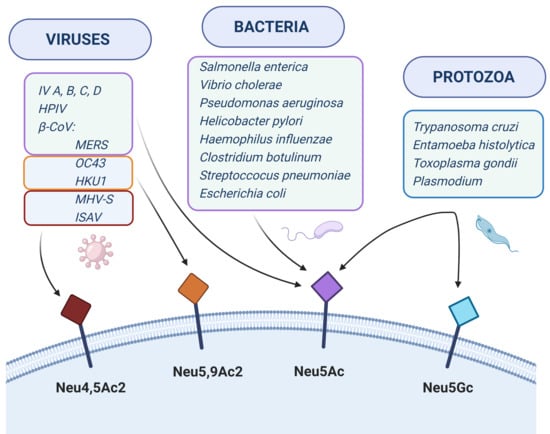
Sialic Acids as Receptors for Pathogens
Abstract
:1. Introduction
1.1. Structure of Sialic Acids
1.2. Neu5Gc vs. Neu5Ac
1.3. Interactions with Pathogens
2. Viruses
2.1. Influenza Virus
2.2. SV40
2.3. Coronaviruses
2.4. Human Parainfluenza Viruses
3. Bacteria
3.1. Salmonella enterica
3.2. Vibrio cholerae
3.3. Pseudomonas aeruginosa
3.4. Helicobacter pylori
3.5. Haemophilus influenzae
3.6. Clostridium botulinum
3.7. Streptoccocus pneumoniae
3.8. Escherichia coli
4. Protozoa
4.1. Trypanosoma cruzi
4.2. Entamoeba histolytica
4.3. Toxoplasma gondii
4.4. Plasmodium
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schauer, R. Sialic acids: Fascinating sugars in higher animals and man. Zoology 2004, 107, 49–64. [Google Scholar] [CrossRef]
- Peri, S.; Kulkarni, A.; Feyertag, F.; Berninsone, P.M.; Alvarez-Ponce, D. Phylogenetic Distribution of CMP-Neu5Ac Hydroxylase (CMAH), the Enzyme Synthetizing the Proinflammatory Human Xenoantigen Neu5Gc. Genome Biol. Evol. 2018, 10, 207–219. [Google Scholar] [CrossRef] [Green Version]
- Schauer, R.; Kamerling, J.P. Exploration of the Sialic Acid World. Adv. Carbohydr. Chem. Biochem. 2018, 75, 1–213. [Google Scholar] [CrossRef]
- Varki, A. Sialic acids in human health and disease. Trends Mol. Med. 2008, 14, 351–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schauer, R. Sialic acids as regulators of molecular and cellular interactions. Curr. Opin. Struct. Biol. 2009, 19, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Padler-Karavani, V. Evolution of sialic acids: Implications in xenotransplant biology. Xenotransplantation 2018, 25, e12424. [Google Scholar] [CrossRef] [PubMed]
- Skarbek, K.; Milewska, M.J. Biosynthetic and synthetic access to amino sugars. Carbohydr. Res. 2016, 434, 44–71. [Google Scholar] [CrossRef]
- Wang, B.; Brand-Miller, J. The role and potential of sialic acid in human nutrition. Eur. J. Clin. Nutr. 2003, 57, 1351–1369. [Google Scholar] [CrossRef] [Green Version]
- Varki, A.; Cummings, R.; Esko, J.; Freeze, H.; Stanley, P.; Bertozzi, C.; Hart, G.; Etzler, M. Essentials of Glycobiology; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2017. [Google Scholar]
- Nguyen, T.; Lee, S.; Yang, Y.-A.; Ahn, C.; Sim, J.H.; Kei, T.G.; Barnard, K.N.; Yu, H.; Millano, S.K.; Chen, X.; et al. The role of 9-O-acetylated glycan receptor moieties in the typhoid toxin binding and intoxication. PLoS Pathog. 2020, 16, e1008336. [Google Scholar] [CrossRef] [Green Version]
- Schauer, R.; Srinivasan, G.V.; Wipfler, D.; Kniep, B.; Schwartz-Albiez, R. O-Acetylated Sialic Acids and Their Role in Immune Defense. Adv. Exp. Med. Biol. 2011, 705, 525–548. [Google Scholar] [CrossRef]
- Park, S.S. Post-Glycosylation Modification of Sialic Acid and Its Role in Virus Pathogenesis. Vaccinces 2019, 7, 171. [Google Scholar] [CrossRef] [Green Version]
- Wasik, B.R.; Barnard, K.N.; Parrish, C.R. Effect of Sialic Acid Modifications on Virus Binding and Infection. Trends Microbiol. 2016, 24, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Rota, P.; La Rocca, P.; Allevi, P.; Pappone, C.; Anastasia, L. Intramolecular Lactones of Sialic Acids. Int. J. Mol. Sci. 2020, 21, 8098. [Google Scholar] [CrossRef] [PubMed]
- Mikolajczyk, K.; Kaczmarek, R.; Czerwinski, M. How glycosylation affects glycosylation: The role of N-glycans in glycosyltransferase activity. Glycobiology 2020, 30, 941–969. [Google Scholar] [CrossRef] [PubMed]
- Carbohydrate-Active enZYmes Database. Available online: http://www.cazy.org (accessed on 1 February 2021).
- Miyagi, T.; Yamaguchi, K. Mammalian sialidases: Physiological and pathological roles in cellular functions. Glycobiology 2012, 22, 880–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sukhorukov, V.N.; Karagodin, V.P.; Zakiev, E.R.; Grechko, A.V.; Orekhov, A.N. Sialidases: Therapeutic and Antiatherogenic Potential. Curr. Pharm. Des. 2017, 23, 4696–4701. [Google Scholar] [CrossRef]
- Kim, S.; Oh, D.B.; Kang, H.A.; Kwon, O. Features and applications of bacterial sialidases. Appl. Microbiol. Biotechnol. 2011, 91, 1–15. [Google Scholar] [CrossRef]
- Air, G.M. Influenza neuraminidase. Influ. Other Respir. Viruses 2012, 6, 245–256. [Google Scholar] [CrossRef]
- Lipnicanova, S.; Chmelova, D.; Ondrejovic, M.; Frecer, V.; Miertus, S. Diversity of sialidases found in the human body—A review. Int. J. Biol. Macromol. 2020, 148, 857–868. [Google Scholar] [CrossRef]
- Glanz, V.Y.; Myasoedova, V.A.; Grechko, A.V.; Orekhov, A.N. Inhibition of sialidase activity as a therapeutic approach. Drug Des. Dev. Ther. 2018, 12, 3431–3437. [Google Scholar] [CrossRef] [Green Version]
- Shie, J.J.; Fang, J.M. Development of effective anti-influenza drugs: Congeners and conjugates—A review. J. Biomed. Sci. 2019, 26. [Google Scholar] [CrossRef] [Green Version]
- Shaw, L.; Schauer, R. The Biosynthesis of N-Glycoloylneuraminic Acid Occurs by Hydroxylation of the CMP-Glycoside of N-Acetylneuraminic Acid. Biol. Chem. Hoppe-Seyler 1988, 369, 477–486. [Google Scholar] [CrossRef]
- Simakov, O.; Kawashima, T.; Marlétaz, F.; Jenkins, J.; Koyanagi, R.; Mitros, T.; Hisata, K.; Bredeson, J.; Shoguchi, E.; Gyoja, F.; et al. Hemichordate genomes and deuterostome origins. Nature 2015, 527, 459–465. [Google Scholar] [CrossRef] [Green Version]
- Kapli, P.; Natsidis, P.; Leite, D.J.; Fursman, M.; Jeffrie, N.; Rahman, I.A.; Philippe, H.; Copley, R.R.; Telford, M.J. Lack of support for Deuterostomia prompts reinterpretation of the first Bilateria. Sci. Adv. 2021, 7, eabe2741. [Google Scholar] [CrossRef]
- Kapli, P.; Telford, M.J. Topology-dependent asymmetry in systematic errors affects phylogenetic placement of Ctenophora and Xenacoelomorpha. Sci. Adv. 2020, 6, eabc5162. [Google Scholar] [CrossRef]
- Ros-Rocher, N.; Pérez-Posada, A.; Leger, M.M.; Ruiz-Trillo, I. The origin of animals: An ancestral reconstruction of the unicellular-to-multicellular transition. Open Biol. 2021, 11, 200359. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geneious 8.1.9. Available online: https://www.geneious.com (accessed on 4 March 2021).
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Hayakawa, T.; Aki, I.; Varki, A.; Satta, Y.; Takahata, N. Fixation of the Human-Specific CMP-N-Acetylneuraminic Acid Hydroxylase Pseudogene and Implications of Haplotype Diversity for Human Evolution. Genetics 2006, 172, 1139–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, H.-H.; Takematsu, H.; Diaz, S.; Iber, J.; Nickerson, E.; Wright, K.L.; Muchmore, E.A.; Nelson, D.L.; Warren, S.T.; Varki, A. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc. Natl. Acad. Sci. USA 1998, 95, 11751–11756. [Google Scholar] [CrossRef] [Green Version]
- Chou, H.-H.; Hayakawa, T.; Diaz, S.; Krings, M.; Indriati, E.; Leakey, M.; Paabo, S.; Satta, Y.; Takahata, N.; Varki, A. Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc. Natl. Acad. Sci. USA 2002, 99, 11736–11741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altman, M.O.; Gagneux, P. Absence of Neu5Gc and Presence of Anti-Neu5Gc Antibodies in Humans—An Evolutionary Perspective. Front. Immunol. 2019, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gagneux, P.; Varki, A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology 1999, 9, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Gagneux, P. Human-specific evolution of sialic acid targets: Explaining the malignant malaria mystery? Proc. Natl. Acad. Sci. USA 2009, 106, 14739–14740. [Google Scholar] [CrossRef] [Green Version]
- Ghaderi, D.; Springer, S.A.; Ma, F.; Cohen, M.; Secrest, P.; Taylor, R.E.; Varki, A.; Gagneux, P. Sexual selection by female immunity against paternal antigens can fix loss of function alleles. Proc. Natl. Acad. Sci. USA 2011, 108, 17743–17748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tangvoranuntakul, P.; Gagneux, P.; Diaz, S.; Bardor, M.; Varki, N.; Varki, A.; Muchmore, E. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc. Natl. Acad. Sci. USA 2003, 100, 12045–12050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardor, M.; Nguyen, D.H.; Diaz, S.; Varki, A. Mechanism of Uptake and Incorporation of the Non-human Sialic Acid N -Glycolylneuraminic Acid into Human Cells. J. Biol. Chem. 2005, 280, 4228–4237. [Google Scholar] [CrossRef] [Green Version]
- Taylor, R.E.; Gregg, C.J.; Padler-Karavani, V.; Ghaderi, D.; Yu, H.; Huang, S.; Sorensen, R.U.; Chen, X.; Inostroza, J.; Nizet, V.; et al. Novel mechanism for the generation of human xeno-autoantibodies against the nonhuman sialic acid N-glycolylneuraminic acid. J. Exp. Med. 2010, 207, 1637–1646. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, N.; Matthews, S.; Soldati-Favre, D. Sialic acids: Key determinants for invasion by the Apicomplexa. Int. J. Parasitol. 2010, 40, 1145–1154. [Google Scholar] [CrossRef] [Green Version]
- Neu, U.; Bauer, J.; Stehle, T. Viruses and sialic acids: Rules of engagement. Curr. Opin. Struct. Biol. 2011, 21, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.; Herrler, G.; Klenk, H.D. Sialic Acid Receptors of Viruses. Top. Curr. Chem. 2015, 367, 1–28. [Google Scholar] [CrossRef]
- Kato, K.; Ishiwa, A. The role of carbohydrates in infection strategies of enteric pathogens. Trop. Med. Health 2015, 43, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Haines-Menges, B.L.; Whitaker, W.B.; Lubin, J.B.; Boyd, E.F. Host Sialic Acids: A Delicacy for the Pathogen with Discerning Taste. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [Green Version]
- Juge, N.; Tailford, L.; Owen, C.D. Sialidases from gut bacteria: A mini-review. Biochem. Soc. Trans. 2016, 44, 166–175. [Google Scholar] [CrossRef] [Green Version]
- Blaum, B.S.; Sthle, T. Sialic Acids in Nonenveloped Virus Infections. Adv. Carbohydr. Chem. Biochem. 2019, 76, 65–111. [Google Scholar] [CrossRef] [PubMed]
- van de Sandt, C.E.; Bodewes, R.; Rimmelzwaan, G.F.; de Vries, R.D. Influenza B viruses: Not to be discounted. Future Microbiol. 2015, 10, 1447–1465. [Google Scholar] [CrossRef] [PubMed]
- de Graaf, M.; Fouchier, R.A.M. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014, 33, 823–841. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Veit, M. Hemagglutinin-esterase-fusion (HEF) protein of influenza C virus. Protein Cell 2016, 7, 28–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, S.; Fu, X.; Li, G.; Kerlin, F.; Veit, M. Novel Influenza D virus: Epidemiology, pathology, evolution and biological characteristics. Virulence 2017, 8, 1580–1591. [Google Scholar] [CrossRef] [Green Version]
- Shah, K.V. Simian Virus 40 and Human Disease. J. Infect. Dis. 2004, 190, 2061–2064. [Google Scholar] [CrossRef] [Green Version]
- Hulswit, R.J.G.; Lang, Y.; Bakkers, M.J.G.; Li, W.; Li, Z.; Schouten, A.; Ophorst, B.; van Kuppeveld, F.J.M.; Boons, G.-J.; Bosch, B.-J.; et al. Human coronaviruses OC43 and HKU1 bind to 9-Oacetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc. Natl. Acad. Sci. USA 2018, 116, 2681–2690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Hulswit, R.J.G.; Widjaja, I.; Raj, V.S.; McBride, R.; Peng, W.; Widagdo, W.; Tortorici, M.A.; van Dieren, B.; Lang, Y.; et al. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. USA 2017, 114, E8508–E8517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amonsen, M.; Smith, D.F.; Cummings, R.D.; Air, G.M. Human parainfluenza viruses hPIV1 ad hPIV3 bind oligosaccharides with alpha2-3-linked sialic acids that are distinct from those bound by H5 avian influenza virus hemagglutinin. J. Virol. 2007, 81, 8341-5. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Portner, A.; Scroggs, R.A.; Uchikawa, M.; Koyama, N.; Matsu, K.; Suzuiki, Y.; Takimoto, T. Receptor Specificities of Human Respiroviruses. J. Virol. 2001, 75, 4604–4613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Bukreyev, A.; Thompson, C.I.; Watson, B.; Peeples, M.E.; Collins, P.L.; Pickles, R.J. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 2005, 79, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Deng, L.; Stack, G.; Yu, H.; Chen, X.; Naito-Matsui, Y.; Varki, A.; Galán, J.E. Evolution of host adaptation in the Salmonella typhoid toxin. Nat. Microbiol. 2017, 2, 1592–1599. [Google Scholar] [CrossRef]
- Kappala, D.; Sarkhel, R.; Dixit, S.K.; Lalsangpuii; Mahawar, M.; Singh, M.; Ramakrishnan, S.; Goswami, T.K. Role of different receptors and actin filaments on Salmonella Typhimurium invasion in chicken macrophages. Immunobiology 2018, 223, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Muanprasat, C.; Chatsudthipong, V. Cholera: Pathophysiology and emerging therapeutic targets. Future Med. Chem. 2013, 5, 781–798. [Google Scholar] [CrossRef]
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018, 7, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Krivan, H.C.; Roberts, D.D.; Ginsburg, V. Many pulmonary pathogenic bacteria bind specifically to the carbohydrate sequence GalNAc beta 1-4Gal found in some glycolipids. Proc. Natl. Acad. Sci. USA 1988, 85, 6157–6161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, B.; Sachdev, G.P.; Cummings, R.D. Pseudomonas aeruginosa mucoid strain 8830 binds glycans containing the sialyl-Lewis x epitope. Glycoconj. J. 2006, 24, 87–95. [Google Scholar] [CrossRef]
- Evans, D.G.; Evans, D.J.; Moulds, J.J.; Graham, D.Y. N-acetylneuraminyllactose-binding fibrillar hemagglutinin of Campylobacter pylori: A putative colonization factor antigen. Infect. Immun. 1988, 56, 2896–2906. [Google Scholar] [CrossRef] [Green Version]
- Mahdavi, J. Helicobacter pylori SabA Adhesin in Persistent Infection and Chronic Inflammation. Science 2002, 297, 573–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roche, N.; Angstrom, J.; Hurtig, M.; Larsson, T.; Boren, T.; Teneberg, S. Helicobacter pylori and Complex Gangliosides. Infect. Immun. 2004, 72, 1519–1529. [Google Scholar] [CrossRef] [Green Version]
- Benktander, J.; Barone, A.; Johansson, M.M.; Teneberg, S. Helicobacter pylori SabA binding gangliosides of human stomach. Virulence 2018, 9, 738–751. [Google Scholar] [CrossRef] [Green Version]
- Bennett, H.J.; Roberts, I.S. Identification of a new sialic acid-binding protein in Helicobacter pylori. FEMS Immunol. Med. Microbiol. 2005, 44, 163–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlsohn, E.; Nystrom, J.; Bolin, I.; Nilsson, C.L.; Svennerholm, A.-M. HpaA is Essential for Helicobacter pylori Colonization in Mice. Infect. Immun. 2006, 74, 920–926. [Google Scholar] [CrossRef] [Green Version]
- Ng, P.S.K.; Day, C.J.; Atack, J.M.; Hartley-Tassell, L.E.; Winter, L.E.; Marshanski, T.; Padler-Karavani, V.; Varki, A.; Barenkamp, S.J.; Apicella, M.A.; et al. Nontypeable Haemophilus influenzae Has Evolved Preferential Use of N- Acetylneuraminic Acid as a Host Adaptation. mBio 2019, 10, e00422-19. [Google Scholar] [CrossRef] [Green Version]
- Greiner, L.L.; Watanabe, H.; Phillips, N.J.; Shao, J.; Morgan, A.; Zaleski, A.; Gibson, B.W.; Apicella, M.A. Nontypeable Haemophilus influenzae Strain 2019 Produces a Biofilm Containing N-Acetylneuraminic Acid That May Mimic Sialylated O-Linked Glycans. Infect. Immun. 2004, 72, 4249–4260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, J.W.; Coussens, N.P.; Allen, S.; Houtman, J.C.D.; Turner, K.H.; Zaleski, A.; Ramaswamy, S.; Gibson, B.W.; Apicella, M.A. Characterization of the N -Acetyl-5-neuraminic Acid-binding Site of the Extracytoplasmic Solute Receptor (SiaP) of Nontypeable Haemophilus influenzae Strain 2019. J. Biol. Chem. 2008, 283, 855–865. [Google Scholar] [CrossRef] [Green Version]
- Fujinaga, Y.; Inoue, K.; Nomura, T.; Sasaki, J.; Marvaud, J.C.; Popoff, M.R.; Kozaki, S.; Oguma, K. Identification and characterization of functional subunits of Clostridium botulinum type A progenitor toxin involved in binding to intestinal microvilli and erythrocytes. FEBS Lett. 2000, 467, 179–183. [Google Scholar] [CrossRef] [Green Version]
- Rummel, A.; Eichner, T.; Weil, T.; Karnath, T.; Gutcaits, A.; Mahrhold, S.; Sandhoff, K.; Proia, R.L.; Acharya, K.R.; Bigalke, H.; et al. Identification of the protein receptor binding site of botulinum neurotoxins B and G proves the double-receptor concept. Proc. Natl. Acad. Sci. USA 2007, 104, 359–364. [Google Scholar] [CrossRef] [Green Version]
- Sagane, Y.; Hayashi, S.; Akiyama, T.; Matsumoto, T.; Hasegawa, K.; Yamano, A.; Suzuki, T.; Niwa, K.; Watanabe, T.; Yajima, S. Conformational divergence in the HA-33/HA-17 trimer of serotype C and D botulinum toxin complex. Biochem. Biophys. Res. Commun. 2016, 476, 280–285. [Google Scholar] [CrossRef]
- Sugawara, Y.; Iwamori, M.; Matsumura, T.; Yutani, M.; Amatsu, S.; Fujinaga, Y. Clostridium botulinum type C hemagglutinin affects the morphology and viability of cultured mammalian cells via binding to the ganglioside GM3. FEBS J. 2015, 282, 3334–3347. [Google Scholar] [CrossRef] [Green Version]
- Hatcher, B.L.; Hale, J.Y.; Briles, D.E. Free Sialic Acid Acts as a Signal That Promotes Streptococcus pneumoniae Invasion of Nasal Tissue and Nonhematogenous Invasion of the Central Nervous System. Infect. Immun. 2016, 84, 2607–2615. [Google Scholar] [CrossRef] [Green Version]
- Coats, M.T.; Murphy, T.; Paton, J.C.; Gray, B.; Briles, D.E. Exposure of Thomsen-Friedenreich antigen in Streptococcus pneumoniae infection is dependent on pneumococcal neuraminidase A. Microb. Pathog. 2011, 50, 343–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byres, E.; Paton, A.W.; Paton, J.C.; Löfling, J.C.; Smith, D.F.; Wilce, M.C.J.; Talbot, U.M.; Chong, D.C.; Yu, H.; Huang, S.; et al. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature 2008, 456, 648–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seyahian, E.A.; Oltra, G.; Ochoa, F.; Melendi, S.; Hermes, R.; Paton, J.C.; Paton, A.W.; Lago, N.; Castro Parodi, M.; Damiano, A.; et al. Systemic effects of Subtilase cytotoxin produced by Escherichia coli O113:H21. Toxicon 2017, 127, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Tolia, N.H.; Enemark, E.J.; Sim, B.K.L.; Joshua-Tor, L. Structural Basis for the EBA-175 Erythrocyte Invasion Pathway of the Malaria Parasite Plasmodium falciparum. Cell 2005, 122, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Rydzak, J.; Kaczmarek, R.; Czerwinski, M.; Lukasiewicz, J.; Tyborowska, J.; Szewczyk, B.; Jaskiewicz, E. The Baculovirus-Expressed Binding Region of Plasmodium falciparum EBA-140 Ligand and Its Glycophorin C Binding Specificity. PLoS ONE 2015, 10, e0115437. [Google Scholar] [CrossRef] [Green Version]
- Zerka, A.; Kaczmarek, R.; Czerwinski, M.; Jaskiewicz, E. Plasmodium reichenowi EBA-140 merozoite ligand binds to glycophorin D on chimpanzee red blood cells, shedding new light on origins of Plasmodium falciparum. Parasites Vectors 2017, 10, 554. [Google Scholar] [CrossRef] [Green Version]
- Freire-de-Lima, L.; Oliveira, I.A.; Neves, J.L.; Penha, L.L.; Alisson-Silva, F.; Dias, W.B.; Todeschini, A.R. Sialic acid: A sweet swing between mammalian host and Trypanosoma cruzi. Front. Immunol. 2012, 3. [Google Scholar] [CrossRef] [Green Version]
- Kato, K.; Takegawa, Y.; Ralston, K.S.; Gilchrist, C.A.; Hamano, S.; Petri, W.A.; Shinohara, Y. Sialic acid-dependent attachment of mucins from three mouse strains to Entamoeba histolytica. Biochem. Biophys. Res. Commun. 2013, 436, 252–258. [Google Scholar] [CrossRef] [Green Version]
- Nishikawa, Y.; Ogiso, A.; Kameyama, K.; Nishimura, M.; Xuan, X.; Ikehara, Y. α2-3 Sialic acid glycoconjugate loss and its effect on infection with Toxoplasma parasites. Exp. Parasitol. 2013, 135, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Jilani, T.N.; Jamil, R.T.; Siddiqui, A.H. H1N1 Influenza; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Gamblin, S.J.; Skehel, J.J. Influenza Hemagglutinin and Neuraminidase Membrane Glycoproteins. J. Biol. Chem. 2010, 285, 28403–28409. [Google Scholar] [CrossRef] [Green Version]
- Overeem, N.J.; Hamming, P.H.E.; Grant, O.C.; Di Iorio, D.; Tieke, M.; Bertolino, M.C.; Li, Z.; Vos, G.; de Vries, R.P.; Woods, R.J.; et al. Hierarchical Multivalent Effects Control Influenza Host Specificity. ACS Cent. Sci. 2020, 6, 2311–2318. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.D.; Ross, T.M. H3N2 influenza viruses in humans: Viral mechanisms, evolution, and evaluation. Hum. Vaccines Immunother. 2018, 14, 1840–1847. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, T.; Takano, M.; Kurebayashi, Y.; Masuda, M.; Kawagishi, S.; Takaguchi, M.; Yamanaka, T.; Minami, A.; Otsubo, T.; Ikeda, K.; et al. N-Glycolylneuraminic Acid on Human Epithelial Cells Prevents Entry of Influenza A Viruses That Possess N-Glycolylneuraminic Acid Binding Ability. J. Virol. 2014, 88, 8445–8456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellebø, A.; Vilas, U.; Falk, K.; Vlasak, R. Infectious Salmon Anemia Virus Specifically Binds to and Hydrolyzes 4-O-Acetylated Sialic Acids. J. Virol. 2004, 78, 3055–3362. [Google Scholar] [CrossRef] [Green Version]
- Vilchez, R.A.; Butel, J.S. Emergent Human Pathogen Simian Virus 40 and Its Role in Cancer. Clin. Microbiol. Rev. 2004, 17, 495–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbone, M.; Gazdar, A.; Butel, J.S. SV40 and human mesothelioma. Transl. Lung Cancer Res. 2020, 9, 47–59. [Google Scholar] [CrossRef]
- Neu, U.; Woellner, K.; Gauglitz, G.; Stehle, T. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc. Natl. Acad. Sci. USA 2008, 105, 5219–5224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campanero-Rhodes, M.A.; Smith, A.; Chai, W.; Sonnino, S.; Mauri, L.; Childs, R.A.; Zhang, Y.; Ewers, H.; Helenius, A.; Imberty, A.; et al. N-Glycolyl GM1 Ganglioside as a Receptor for Simian Virus 40. J. Virol. 2007, 81, 12846–12858. [Google Scholar] [CrossRef] [Green Version]
- Rotondo, J.C.; Mazzoni, E.; Bononi, I.; Tognon, M.; Martini, F. Association Between Simian Virus 40 and Human Tumors. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Harapan, H.; Itoh, N.; Yufika, A.; Winardi, W.; Keam, S.; Te, H.; Megawati, D.; Hayati, Z.; Wagner, A.L.; Mudatsir, M. Coronavirus disease 2019 (COVID-19): A literature review. J. Infect. Public Health 2020, 13, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Hu, B.; Huang, X.; Chai, Y.; Zhou, D.; Wang, Y.; Shuai, H.; Yang, D.; Hou, Y.; Zhang, X.; et al. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat. Commun. 2021, 12, 134. [Google Scholar] [CrossRef]
- Sun, X.-L. The role of cell surface sialic acids for SARS-CoV-2 infection. Glycobiology 2021. [Google Scholar] [CrossRef]
- Huang, X.; Dong, W.; Milewska, A.; Golda, A.; Qi, Y.; Zhu, Q.K.; Marasco, W.A.; Baric, R.S.; Sims, A.C.; Pyrc, K.; et al. Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J. Virol. 2015, 89, 7202–7213. [Google Scholar] [CrossRef] [Green Version]
- Regl, G.; Kaser, A.; Iwersen, M.; Schmid, H.; Kohla, G.; Strobl, B.; Vilas, U.; Schauer, R.; Vlasak, R. The Hemagglutinin-Esterase of Mouse Hepatitis Virus Strain S is a Sialate-4-O-Acetylesterase. J. Virol. 1999, 73, 4721–4727. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, K.; Kasai, M.; Kato, S.; Kasai, H.; Hatakeyama, K. Haemagglutinin-esterase protein (HE) of murine corona virus: DVIMD (diarrhea virus of infant mice). Arch. Virol. 1998, 143, 1523–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawełczyk, M.; Kowalski, M.L. The Role of Human Parainfluenza Virus Infections in the Immunopathology of teh Respiratory Tract. Curr. Allergy Asthma Rep. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Farahmans, M.; Malekshahi, S.S.; Jebbari, M.R.; Shayestehpour, M. The landscape of extrapulmonay manifestations of human parainfluenza viruses: A systematic narrative review. Microbiol. Immunol. 2020, 65, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Branche, A.E.; Falsey, A.R. Parainfluenza Virus Infection. Semin. Respir. Crit. Care Med. 2016, 37, 538–554. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same species, different diseases: How and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spano, S.; Ugalde, J.E.; Galan, J.E. Delivery of a Salmonella Typhi Exotoxin from a Host Intracellular Compartment. Cell Host Microbe 2008, 3, 30–38. [Google Scholar] [CrossRef] [Green Version]
- Acheson, D.; Hohmann, E.L. Nontyphoidal Salmonellosis. Clin. Infect. Dis. 2001, 32, 263–269. [Google Scholar] [CrossRef]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [Green Version]
- Saitoh, M.; Tanaka, K.; Nishimori, K.; Makino, S.; Kanno, T.; Ishihara, R.; Hatama, S.; Kitano, R.; Kishima, M.; Sameshima, T.; et al. The artAB genes encode a putative ADP-ribosyltransferase toxin homologue associated with Salmonella enterica serovar Typhimurium DT104. Microbiology 2005, 151, 3089–3096. [Google Scholar] [CrossRef] [Green Version]
- Minami, A.; Ishibashi, S.; Ikeda, K.; Ishitsubo, E.; Hori, T.; Tokiwa, H.; Taguchi, R.; Ieno, D.; Otsubo, T.; Matsuda, Y.; et al. Catalytic preference of Salmonella typhimurium LT2 sialidase for N -acetylneuraminic acid residues over N-glycolylneuraminic acid residues. FEBS Open Bio 2013, 3, 231–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alisson-Silva, F.; Liu, J.Z.; Diaz, S.L.; Deng, L.; Gareau, M.G.; Marchelletta, R.; Chen, X.; Nizet, V.; Varki, N.; Barrett, K.E.; et al. Human evolutionary loss of epithelial Neu5Gc expression and species-specific susceptibility to cholera. PLoS Pathog. 2018, 14, e1007133. [Google Scholar] [CrossRef] [Green Version]
- Van den Broeck, D.; Horvath, C.; De Wolf, M.J.S. Vibrio cholerae: Cholera toxin. Int. J. Biochem. Cell Biol. 2007, 39, 1771–1775. [Google Scholar] [CrossRef]
- Alonso, A.; Rojo, F.; Martinez, J.L. Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ. Microbiol. 1999, 1, 421–430. [Google Scholar] [CrossRef]
- Breidenstein, E.B.M.; de la Fuente-Núñez, C.; Hancock, R.E.W. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [Green Version]
- Chua, S.L.; Yam, J.K.H.; Hao, P.; Adav, S.S.; Salido, M.M.; Liu, Y.; Givskov, M.; Sze, S.K.; Tolker-Nielsen, T.; Yang, L. Selective labelling and eradication of antibiotic-tolerant bacterial populations in Pseudomonas aeruginosa biofilms. Nat. Commun. 2016, 7, 10750. [Google Scholar] [CrossRef] [Green Version]
- Bucior, I.; Pielage, J.F.; Engel, J.N. Pseudomonas aeruginosa Pili and Flagella Mediate Distinct Binding and Signaling Events at the Apical and Basolateral Surface of Airway Epithelium. PLoS Pathog. 2012, 8, e1002616. [Google Scholar] [CrossRef] [Green Version]
- Lau, G.W.; Hassett, D.J.; Ran, H.; Kong, F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Microbiol. 2004, 10, 599–606. [Google Scholar] [CrossRef]
- Blier, A.-S.; Veron, W.; Bazire, A.; Gerault, E.; Taupin, L.; Vieillard, J.; Rehel, K.; Dufour, A.; Le Derf, F.; Orange, N.; et al. C-type natriuretic peptide modulates quorum sensing molecule and toxin production in Pseudomonas aeruginosa. Microbiology 2011, 157, 1929–1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef] [PubMed]
- Adamo, R.; Sokol, S.; Soong, G.; Gomez, M.I.; Prince, A. Pseudomonas aeruginosa Flagella Activate Airway Epithelial Cells through asialoGM1 and Toll-Like Receptor 2 as well as Toll-Like Receptor 5. Am. J. Respir. Cell Mol. Biol. 2004, 30, 627–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, S.K.; Ritchings, B.W.; Almira, E.C.; Lory, S.; Ramphal, R. The Pseudomonas aeruginosa Flagellar Cap Protein, FliD, is Responsible for Mucin Adhesion. Infect. Immun. 1998, 66, 1000–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plotkowski, M.-C.; Costa, A.O.; Morandi, V.; Barbosa, H.S.; Nader, H.B.; Bentzmann, S.D.E.; Puchelle, E. Role of heparan sulphate proteoglycans as potential receptors for non-piliated Pseudomonas aeruginosa adherence to non-polarised airway epithelial cells. Infect. Immun. 2001, 50, 183–190. [Google Scholar] [CrossRef] [Green Version]
- Bucior, I.; Mostov, K.; Engel, J.N. Pseudomonas Aeruginosa-Mediated Damage Requires Distinct Receptors at the Apical and Basolateral Surfaces of the Polarized Epithelium. Infect. Immun. 2010, 78, 939–953. [Google Scholar] [CrossRef] [Green Version]
- Plotkowski, M.-C.; Tournier, J.-M.; Puchelle, E. Pseudomonas aeruginosa Strains Possess Specific Adhesins for Laminin. Infect. Immun. 1996, 64, 600–605. [Google Scholar] [CrossRef] [Green Version]
- Paulsson, M.; Su, Y.-C.; Ringwood, T.; Uddén, F.; Riesbeck, K. Pseudomonas aeruginosa uses multiple receptors for adherence to laminin during infection of the respiratory tract and skin wounds. Sci. Rep. 2019, 9, 18168. [Google Scholar] [CrossRef]
- Leroy-Dudal, J.; Gagnière, H.; Cossard, E.; Carreiras, F.; Di Martino, P. Role of αvβ5 integrins and vitronectin in Pseudomonas aeruginosa PAK interaction with A549 respiratory cells. Microbes Infect. 2004, 6, 875–881. [Google Scholar] [CrossRef]
- Liu, W.; Sun, T.; Wang, Y. Integrin αvβ6 mediates epithelial-mesenchymal transition in human bronchial epithelial cells induced by lipopolysaccharides of Pseudomonas aeruginosa via TGF-β1-Smad2/3 signaling pathway. Folia Microbiol. 2020, 65, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Thuenauer, R.; Landi, A.; Trefzer, A.; Altmann, S.; Wehrum, S.; Eierhoff, T.; Diedrich, B.; Dengjel, J.; Nyström, A.; Imberty, A.; et al. The Pseudomonas aeruginosa Lectin LecB Causes Integrin Internalization and Inhibits Epithelial Wound Healing. mBio 2020, 11, e03260-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saiman, L.; Prince, A. Pseudomonas aeruginosa pili bind to asialoGM1 which is increased on the surface of cystic fibrosis epithelial cells. J. Clin. Investig. 1993, 92, 1875–1880. [Google Scholar] [CrossRef] [Green Version]
- Kidd, M.; Modlin, I.M. A Century of Helicobacter pylori. Digestion 1998, 59, 1–15. [Google Scholar] [CrossRef]
- Alfarouk, K.O.; Bashir, A.H.H.; Aljarbou, A.N.; Ramadan, A.M.; Muddathir, A.K.; AlHoufie, S.T.S.; Hifny, A.; Elhassan, G.O.; Ibrahim, M.E.; Alqahtani, S.S.; et al. The Possible Role of Helicobacter pylori in Gastric Cancer and Its Management. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Gu, H. Role of Flagella in the Pathogenesis of Helicobacter pylori. Curr. Microbiol. 2017, 74, 863–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zambelli, B.; Musiani, F.; Benini, S.; Ciurli, S. Chemistry of Ni 2+ in Urease: Sensing, Trafficking, and Catalysis. Acc. Chem. Res. 2011, 44, 520–530. [Google Scholar] [CrossRef] [PubMed]
- King, P. Haemophilus influenzae and the lung (Haemophilus and the lung). Clin. Transl. Med. 2012, 1. [Google Scholar] [CrossRef] [Green Version]
- Zarei, A.E.; Linjawi, M.H.; Redwan, E.M. Circulating innate and adaptive immunity against anti-Haemophilus influenzae type b. Hum. Antibodies 2019, 27, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Forstner, C.; Rohde, G.; Rupp, J.; Schuette, H.; Ott, S.R.; Hagel, S.; Harrison, N.; Thalhammer, F.; von Baum, H.; Suttorp, N.; et al. Community-acquired Haemophilus influenzae pneumonia—New insights from the CAPNETZ study. J. Infect. 2016, 72, 554–563. [Google Scholar] [CrossRef]
- Livorsi, D.J.; MacNeil, J.R.; Cohn, A.C.; Bareta, J.; Zansky, S.; Petit, S.; Gershman, K.; Harrison, L.H.; Lynfield, R.; Reingold, A.; et al. Invasive Haemophilus influenzae in the United States, 1999–2008: Epidemiology and outcomes. J. Infect. 2012, 65, 496–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanderson, A.R.; Leid, J.G.; Hunsaker, D. Bacterial Biofilms on the Sinus Mucosa of Human Subjects with Chronic Rhinosinusitis. Laryngoscope 2006, 116, 1121–1126. [Google Scholar] [CrossRef]
- Epling, J. Bacterial conjuctivitis. BMJ Clin. Evid. 2012, 2, 1–21. [Google Scholar]
- Hall-Stoodley, L.; Hu, F.Z.; Gieseke, A.; Nistico, L.; Nguyen, D.; Hayes, J.; Forbes, M.; Greenberg, D.P.; Dice, B.; Burrows, A.; et al. Direct Detection of Bacterial Biofilms on the Middle-Ear Mucosa of Children with Chronic Otitis Media. JAMA 2006, 296, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, N.; Bakaletz, L.O. Selective adherence of non-typeable Haemophilus influenzae(NTHi) to mucus or epithelial cells in the chinchilla Eustachian tube and middle ear. Microb. Pathog. 1996, 21, 343–356. [Google Scholar] [CrossRef]
- Reddy, M.; Murphy, T.; Faden, H.; Bernstein, J. Middle ear mucin glycoprotein: Purification and interaction with nontypable and. Otolaryngol. Head Neck Surg. 1997, 116, 175–180. [Google Scholar] [CrossRef]
- Hallström, T.; Singh, B.; Resman, F.M.; Blom, A.; Mörgelin, M.; Riesbeck, K. Haemophilus influenzae Protein E Binds to the Extracellular Matrix by Concurrently Interacting with Laminin and Vitronectin. J. Infect. Dis. 2011, 204, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-C.; Halang, P.; Fleury, C.; Jalalvand, F.; Mörgelin, M.; Riesbeck, K. Haemophilus Protein F Orthologs of Pathogens Infecting the Airways: Exploiting Host Laminin at Heparin-Binding Sites for Maximal Adherence to Epithelial Cells. J. Infect. Dis. 2017, 216, 1303–1307. [Google Scholar] [CrossRef]
- Jalalvand, F.; Su, Y.-C.; Mörgelin, M.; Brant, M.; Hallgren, O.; Westergren-Thorsson, G.; Singh, B.; Riesbeck, K. Haemophilus influenzae Protein F Mediates Binding to Laminin and Human Pulmonary Epithelial Cells. J. Infect. Dis. 2013, 207, 803–813. [Google Scholar] [CrossRef] [Green Version]
- St Geme, J.W.; Falkow, S.; Barenkamp, S.J. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA 1993, 90, 2875–2879. [Google Scholar] [CrossRef] [Green Version]
- Barenkamp, S.J.; St Geme, J.W. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 1996, 19, 1215–1223. [Google Scholar] [CrossRef]
- Gilsdorf, J.R.; McCrea, K.W.; Marrs, C.F. Role of pili in Haemophilus influenzae adherence and colonization. Infect. Immun. 1997, 65, 2997–3002. [Google Scholar] [CrossRef] [Green Version]
- Mu, X.-Q.; Egelman, E.H.; Bullitt, E. Structure and Function of Hib Pili from Haemophilus influenzae Type b. J. Bacteriol. 2002, 184, 4868–4874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clementi, C.F.; Håkansson, A.P.; Murphy, T.F. Internalization and Trafficking of Nontypeable Haemophilus influenzae in Human Respiratory Epithelial Cells and Roles of IgA1 Proteases for Optimal Invasion and Persistence. Infect. Immun. 2014, 82, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Morey, P.; Viadas, C.; Euba, B.; Hood, D.W.; Barberan, M.; Gil, C.; Grillo, M.J.; Bengoechea, J.A.; Garmendia, J. Relative Contributions of Lipooligosaccharide Inner and Outer Core Modifications to Nontypeable Haemophilus influenzae Pathogenesis. Infect. Immun. 2013, 81, 4100–4111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrillo-Marquez, M.A. Botulism. Pediatrics Rev. 2016, 37, 183–192. [Google Scholar] [CrossRef]
- Connan, C.; Popoff, M.R. Two-component systems and toxinogenesis regulation in Clostridium botulinum. Res. Microbiol. 2015, 166, 332–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosow, L.K.; Strober, J.B. Infant Botulism: Review and Clinical Update. Pediatric Neurol. 2015, 52, 487–492. [Google Scholar] [CrossRef]
- Yamashita, S.; Yoshida, H.; Uchiyama, N.; Nakakita, Y.; Nakakita, S.; Tonozuka, T.; Oguma, K.; Nishikawa, A.; Kamitori, S. Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides. FEBS Lett. 2012, 586, 2404–2410. [Google Scholar] [CrossRef] [Green Version]
- Henriques-Normark, B.; Tuomanen, E.I. The Pneumococcus: Epidemiology, Microbiology, and Pathogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a010215. [Google Scholar] [CrossRef]
- Engholm, D.H.; Kilian, M.; Goodsell, D.S.; Andersen, E.S.; Kjærgaard, R.S. A visual review of the human pathogen Streptococcus pneumoniae. FEMS Microbiol. Rev. 2017, 41, 854–879. [Google Scholar] [CrossRef]
- Mitchell, A.M.; Mitchell, T.J. Streptococcus pneumoniae: Virulence factors and variation. Clin. Microbiol. Infect. 2010, 16, 411–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaper, J.B. Pathogenic Escherichia coli. Int. J. Med. Microbiol. 2005, 295, 355–356. [Google Scholar] [CrossRef]
- Figler, H.M.; Dudley, E.G. The interplay of Escherichia coli O157:H7 and commensal E. coli: The importance of strain-level identification. Expert Rev. Gastroenterol. Hepatol. 2016, 10, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Capitani, G.; Eidam, O.; Glockshuber, R.; Grütter, M.G. Structural and functional insights into the assembly of type 1 pili from Escherichia coli. Microbes Infect. 2006, 8, 2284–2290. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.W.; Kaijser, B. The local immune response to Escherichia coli O and K antigen in experimental pyelonephritis. J. Clin. Investig. 1976, 58, 276–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vo, J.L.; Martínez Ortiz, G.C.; Subedi, P.; Keerthikumar, S.; Mathivanan, S.; Paxman, J.J.; Heras, B. Autotransporter Adhesins in Escherichia coli Pathogenesis. Proteomics 2017, 17, 1600431. [Google Scholar] [CrossRef]
- Yu, F.; Mizushima, S. Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J. Bacteriol. 1982, 151, 718–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nhu, N.T.K.; Phan, M.-D.; Forde, B.M.; Murthy, A.M.V.; Peters, K.M.; Day, C.J.; Poole, J.; Kidd, T.J.; Welch, R.A.; Jennings, M.P.; et al. Complex Multilevel Control of Hemolysin Production by Uropathogenic Escherichia coli. mBio 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Molina, J.A.; Molina, I. Chagas disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef]
- Giorgi, M.E.; de Lederkremer, R.M. Trans-sialidase and mucins of Trypanosoma cruzi: An important interplay for the parasite. Carbohydr. Res. 2011, 346, 1389–1393. [Google Scholar] [CrossRef]
- Ruiz Díaz, P.; Mucci, J.; Meira, M.A.; Bogliotti, Y.; Musikant, D.; Leguizamón, M.S.; Campetella, O. Trypanosoma cruzi trans -Sialidase Prevents Elicitation of Th1 Cell Response via Interleukin 10 and Downregulates Th1 Effector Cells. Infect. Immun. 2015, 83, 2099–2108. [Google Scholar] [CrossRef] [Green Version]
- Agustí, R.; Giorgi, M.E.; de Lederkremer, R.M. The trans-sialidase from Trypanosoma cruzi efficiently transfers α-(2→3)-linked N-glycolylneuraminic acid to terminal β-galactosyl units. Carbohydr. Res. 2007, 342, 2465–2469. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Simpson, A.G.B.; Lane, C.E.; Lukeš, J.; Bass, D.; Bowser, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The Revised Classification of Eukaryotes. J. Eukaryot. Microbiol. 2012, 59, 429–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betanzos, A.; Bañuelos, C.; Orozco, E. Host Invasion by Pathogenic Amoebae: Epithelial Disruption by Parasite Proteins. Genes 2019, 10, 618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marie, C.; Petri, W.A. Regulation of Virulence of Entamoeba histolytica. Annu. Rev. Microbiol. 2014, 68, 493–520. [Google Scholar] [CrossRef]
- Rawat, A.; Singh, P.; Jyoti, A.; Kaushik, S.; Srivastava, V.K. Averting transmission: A pivotal target to manage amoebiasis. Chem. Biol. Drug Des. 2020, 96, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Udezulu, I.A.; Leitch, G.J. A membrane-associated neuraminidase in Entamoeba histolytica trophozoites. Infect. Immun. 1987, 55, 181–186. [Google Scholar] [CrossRef] [Green Version]
- Nok, A.J.; Rivera, W. Characterization of sialidase from Entamoaeba hystolitica and possible pathogenic role in amebiasis. Parasitol. Res. 2003, 89, 302–307. [Google Scholar] [CrossRef]
- Kochanowsky, J.A.; Koshy, A.A. Toxoplasma gondii. Curr. Biol. 2018, 28, R770–R771. [Google Scholar] [CrossRef] [Green Version]
- Lima, T.S.; Lodoen, M.B. Mechanisms of Human Innate Immune Evasion by Toxoplasma gondii. Front. Cell. Infect. Microbiol. 2019, 9. [Google Scholar] [CrossRef]
- Baba, M.; Sato, M.; Kitoh, K.; Takashima, Y. The distribution pattern of α2,3- and α2,6-linked sialic acids affects host cell preference in Toxoplasma gondii. Exp. Parasitol. 2015, 155, 74–81. [Google Scholar] [CrossRef]
- Friedrich, N.; Santos, J.M.; Liu, Y.; Palma, A.S.; Leon, E.; Saouros, S.; Kiso, M.; Blackman, M.J.; Matthews, S.; Feizi, T.; et al. Members of a Novel Protein Family Containing Microneme Adhesive Repeat Domains Act as Sialic Acid-binding Lectins during Host Cell Invasion by Apicomplexan Parasites. J. Biol. Chem. 2010, 285, 2064–2076. [Google Scholar] [CrossRef] [Green Version]
- Yang, N.; Xing, M.; Ding, Y.; Wang, D.; Guo, X.; Sang, X.; Li, J.; Li, C.; Wang, Y.; Feng, Y.; et al. The Putative TCP-1 Chaperonin Is an Important Player Involved in Sialic Acid-Dependent Host Cell Invasion by Toxoplasma gondii. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Xing, M.; Yang, N.; Jiang, N.; Wang, D.; Sang, X.; Feng, Y.; Chen, R.; Wang, X.; Chen, Q. A Sialic Acid-Binding Protein SABP1 of Toxoplasma gondii Mediates Host Cell Attachment and Invasion. J. Infect. Dis. 2020, 222, 126–135. [Google Scholar] [CrossRef]
- Liu, W.; Li, Y.; Learn, G.H.; Rudicell, R.S.; Robertson, J.D.; Keele, B.F.; Ndjango, J.-B.N.; Sanz, C.M.; Morgan, D.B.; Locatelli, S.; et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature 2010, 467, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Prugnolle, F.; Durand, P.; Neel, C.; Ollomo, B.; Ayala, F.J.; Arnathau, C.; Etienne, L.; Mpoudi-Ngole, E.; Nkoghe, D.; Leroy, E.; et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 2010, 107, 1458–1463. [Google Scholar] [CrossRef] [Green Version]
- Gaur, D.; Chitnis, C.E. Molecular interactions and signaling mechanisms during erythrocyte invasion by malaria parasites. Curr. Opin. Microbiol. 2011, 14, 422–428. [Google Scholar] [CrossRef]
- Tham, W.-H.; Healer, J.; Cowman, A.F. Erythrocyte and reticulocyte binding-like proteins of Plasmodium falciparum. Trends Parasitol. 2012, 28, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.H.; Blair, P.L.; Kaneko, O.; Peterson, D.S. An expanding ebl family of Plasmodium falciparum. Trends Parasitol. 2001, 17, 297–299. [Google Scholar] [CrossRef]
- Wanaguru, M.; Crosnier, C.; Johnson, S.; Rayner, J.C.; Wright, G.J. Biochemical Analysis of the Plasmodium falciparum Erythrocyte-binding Antigen-175 (EBA175)-Glycophorin-A Interaction. J. Biol. Chem. 2013, 288, 32106–32117. [Google Scholar] [CrossRef] [Green Version]
- Salinas, N.D.; Paing, M.M.; Tolia, N.H. Critical Glycosylated Residues in Exon Three of Erythrocyte Glycophorin a Engage Plasmodium falciparum EBA-175 and Define Receptor Specificity. mBio 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.K.; Triglia, T.; Reed, M.B.; Cowman, A.F. A novel ligand from Plasmodium falciparum that binds to a sialic acid-containing receptor on the surface of human erythrocytes. Mol. Microbiol. 2001, 41, 47–58. [Google Scholar] [CrossRef]
- Narum, D.L.; Fuhrmann, S.R.; Luu, T.; Sim, B.K.L. A novel Plasmodium falciparum erythrocyte binding protein-2 (EBP2/BAEBL) involved in erythrocyte receptor binding. Mol. Biochem. Parasitol. 2002, 119, 159–168. [Google Scholar] [CrossRef]
- Gilberger, T.-W.; Thompson, J.K.; Triglia, T.; Good, R.T.; Duraisingh, M.T.; Cowman, A.F. A Novel Erythrocyte Binding Antigen-175 Paralogue from Plasmodium falciparum Defines a New Trypsin-resistant Receptor on Human Erythrocytes. J. Biol. Chem. 2003, 278, 14480–14486. [Google Scholar] [CrossRef] [Green Version]
- Lobo, C.-A.; Rodriguez, M.; Reid, M.; Lustigman, S. Glycophorin C is the receptor for the Plasmodium falciparum erythrocyte binding ligand PfEBP-2 (baebl). Blood 2003, 101, 4628–4631. [Google Scholar] [CrossRef]
- Maier, A.G.; Duraisingh, M.T.; Reeder, J.C.; Patel, S.S.; Kazura, J.W.; Zimmerman, P.A.; Cowman, A.F. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat. Med. 2003, 9, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Duriseti, S.; Sun, P.; Miller, L.H. Molecular basis of binding of the Plasmodium falciparum receptor BAEBL to erythrocyte receptor glycophorin C. Mol. Biochem. Parasitol. 2009, 168, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Jaskiewicz, E.; Peyrard, T.; Kaczmarek, R.; Zerka, A.; Jodlowska, M.; Czerwinski, M. The Gerbich blood group system: Old knowledge, new importance. Transfus. Med. Rev. 2018, 32, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Mayer, D.C.G.; Jiang, L.; Achur, R.N.; Kakizaki, I.; Gowda, D.C.; Miller, L.H. The glycophorin C N-linked glycan is a critical component of the ligand for the Plasmodium falciparum erythrocyte receptor BAEBL. Proc. Natl. Acad. Sci. USA 2006, 103, 2358–2362. [Google Scholar] [CrossRef] [Green Version]
- Ashline, D.J.; Duk, M.; Lukasiewicz, J.; Reinhold, V.N.; Lisowska, E.; Jaskiewicz, E. The structures of glycophorin C N-glycans, a putative component of the GPC receptor site for Plasmodium falciparum EBA-140 ligand. Glycobiology 2015, 25, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.H.; Malpede, B.M.; Batchelor, J.D.; Tolia, N.H. Crystal and Solution Structures of Plasmodium falciparum Erythrocyte-binding Antigen 140 Reveal Determinants of Receptor Specificity during Erythrocyte Invasion. J. Biol. Chem. 2012, 287, 36830–36836. [Google Scholar] [CrossRef] [Green Version]
- Malpede, B.M.; Lin, D.H.; Tolia, N.H. Molecular Basis for Sialic Acid-dependent Receptor Recognition by the Plasmodium falciparum Invasion Protein Erythrocyte-binding Antigen-140/BAEBL. J. Biol. Chem. 2013, 288, 12406–12415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaskiewicz, E.; Jodłowska, M.; Kaczmarek, R.; Zerka, A. Erythrocyte glycophorins as receptors for Plasmodium merozoites. Parasites Vectors 2019, 12, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNicholas, S.; Potterton, E.; Wilson, K.S.; Noble, M.E. Presenting Your Structures: The CCP4mg Molecular-Graphics Software. Acta Crystallogr. Sect. D 2011, 67 Pt. 4, 386–394. [Google Scholar] [CrossRef] [Green Version]
- Rayner, J.C.; Huber, C.S.; Barnwell, J.W. Conservation and divergence in erythrocyte invasion ligands: Plasmodium reichenowi EBL genes. Mol. Biochem. Parasitol. 2004, 138, 243–247. [Google Scholar] [CrossRef]
- Martin, M.J.; Rayner, J.C.; Gagneux, P.; Barnwell, J.W.; Varki, A. Evolution of human-chimpanzee differences in malaria susceptibility: Relationship to human genetic loss of N-glycolylneuraminic acid. Proc. Natl. Acad. Sci. USA 2005, 102, 12819–12824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanaguru, M.; Liu, W.; Hahn, B.H.; Rayner, J.C.; Wright, G.J. RH5-Basigin interaction plays a major role in the host tropism of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 2013, 110, 20735–20740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dankwa, S.; Lim, C.; Bei, A.K.; Jiang, R.H.Y.; Abshire, J.R.; Patel, S.D.; Goldberg, J.M.; Moreno, Y.; Kono, M.; Niles, J.C.; et al. Ancient human sialic acid variant restricts an emerging zoonotic malaria parasite. Nat. Commun. 2016, 7, 11187. [Google Scholar] [CrossRef] [Green Version]
- Proto, W.R.; Siegel, S.V.; Dankwa, S.; Liu, W.; Kemp, A.; Marsden, S.; Zenonos, Z.A.; Unwin, S.; Sharp, P.M.; Wright, G.J.; et al. Adaptation of Plasmodium falciparum to humans involved the loss of an ape-specific erythrocyte invasion ligand. Nat. Commun. 2019, 10, 4512. [Google Scholar] [CrossRef] [Green Version]


| Pathogen/Toxin | Disease | Ligand on Pathogen | Type of Sialic AcidBound/Incorporated | Symptoms in Humans | References |
|---|---|---|---|---|---|
| Avian influenza virus A | Flu | HA | α-2,3-Neu5Ac | Mild symptoms, fever, cough, sore throat, muscle anches | Van de Sandt et al., 2015 [50] |
| Human influenza virus A and B | Flu | HA | α-2,6-Neu5Ac/Neu5Gc 1 | Fever, cough, runny or stuffy nose, tiredness, sore throat, muscle anches | de Graaf and Fouchier 2014 [51] |
| Human influenza virus C | Flu | HEF | O-acetyl form of α-2,3- or α-2,6-Neu5Ac | Mild cough, fever, malaise | Wang and Veit 2016 [52] |
| Human influenza virus D | Flu in animals | HEF | O-acetyl form of α-2,3- or α-2,6-Neu5Ac | No symptoms detected in humans | Su et al., 2017 [53] |
| SV40 virus | Carcinogenic in humans | VP1 | NeuAc (weak binding in humans) and NeuGc (strong binding in simians) on GM1 glycolipid | Mesothelioma, osteosarcoma, pediatric and adult brain tumors and non-Hodgkin lymphomas | Shah 2004 [54] |
| Human betacoronaviruses - OC43 and HKU1 - MERS | Colds, human pulmonary infectious, lethal encephalitis (only OC43) Middle East respiratory syndrome | Spike protein | 9-O-acetylated Neu5Ac α-2,3-Neu5Ac | Fever, weakness, abdominal pain, rhinitis, and sore throat, sometimes severe pneumonia (HKU1), vomiting, diarrhea, abdominal pain (OC43) Fever, cough, breath shortness, pneumonia, sometimes diarrhea | Hulswit et al., 2018 [55] Li et al., 2017 [56] |
| Human parainfluenza viruses | Respiratory tract infections | HN | α-2,3-Neu5Ac α-2,6-Sias (HPIV-3) | cold-like, bronchiolitis, tracheobronchitis, laryngotracheobronchitis, pneumonia | Amonsen et al., 2007 [57]; Suzuki et al., 2001 [58]; Zhang et al., 2005 [59] |
| Salmonella enterica - S. Typhi - S. Typhimurium (LT2) | Typhoid fever Bacteremia and acute gastroenteritis | B subunit of toxin | α-2,3-Neu5Ac α-2,3-Neu5Ac > α-2,6-Neu5Ac and low ability to Neu5Gc | Fever, headache, dry cough, loss of appetite fever, nausea, vomiting, diarrhea diseases | Gao et al., 2017 [60] Kappala et al., 2018 [61] |
| Vibrio cholerae | Cholera | B subunit | Neu5Ac and Neu5Gc on GM1 | Watery diarrhea, vomiting, rapid heart rate, low blood pressure | Muanprasat and Chatsudthipong 2013 [62] |
| Pseudomonas aeruginosa | Pneumonia, meningoencephalitis and sepsis | Pili and flagellum Flagellum | asialo-GM1 and asialo-GM2 with GalNAcβ1-4Gal moiety sialyl-Lex epitope with terminal α-2,3-Neu5Ac | Pneumonia and urinary tracts-related symptoms, such as cough, fever, difficulty breathing, swelling | Bassetti et al., 2018 [63]; Krivan et al., 1988 [64]; Xia et al., 2006 [65] |
| Helicobacter pylori | Ulcer, gastritis and gastric cancer | SabA adhesion HpaA | N-acetyllactosamine-based gangliosides with terminal α-2,3-linked Neu5Ac > Neu5Gc and α-2,6-linked Neu5Ac Sialic acid | Ache or burning pain in abdomen, nausea, loss of appetite, bloating | Evans et al., 1988 [66]; Mahdavi 2002 [67]; Roche et al., 2004 [68]; Benktander et al., 2018 [69]; Bennett and Roberts 2005 [70]; Carlsohn et al., 2006 [71] |
| Haemophilus influenzae | Respiratory tract infections: pneumonia, sinusitis; conjunctivitis and otitis media | Lipooligosaccharide (LOS) | Incorporation of α-2,6-Neu5Ac (from human host) and/or dietary Neu5Gc into LOS | Fever, chills, diarrhea, anxiety, difficulty breathing, cough, muscle pain | Ng et al., 2019 [72]; Greiner et al., 2004 [73]; Johnston et al., 2008 [74] |
| Clostridium botulinum | Botulism | Botulin/HA complex | Gangliosides, prefer α-2,3-Neu5Ac > α-2,6-Neu5Ac | Double and blurred vision, dropping eyelids, difficulty swallowing, dry mouth, a thick-feeling tongue | Fujinaga et al., 2000 [75]; Rummel et al., 2007 [76]; Sagane et al., 2016 [77]; Sugawara et al., 2015 [78] |
| Streptococcus pneumoniae | pneumonia, otitis media, sinusitis, meningitis and bronchitis | Sialidases: NanA-C | Cleavage of α-2,3- and α-2,6-Neu5Ac | Fever, cough, shortness of breath, increased sensitivity to light, ear pain, hearing loss | Hatcher et al., 2016 [79]; Coats et al., 2011 [80] |
| SubAB toxin produced by Shiga toxin-produced Escherichia coli | Hemolytic-uremic syndrome (HUS) | B subunit | α-2,3-Neu5Gc > α-2,3-Neu5Ac > α-2,6-Neu5Gc | Extreme fatigue, decreased urination or/and blood in the urine, swelling of the legs, feet, ankles, high blood pressure | Byres et al., 2008 [81]; Seyahian et al., 2017 [82] |
| Plasmodium falciparum | Malaria | EBA-175, EBA-140 | α-2,3-Neu5Ac on GPA and GPC, respectively | Fever, headache, vomiting, tiredness | Tolia et al., 2005 [83]; Rydzak et al., 2015 [84] |
| Plasmodium reichenowi | Malaria in chimpanzee | EBA-140 | Neu5Gc on chimpanzee GPD | Malaria-like symptoms | Zerka et al., 2017 [85] |
| Trypanosoma cruzi | Chagas disease | No ligand, trans-sialidase | Trans-sialidase transfers α-2,3-linked Sias from host to surface mucin-like glycoproteins | Fever, malaise, headache, enlargement of the liver, spleen, lymph nodes | Freire-de-Lima et al., 2012 [86] |
| Entamoeba histolytica | Amoebiasis | Sialoglycoproteins | α-2,3-Neu5Ac | Stomach pain and cramping, loose feces, in severe form bloody feces and fever | Kato et al., 2013 [87] |
| Toxoplasma gondii | Toxoplasmosis | TgMIC1 TgMIC13 | α-2,3-linked Sias 4-O-acetylated Sias and α-2,9-linked Sias | Influenza-like symptoms with swollen lymph glands or muscle aches, damage of brain or eye, reduced vision | Nishikawa et al., 2013 [88] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burzyńska, P.; Sobala, Ł.F.; Mikołajczyk, K.; Jodłowska, M.; Jaśkiewicz, E. Sialic Acids as Receptors for Pathogens. Biomolecules 2021, 11, 831. https://doi.org/10.3390/biom11060831
Burzyńska P, Sobala ŁF, Mikołajczyk K, Jodłowska M, Jaśkiewicz E. Sialic Acids as Receptors for Pathogens. Biomolecules. 2021; 11(6):831. https://doi.org/10.3390/biom11060831
Chicago/Turabian StyleBurzyńska, Patrycja, Łukasz F. Sobala, Krzysztof Mikołajczyk, Marlena Jodłowska, and Ewa Jaśkiewicz. 2021. "Sialic Acids as Receptors for Pathogens" Biomolecules 11, no. 6: 831. https://doi.org/10.3390/biom11060831
APA StyleBurzyńska, P., Sobala, Ł. F., Mikołajczyk, K., Jodłowska, M., & Jaśkiewicz, E. (2021). Sialic Acids as Receptors for Pathogens. Biomolecules, 11(6), 831. https://doi.org/10.3390/biom11060831