz, P.; Kern, W. Hematologist-Level Classification of Mature B-Cell Neoplasm Using Deep Learning on Multiparameter Flow Cytometry Data. Cytom. Part A 2020, 97, 1073–1080. [Google Scholar] [CrossRef]
- Bucinski, A.; Marszall, M.P.; Krysinski, J.; Lemieszek, A.; Zaluski, J. Contribution of artificial intelligence to the knowledge of prognostic factors in Hodgkin’s lymphoma. Eur. J. Cancer Prev. 2010, 19, 308–312. [Google Scholar] [CrossRef]
- deAndres-Galiana, E.J.; Fernandez-Martinez, J.L.; Luaces, O.; Del Coz, J.J.; Fernandez, R.; Solano, J.; Nogues, E.A.; Zanabilli, Y.; Alonso, J.M.; Payer, A.R.; et al. On the prediction of Hodgkin lymphoma treatment response. Clin. Transl. Oncol. 2015, 17, 612–619. [Google Scholar] [CrossRef]
- Xu-Monette, Z.Y.; Zhang, H.; Zhu, F.; Tzankov, A.; Bhagat, G.; Visco, C.; Dybkaer, K.; Chiu, A.; Tam, W.; Zu, Y.; et al. A refined cell-of-origin classifier with targeted NGS and artificial intelligence shows robust predictive value in DLBCL. Blood Adv. 2020, 4, 3391–3404. [Google Scholar] [CrossRef]
- Cobo, M.J.; Lopez-Herrera, A.G.; Herrera-Viedma, E.; Herrera, F. SciMAT: A new science mapping analysis software tool. J. Am. Soc. Inf. Sci. Technol. 2012, 63, 1609–1630. [Google Scholar] [CrossRef]
- Van Eck, N.J.; Waltman, L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 2010, 84, 523–538. [Google Scholar] [CrossRef]
- Cobo, M.J.; López-Herrera, A.G.; Herrera-Viedma, E.; Herrera, F. An approach for detecting, quantifying, and visualizing the evolution of a research field: A practical application to the Fuzzy Sets Theory field. J. Informetr. 2011, 5, 146–166. [Google Scholar] [CrossRef]
- Moral-Munoz, J.A.; Herrera-Viedma, E.; Santisteban-Espejo, A.; Cobo, M.J. Software tools for conducting bibliometric analysis in science: An up-to-date review. Prof. Inf. 2020, 29, e290103. [Google Scholar] [CrossRef]
- Callon, M.; Courtial, J.P.; Laville, F. Co-Word Analysis as a Tool for Describing the Network of Interactions between Basic and Technological Research—The Case of Polymer Chemistry. Scientometrics 1991, 22, 155–205. [Google Scholar] [CrossRef]
- Callon, M.; Courtial, J.-P.; Turner, W.A.; Bauin, S. From translations to problematic networks: An introduction to co-word analysis. Soc. Sci. Inf. 1983, 22, 191–235. [Google Scholar] [CrossRef]
- Coulter, N.; Monarch, I.; Konda, S. Software engineering as seen through its research literature: A study in co-word analysis. J. Am. Soc. Inf. Sci. 1998, 49, 1206–1223. [Google Scholar] [CrossRef]
- Muñoz-Leiva, F.; Viedma-del-Jesús, M.I.; Sánchez-Fernández, J.; López-Herrera, A.G. An application of co-word analysis and bibliometric maps for detecting the most highlighting themes in the consumer behaviour research from a longitudinal perspective. Qual. Quant. 2012, 46, 1077–1095. [Google Scholar] [CrossRef]
- Orts, F.; Filatovas, E.; Ortega, G.; Kurasova, O.; Garzón, E.M. Improving the energy efficiency of SMACOF for multidimensional scaling on modern architectures. J. Supercomput. 2019, 75, 1038–1050. [Google Scholar] [CrossRef]
- Kessler, M.M. Bibliographic coupling between scientific papers. Am. Doc. 1963, 14, 10–25. [Google Scholar] [CrossRef]
- Goffman, W.; Morris, T.G. Bradford’s law and library acquisitions. Nature 1970, 226, 922–923. [Google Scholar] [CrossRef] [PubMed]
- Kiechle, F.L.; Zhang, X.; Holland-Staley, C.A. The -omics era and its impact. Arch. Pathol. Lab. Med. 2004, 128, 1337–1345. [Google Scholar] [CrossRef]
- van Karnebeek, C.D.M.; Wortmann, S.B.; Tarailo-Graovac, M.; Langeveld, M.; Ferreira, C.R.; van de Kamp, J.M.; Hollak, C.E.; Wasserman, W.W.; Waterham, H.R.; Wevers, R.A.; et al. The role of the clinician in the multi-omics era: Are you ready? J. Inherit. Metab. Dis. 2018, 41, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Mallampati, S.; An, G.; Wang, J. Targeted Therapy in Hematological Malignancies: From Basic Research to Clinical Practice. Biomed. Res. Int. 2015, 2015, 157570. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Wei, A.H. How I treat acute myeloid leukemia in the era of new drugs. Blood 2020, 135, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qin, W.; Huo, Y.J.; Li, X.; Shi, Q.; Rasko, J.E.J.; Janin, A.; Zhao, W.L. Advances in targeted therapy for malignant lymphoma. Signal. Transduct. Target. Ther. 2020, 5, 15. [Google Scholar] [CrossRef]
- El Achi, H.; Khoury, J.D. Artificial Intelligence and Digital Microscopy Applications in Diagnostic Hematopathology. Cancers 2020, 12, 797. [Google Scholar] [CrossRef]
- Niazi, M.K.K.; Parwani, A.V.; Gurcan, M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019, 20, e253–e261. [Google Scholar] [CrossRef]
- Acs, B.; Rantalainen, M.; Hartman, J. Artificial intelligence as the next step towards precision pathology. J. Intern. Med. 2020, 288, 62–81. [Google Scholar] [CrossRef]
- Robertson, S.; Azizpour, H.; Smith, K.; Hartman, J. Digital image analysis in breast pathology-from image processing techniques to artificial intelligence. Transl. Res. 2018, 194, 19–35. [Google Scholar] [CrossRef]
- Chang, H.Y.; Jung, C.K.; Woo, J.I.; Lee, S.; Cho, J.; Kim, S.W.; Kwak, T.Y. Artificial Intelligence in Pathology. J. Pathol. Transl. Med. 2019, 53, 1–12. [Google Scholar] [CrossRef]
- Price, D.D.S. A general theory of bibliometric and other cumulative advantage processes. J. Am. Soc. Inform. Sci 1976, 27, 292–306. [Google Scholar] [CrossRef]
- Kuhn, T.S. The Structure of Scientific Revolutions; University of Chicago Press: Chicago, IL, USA, 1962. [Google Scholar]
- Ostertagová, E. Modelling using Polynomial Regression. Procedia Eng. 2012, 48, 500–506. [Google Scholar] [CrossRef]
- Roig-Tierno, N.; Gonzalez-Cruz, T.F.; Llopis-Martinez, J. An overview of qualitative comparative analysis: A bibliometric analysis. J. Innov. Knowl. 2017, 2, 15–23. [Google Scholar] [CrossRef]
- Weisman, A.J.; Kim, J.; Lee, I.; McCarten, K.M.; Kessel, S.; Schwartz, C.L.; Kelly, K.M.; Jeraj, R.; Cho, S.Y.; Bradshaw, T.J. Automated quantification of baseline imaging PET metrics on FDG PET/CT images of pediatric Hodgkin lymphoma patients. EJNMMI Phys. 2020, 7, 76. [Google Scholar] [CrossRef]
- Blanc-Durand, P.; Jegou, S.; Kanoun, S.; Berriolo-Riedinger, A.; Bodet-Milin, C.; Kraeber-Bodere, F.; Carlier, T.; Le Gouill, S.; Casasnovas, R.O.; Meignan, M.; et al. Fully automatic segmentation of diffuse large B cell lymphoma lesions on 3D FDG-PET/CT for total metabolic tumour volume prediction using a convolutional neural network. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Lartizien, C.; Rogez, M.; Niaf, E.; Ricard, F. Computer-aided staging of lymphoma patients with FDG PET/CT imaging based on textural information. IEEE J. Biomed. Health Inform. 2014, 18, 946–955. [Google Scholar] [CrossRef]
- Birkle, C.; Pendlebury, D.A.; Schnell, J.; Adams, J. Web of Science as a data source for research on scientific and scholarly activity. Quant. Sci. Stud. 2020, 1, 363–376. [Google Scholar] [CrossRef]
- Santisteban-Espejo, A.; Campos, F.; Martin-Piedra, L.; Durand-Herrera, D.; Moral-Munoz, J.A.; Campos, A.; Martin-Piedra, M.A. Global tissue engineering trends: A scientometric and evolutive study. Tissue Eng. Part A 2018, 24, 1504–1517. [Google Scholar] [CrossRef]
- Santisteban-Espejo, A.; Campos, F.; Chato-Astrain, J.; Durand-Herrera, D.; Garcia-Garcia, O.; Campos, A.; Martin-Piedra, M.A.; Moral-Munoz, J.A. Identification of cognitive and social framework of tissue engineering by science mapping analysis. Tissue Eng. Part C Methods 2019, 25, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.H.; Hew, J.J.; Loke, S.P. Evaluating and comparing ten-year (2006–2015) research performance between Malaysian public and private higher learning institutions: A bibliometric approach. Int. J. Innov. Learn. 2018, 23, 145–165. [Google Scholar] [CrossRef]
- Rosas, S.R.; Kagan, J.M.; Schouten, J.T.; Slack, P.A.; Trochim, W.M. Evaluating research and impact: A bibliometric analysis of research by the NIH/NIAID HIV/AIDS clinical trials networks. PLoS ONE 2011, 6, e17428. [Google Scholar] [CrossRef] [PubMed]
- Abramo, G.; D’Angelo, C.A.; Caprasecca, A. Allocative efficiency in public research funding: Can bibliometrics help? Res. Policy 2009, 38, 206–215. [Google Scholar] [CrossRef]
- Sagi, O.; Rokach, L. Ensemble learning: A survey. WIREs Data Min. Knowl. Discov. 2018, 8, e1249. [Google Scholar] [CrossRef]
- Shouval, R.; Labopin, M.; Unger, R.; Giebel, S.; Ciceri, F.; Schmid, C.; Esteve, J.; Baron, F.; Gorin, N.C.; Savani, B.; et al. Prediction of Hematopoietic Stem Cell Transplantation Related Mortality- Lessons Learned from the In-Silico Approach: A European Society for Blood and Marrow Transplantation Acute Leukemia Working Party Data Mining Study. PLoS ONE 2016, 11, e0150637. [Google Scholar] [CrossRef]
- Zhao, S.; Dong, X.; Shen, W.; Ye, Z.; Xiang, R. Machine learning-based classification of diffuse large B-cell lymphoma patients by eight gene expression profiles. Cancer Med. 2016, 5, 837–852. [Google Scholar] [CrossRef]
- Gaidano, V.; Tenace, V.; Santoro, N.; Varvello, S.; Cignetti, A.; Prato, G.; Saglio, G.; De Rosa, G.; Geuna, M. A Clinically Applicable Approach to the Classification of B-Cell Non-Hodgkin Lymphomas with Flow Cytometry and Machine Learning. Cancers 2020, 12, 1684. [Google Scholar] [CrossRef]
- Deeb, S.J.; Tyanova, S.; Hummel, M.; Schmidt-Supprian, M.; Cox, J.; Mann, M. Machine Learning-based Classification of Diffuse Large B-cell Lymphoma Patients by Their Protein Expression Profiles. Mol. Cell Proteom. 2015, 14, 2947–2960. [Google Scholar] [CrossRef]
- Mohlman, J.; Leventhal, S.; Venkat, A.; Gyulassy, A.; Pascucci, V.; Salama, M. Application of a Convolutional Neural Network to Distinguish Burkitt Lymphoma From Diffuse Large B-Cell Lymphoma. Am. J. Clin. Pathol. 2018, 150, S119. [Google Scholar] [CrossRef]
- Ando, T.; Suguro, M.; Kobayashi, T.; Seto, M.; Honda, H. Multiple fuzzy neural network system for outcome prediction and classification of 220 lymphoma patients on the basis of molecular profiling. Cancer Sci. 2003, 94, 906–913. [Google Scholar] [CrossRef]
- Chen, H.; Engkvist, O.; Wang, Y.; Olivecrona, M.; Blaschke, T. The rise of deep learning in drug discovery. Drug Discov. Today 2018, 23, 1241–1250. [Google Scholar] [CrossRef]
- Turki, T.; Wei, Z.; Wang, J.T.L. Transfer Learning Approaches to Improve Drug Sensitivity Prediction in Multiple Myeloma Patients. IEEE Access 2017, 5, 7381–7393. [Google Scholar] [CrossRef]
- Davis, N.; McKinney, M.S.; Reddy, A.; Love, C.; Smith, E.; Happ, L.; Dave, S. Novel Mechanisms for Resistance to Targeted Therapy Identified through Machine Learning Approaches in 1167 RNA-Seq Drug Exposure Profiles in Lymphoma. Blood 2018, 132, 1370. [Google Scholar] [CrossRef]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef]
- Sibille, L.; Seifert, R.; Avramovic, N.; Vehren, T.; Spottiswoode, B.; Zuehlsdorff, S.; Schafers, M. (18)F-FDG PET/CT Uptake Classification in Lymphoma and Lung Cancer by Using Deep Convolutional Neural Networks. Radiology 2020, 294, 445–452. [Google Scholar] [CrossRef]
- Capobianco, N.; Meignan, M.; Cottereau, A.S.; Vercellino, L.; Sibille, L.; Spottiswoode, B.; Zuehlsdorff, S.; Casasnovas, O.; Thieblemont, C.; Buvat, I. Deep-Learning (18)F-FDG Uptake Classification Enables Total Metabolic Tumor Volume Estimation in Diffuse Large B-Cell Lymphoma. J. Nucl. Med. 2021, 62, 30–36. [Google Scholar] [CrossRef]
- Agius, R.; Brieghel, C.; Andersen, M.A.; Pearson, A.T.; Ledergerber, B.; Cozzi-Lepri, A.; Louzoun, Y.; Andersen, C.L.; Bergstedt, J.; von Stemann, J.H.; et al. Machine learning can identify newly diagnosed patients with CLL at high risk of infection. Nat. Commun. 2020, 11, 363. [Google Scholar] [CrossRef]
- Shaabanpour Aghamaleki, F.; Mollashahi, B.; Nosrati, M.; Moradi, A.; Sheikhpour, M.; Movafagh, A. Application of an Artificial Neural Network in the Diagnosis of Chronic Lymphocytic Leukemia. Cureus 2019, 11, e4004. [Google Scholar] [CrossRef]
- Parodi, S.; Manneschi, C.; Verda, D.; Ferrari, E.; Muselli, M. Logic Learning Machine and standard supervised methods for Hodgkin’s lymphoma prognosis using gene expression data and clinical variables. Health Inform. J. 2018, 24, 54–65. [Google Scholar] [CrossRef]
- Beaulac, C.; Rosenthal, J.S.; Pei, Q.; Friedman, D.; Wolden, S.; Hodgson, D. An evaluation of machine learning techniques to predict the outcome of children treated for Hodgkin-Lymphoma on the AHOD0031 trial. Appl. Artif. Intell. 2020, 34, 1100–1114. [Google Scholar] [CrossRef]
| Institution | 1990–2005 | Institution | 2006–2014 | Institution | 2015–2020 | Institution | TOTAL | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C | % | C | % | C | % | C | % | ||||
| NAGOYA UNIVERSITY | 5 | 7.81 | CORNELL UNIV | 4 | 3.50 | CHINESE ACAD SCI | 10 | 2.87 | UNIV OF TEXAS SYSTEM | 19 | 3.61 |
| AICHI CANC CTR | 4 | 6.25 | INDIAN STAT INST | 4 | 3.50 | EMORY UNIV | 10 | 2.87 | INSERM | 15 | 2.85 |
| ST JOHNS HOSP | 3 | 4.68 | JADAVPUR UNIV | 4 | 3.50 | UNIV TEXAS MD ANDERSON | 8 | 2.29 | HARVARD UNIV | 13 | 2.47 |
| CENT MED LABS | 2 | 3.12 | NANYANG TECHNOL UNIV | 3 | 2.63 | MEM SLOAN KETTERING CANC CTR | 7 | 2.01 | UNIV CALIFORNIA SYSTEM | 13 | 2.47 |
| FLORIDA INT UNIV | 2 | 3.12 | NCI | 3 | 2.63 | UNIV PENN | 7 | 2.01 | CHINESE ACAD SCI | 12 | 2.28 |
| HARVARD UNIV | 2 | 3.12 | NIH | 3 | 2.63 | ICAHN SCH MED MT SINAI | 6 | 1.72 | CORNELL UNIV | 12 | 2.28 |
| NANYANG TECHNOL UNIV | 2 | 3.12 | RUTGERS STATE UNIV | 3 | 2.63 | MAYO CLIN | 6 | 1.72 | UTMD ANDERSON CANCER CENTER | 11 | 2.09 |
| OHIO STATE UNIV | 2 | 3.12 | TONGJI UNIV | 3 | 2.63 | TECH UNIV MUNICH | 6 | 1.72 | EMORY UNIV | 10 | 1.90 |
| THOMAS JEFFERSON UNIV | 2 | 3.12 | UNIV MICHIGAN | 3 | 2.63 | CHINA UNIV MIN TECHNOL | 5 | 1.43 | MEM SLOAN KATTERING CANC CTR | 10 | 1.90 |
| UNIV BIRMINGHAMN | 2 | 3.12 | UNIV OXFORD | 3 | 2.63 | COLUMBIA UNIV | 5 | 1.43 | UNIV PENNSYLVANIA | 10 | 1.90 |
| UNIV MARYLAND | 2 | 3.12 | UNIV TOKYO | 3 | 2.63 | GEORGIA INST TECHNOL | 5 | 1.43 | APHP PARIS | 9 | 1.71 |
| UNIV ROMA LA SAPIENZA | 2 | 3.12 | UNIV TURIN | 3 | 2.63 | MASSACHUSETTS GEN HOSP | 5 | 1.43 | CENT NAT DE LA RECHER SCIENTIFIQUE | 9 | 1.71 |
| UNIV ROMA TOR VERGATA | 2 | 3.12 | UNIV ZAGREB | 3 | 2.63 | NEW JERSEY INST TECHNOL | 5 | 1.43 | SCHOOL OF MED MOUNT SINAI | 9 | 1.71 |
| UNIV SO CALIF | 2 | 3.12 | CHARITE | 2 | 1.75 | OHIO STATE UNIV | 5 | 1.43 | NIH | 9 | 1.71 |
| UNIV TURIN | 2 | 3.12 | DANA FARBER CANC INST | 2 | 1.75 | SHANGAI JIAO TONG UNIV | 5 | 1.43 | MAYO CLINIC | 8 | 1.52 |
| YONSEI UNIV | 2 | 3.12 | FLORIDA INT UNIV | 2 | 1.75 | SICHUAN UNIV | 5 | 1.43 | STATE UNIV SYSTEM OF FLORIDA | 8 | 1.52 |
| BETHESDA HOSP | 1 | 1.56 | GOETHE UNIV FRANKFURT | 2 | 1.75 | UNIV LEIPZIG | 5 | 1.43 | TECH UNIV OF MUNICH | 8 | 1.52 |
| CEDARS SINAI MED CTR | 1 | 1.56 | HARVARD UNIV | 2 | 1.75 | UNIV SYDNEY | 5 | 1.43 | YONSEI UNIV | 8 | 1.52 |
| CENTROL NACL INVEST ONCOL | 1 | 1.56 | HOP LYON SUD | 2 | 1.75 | YONSEI UNIV | 5 | 1.43 | COLUMBIA UNIV | 7 | 1.33 |
| CHINESE PEOPLES LIBERAT ARMY GEN HOPS | 1 | 1.56 | INDIAN INST TECHNOL | 2 | 1.75 | CHB HOSP | 4 | 1.14 | GOETHE UNIV FRANKFURT | 7 | 1.33 |
| Source Title | 1990–2005 | Source Title | 2006–2014 | Source Title | 2015–2020 | Source Title | TOTAL | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C | % | C | % | C | % | C | % | ||||
| ARTIFICIAL INTELLIGENCE IN MEDICINE | 3 | 4.68 | LECTURE NOTES IN COMPUTER SCIENCE | 5 | 4.38 | EUROP JOURN NUCL MED MOL IMAG | 10 | 2.87 | LECTURE NOTES IN COMPUTER SCIENCE | 14 | 2.66 |
| HUMAN PATHOLOGY | 3 | 4.68 | BMC BIOINFORMATICS | 4 | 3.50 | BLOOD | 9 | 2.25 | BLOOD | 12 | 2.28 |
| LECTURE NOTES IN COMPUTER SCIENCE | 3 | 4.68 | PLOS ONE | 3 | 2.63 | SCIENTIFIC REPORTS | 8 | 2.29 | EUROP JOURN NUCL MED MOL IMAG | 10 | 1.90 |
| PROCEEDINGS OF ANNUAL ICIEE-EMBS | 3 | 4.68 | ANALYTICAL CELLULAR PATHOLOGY | 2 | 1.75 | PROCEEDINGS OF THE SPIE | 7 | 2.01 | PROCEEDINGS OF THE SPIE | 9 | 1.71 |
| BLOOD | 2 | 3.12 | ARTIFICIAL INTELLIGENCE IN MEDICINE | 2 | 1.75 | JOURNAL OF NUCLEAR MEDICINE | 6 | 1.72 | SCIENTIFIC REPORTS | 8 | 1.52 |
| COMPUTATIONAL BIOLOGY AND CHEMISTRY | 2 | 3.12 | BMC GENOMICS | 2 | 1-75 | LECTURE NOTES IN COMPUTER SCIENCE | 6 | 1.72 | BMC BIOINFORMATICS | 7 | 1.33 |
| CYTOMETRY | 2 | 3.12 | COMPUTERS IN BIOLOGY AND MEDICINE | 2 | 1.75 | COMPUTER METHODS AND PROGRAMS IN BIOMEDICINE | 5 | 1.43 | PLOS ONE | 7 | 1.33 |
| JOURNAL OF BIOSCIENCE AND BIOENGINEERING | 2 | 3.12 | HEMATOLOGY | 2 | 1.75 | FRONTIERS IN ONCOLOGY | 5 | 1.43 | ARTIFICIAL INTELLIGENCE IN MEDICINE | 6 | 1.14 |
| NEUROCOMPUTING | 2 | 3.12 | IEEE ENGINEERING MBSCP | 2 | 1.75 | IEEE ACCESS | 5 | 1.43 | JOURNAL OF NUCLEAR MEDICINE | 6 | 1.14 |
| PROCEEDING OF THE 2005 IEE SCIBCB | 2 | 3.12 | LEUKEMIA | 2 | 1.75 | LABORATORY INVESTIGATION | 5 | 1.43 | COMPUTER METHODS AND PROGRAMS IN BIOMEDICINE | 5 | 0.95 |
| 2000 IEEE EMBS ICITABMP | 1 | 1.56 | PROCEEDING OF THE SPIE | 2 | 1.75 | AMERICAN JOURNAL OF CLINICAL PATHOLOGY | 4 | 1.14 | FRONTIERS IN ONCOLOY | 5 | 0.95 |
| 2001 IEE NUCLEAR SCIENCE SCR | 1 | 1.56 | 2006 IEEE IJCNNP | 1 | 0.87 | BLOOD ADVANCES | 4 | 1.14 | IEEE ACCESS | 5 | 0.95 |
| 2004 IEE SCBCP | 1 | 1.56 | 2008 IEEE WORSHOP ON MLSP | 1 | 0.87 | CANCERS | 4 | 1.14 | JOURNAL OF BIOMEDICAL INFORMATICS | 5 | 0.95 |
| 2005 27TH ANNUAL IC-IEE E-EMBS | 1 | 1.56 | 2008 INTERNATIONAL STCITAB | 1 | 0.87 | IEEE ICBB | 4 | 1.14 | LABORATORY INVESTIGATION | 5 | 0.95 |
| 2005 IEE CSBCP | 1 | 1.56 | 2009 ANNUAL IC-IEEE-EMBS | 1 | 0.87 | JOURNAL OF BIOMEDICAL INFORMATICS | 4 | 1.14 | AMERICAN JOURNAL OF CLINICAL PATHOLOGY | 4 | 0.76 |
| 2005 IEE NETWORKING SCP | 1 | 1.56 | 2009 IEEE CONGRESS ON EC | 1 | 0.87 | MEDICAL PHYSICS | 4 | 1.14 | BLOOD ADVANCES | 4 | 0.76 |
| 7TH WORLD CULTICONFERENCE ON SCI. | 1 | 1.56 | 2010 7TH IEEE ISBINM | 1 | 0.87 | PLOS ONE | 4 | 1.14 | CANCERS | 4 | 0.76 |
| AMERICAN JOURNAL OF DERMATOPATHOLOGY | 1 | 1.56 | 2012 7TH ICCCT | 1 | 0.87 | CLINICAL CANCER RESEARCH | 3 | 0.86 | COMPUTERS IN BIOLOGY AND MEDICINE | 4 | 0.76 |
| AMERICAN JOURNAL OF HEMATOLOGY | 1 | 1.56 | 2012 9TH IEEE ISBI | 1 | 0.87 | GENOME MEDICINE | 3 | 0.86 | IEE ICBB | 4 | 0.76 |
| AMIA 2002 SYMPOSIUM PROCEEDINGS | 1 | 1.56 | 2013 12TH ICMLA | 1 | 0.87 | INTERNATIONAL JOURNAL OF LABORATORY HEMATOLOGY | 3 | 0.86 | LEUKEMIA | 4 | 0.76 |
| Country | 1990–2005 | Country | 2006–2014 | Country | 2015–2020 | Country | TOTAL | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C | % | C | % | C | % | C | % | ||||
| USA | 25 | 39.06 | USA | 38 | 33.33 | USA | 127 | 36.49 | USA | 190 | 36.19 |
| ENGLAND | 6 | 9.37 | PEOPLE’S R CHINA | 14 | 12.28 | PEOPLE’S R CHINA | 57 | 16.37 | PEOPLE’S R CHINA | 72 | 13.71 |
| GERMANY | 6 | 9.37 | INDIA | 12 | 10.52 | GERMANY | 33 | 9.48 | GERMANY | 44 | 8.38 |
| JAPAN | 5 | 7.81 | ENGLAND | 11 | 9.64 | FRANCE | 23 | 6.60 | INDIA | 35 | 6.66 |
| ITALY | 4 | 6.25 | ITALY | 8 | 7.01 | INDIA | 22 | 6.32 | FRANCE | 31 | 5.90 |
| CANADA | 3 | 4.68 | FRANCE | 7 | 6.14 | SPAIN | 19 | 5.46 | ENGLAND | 30 | 5.71 |
| IRELAND | 3 | 4.68 | JAPAN | 6 | 5.26 | ITALY | 18 | 5.17 | ITALY | 30 | 5.71 |
| SINGAPORE | 3 | 4.68 | GERMANY | 5 | 4.38 | AUSTRALIA | 14 | 4.02 | JAPAN | 23 | 4,38 |
| AUSTRALIA | 2 | 3.12 | IRAN | 5 | 4.38 | ENGLAND | 13 | 3.73 | SPAIN | 23 | 4.38 |
| NETHERLANDS | 2 | 3.12 | POLAND | 4 | 3.50 | JAPAN | 12 | 3.44 | AUSTRALIA | 17 | 3.23 |
| NEW ZEALAND | 2 | 3.12 | SINGAPORE | 4 | 3.50 | SOUTH KOREA | 12 | 3.44 | SOUTH KOREA | 16 | 3.04 |
| SOUTH KOREA | 2 | 3.12 | CROATIA | 3 | 2.63 | CANADA | 10 | 2.87 | CANADA | 14 | 2.66 |
| WALES | 2 | 3.12 | SPAIN | 3 | 2.63 | SWITZERLAND | 10 | 2.87 | NETHERLANDS | 12 | 2.28 |
| BARBADOS | 1 | 1.56 | AUSTRIA | 2 | 1.75 | NETHERLANDS | 9 | 2.58 | SWITZERLAND | 12 | 2,28 |
| CROATIA | 1 | 1.56 | BRAZIL | 2 | 1.75 | AUSTRIA | 8 | 2.29 | AUSTRIA | 10 | 1.90 |
| FRANCE | 1 | 1.56 | MEXICO | 2 | 1.75 | BRAZIL | 8 | 2.29 | BRAZIL | 10 | 1.90 |
| INDIA | 1 | 1.56 | SLOVENIA | 2 | 1.75 | DENMARK | 8 | 2.29 | IRAN | 10 | 1.90 |
| ISRAEL | 1 | 1.56 | SOUTH KOREA | 2 | 1.75 | SWEDEN | 7 | 2.01 | DENMARK | 9 | 1.71 |
| PEOPLES R CHINA | 1 | 1.56 | AUSTRALIA | 1 | 9.87 | SAUDI ARABIA | 6 | 1.72 | SINGAPORE | 8 | 1.52 |
| POLAND | 1 | 1.56 | BELGIUM | 1 | 0.87 | EGYPT | 5 | 1.43 | SWEDEN | 8 | 1.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moran-Sanchez, J.; Santisteban-Espejo, A.; Martin-Piedra, M.A.; Perez-Requena, J.; Garcia-Rojo, M. Translational Applications of Artificial Intelligence and Machine Learning for Diagnostic Pathology in Lymphoid Neoplasms: A Comprehensive and Evolutive Analysis. Biomolecules 2021, 11, 793. https://doi.org/10.3390/biom11060793
Moran-Sanchez J, Santisteban-Espejo A, Martin-Piedra MA, Perez-Requena J, Garcia-Rojo M. Translational Applications of Artificial Intelligence and Machine Learning for Diagnostic Pathology in Lymphoid Neoplasms: A Comprehensive and Evolutive Analysis. Biomolecules. 2021; 11(6):793. https://doi.org/10.3390/biom11060793
Chicago/Turabian StyleMoran-Sanchez, Julia, Antonio Santisteban-Espejo, Miguel Angel Martin-Piedra, Jose Perez-Requena, and Marcial Garcia-Rojo. 2021. "Translational Applications of Artificial Intelligence and Machine Learning for Diagnostic Pathology in Lymphoid Neoplasms: A Comprehensive and Evolutive Analysis" Biomolecules 11, no. 6: 793. https://doi.org/10.3390/biom11060793
APA StyleMoran-Sanchez, J., Santisteban-Espejo, A., Martin-Piedra, M. A., Perez-Requena, J., & Garcia-Rojo, M. (2021). Translational Applications of Artificial Intelligence and Machine Learning for Diagnostic Pathology in Lymphoid Neoplasms: A Comprehensive and Evolutive Analysis. Biomolecules, 11(6), 793. https://doi.org/10.3390/biom11060793






