Integrated Multi-Class Classification and Prediction of GPCR Allosteric Modulators by Machine Learning Intelligence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection and Preparation
2.2. Molecular Fingerprint and Descriptor Calculation
2.3. Model Building
2.4. Chemical Space Analysis
2.5. Model Evaluation
3. Results and Discussion
3.1. Overall Workflow
3.2. Data Set Analysis
3.3. Performance Evaluation on Different Feature Types
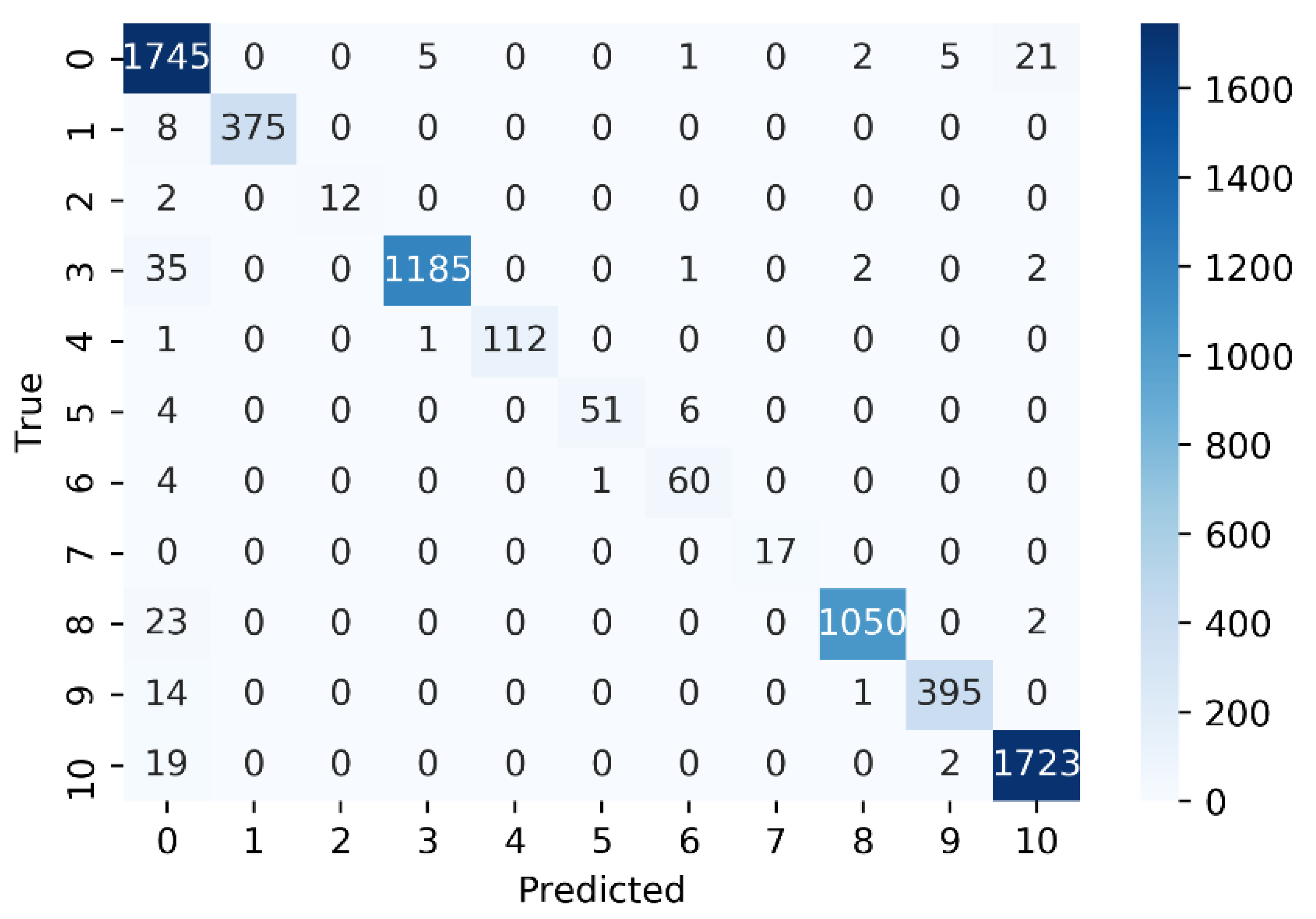
3.4. Performance Evaluation on Individual GPCR Classes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ritter, S.L.; Hall, R.A. Fine-tuning of GPCR activity by receptor-interacting proteins. Nat. Rev. Mol. Cell Biol. 2009, 10, 819–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raschka, S.; Kaufman, B. Machine learning and AI-based approaches for bioactive ligand discovery and GPCR-ligand recognition. Methods 2020, 180, 89–110. [Google Scholar] [CrossRef]
- Congreve, M.; de Graaf, C.; Swain, N.A.; Tate, C.G. Impact of GPCR Structures on Drug Discovery. Cell 2020, 181, 81–91. [Google Scholar] [CrossRef]
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schioth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Bridges, T.M.; Lindsley, C.W. G-protein-coupled receptors: From classical modes of modulation to allosteric mechanisms. ACS Chem. Biol. 2008, 3, 530–541. [Google Scholar] [CrossRef]
- Feng, Z.; Hu, G.; Ma, S.; Xie, X.-Q. Computational Advances for the Development of Allosteric Modulators and Bitopic Ligands in G Protein-Coupled Receptors. AAPS J. 2015, 17, 1080–1095. [Google Scholar] [CrossRef] [Green Version]
- Sloop, K.W.; Emmerson, P.J.; Statnick, M.A.; Willard, F.S. The current state of GPCR-based drug discovery to treat metabolic disease. Br. J. Pharmacol. 2018, 175, 4060–4071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conn, P.J.; Lindsley, C.W.; Meiler, J.; Niswender, C.M. Opportunities and challenges in the discovery of allosteric modulators of GPCRs for treating CNS disorders. Nat. Rev. Drug Discov. 2014, 13, 692–708. [Google Scholar] [CrossRef] [Green Version]
- Leach, K.; Sexton, P.M.; Christopoulos, A. Allosteric GPCR modulators: Taking advantage of permissive receptor pharmacology. Trends Pharmacol. Sci. 2007, 28, 382–389. [Google Scholar] [CrossRef]
- Lindsley, C.W.; Emmitte, K.A.; Hopkins, C.R.; Bridges, T.M.; Gregory, K.J.; Niswender, C.M.; Conn, P.J. Practical Strategies and Concepts in GPCR Allosteric Modulator Discovery: Recent Advances with Metabotropic Glutamate Receptors. Chem. Rev. 2016, 116, 6707–6741. [Google Scholar] [CrossRef] [Green Version]
- Nickols, H.H.; Conn, P.J. Development of allosteric modulators of GPCRs for treatment of CNS disorders. Neurobiol. Dis. 2014, 61, 55–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bian, Y.; Jun, J.J.; Cuyler, J.; Xie, X.-Q. Covalent allosteric modulation: An emerging strategy for GPCRs drug discovery. Eur. J. Med. Chem. 2020, 206, 112690. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Jing, Y.; Wang, L.; Ma, S.; Jun, J.J.; Xie, X.-Q. Prediction of Orthosteric and Allosteric Regulations on Cannabinoid Receptors Using Supervised Machine Learning Classifiers. Mol. Pharm. 2019, 16, 2605–2615. [Google Scholar] [CrossRef] [PubMed]
- Laprairie, R.B.; Bagher, A.M.; Kelly, M.E.; Denovan-Wright, E.M. Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor. Br. J. Pharmacol. 2015, 172, 4790–4805. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Tong, J.; Ding, K.; Zhou, Q.; Zhao, S. GPCR Allosteric Modulator Discovery. Adv. Exp. Med. Biol. 2019, 1163, 225–251. [Google Scholar] [CrossRef]
- Schneider, P.; Schneider, G. De Novo Design at the Edge of Chaos. J. Med. Chem. 2016, 59, 4077–4086. [Google Scholar] [CrossRef]
- Zhang, R.; Xie, X. Tools for GPCR drug discovery. Acta Pharmacol. Sin. 2012, 33, 372–384. [Google Scholar] [CrossRef] [Green Version]
- Finak, G.; Gottardo, R. Promises and Pitfalls of High-Throughput Biological Assays. Methods Mol. Biol. 2016, 1415, 225–243. [Google Scholar] [CrossRef]
- Evers, A.; Klabunde, T. Structure-based drug discovery using GPCR homology modeling: Successful virtual screening for antagonists of the alpha1A adrenergic receptor. J. Med. Chem. 2005, 48, 1088–1097. [Google Scholar] [CrossRef]
- Liu, L.; Jockers, R. Structure-Based Virtual Screening Accelerates GPCR Drug Discovery. Trends Pharmacol. Sci. 2020, 41, 382–384. [Google Scholar] [CrossRef]
- Petrucci, V.; Chicca, A.; Glasmacher, S.; Paloczi, J.; Cao, Z.; Pacher, P.; Gertsch, J. Pepcan-12 (RVD-hemopressin) is a CB2 receptor positive allosteric modulator constitutively secreted by adrenals and in liver upon tissue damage. Sci. Rep. 2017, 7, 9560. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Duffy, R.A.; Boykow, G.C.; Chackalamannil, S.; Madison, V.S. Identification of novel cannabinoid CB1 receptor antagonists by using virtual screening with a pharmacophore model. J. Med. Chem. 2008, 51, 2439–2446. [Google Scholar] [CrossRef] [PubMed]
- Bemister-Buffington, J.; Wolf, A.J.; Raschka, S.; Kuhn, L.A. Machine Learning to Identify Flexibility Signatures of Class A GPCR Inhibition. Biomolecules 2020, 10, 454. [Google Scholar] [CrossRef] [Green Version]
- Bian, Y.; Wang, J.; Jun, J.J.; Xie, X.-Q. Deep Convolutional Generative Adversarial Network (dcGAN) Models for Screening and Design of Small Molecules Targeting Cannabinoid Receptors. Mol. Pharm. 2019, 16, 4451–4460. [Google Scholar] [CrossRef]
- Liang, G.; Fan, W.; Luo, H.; Zhu, X. The emerging roles of artificial intelligence in cancer drug development and precision therapy. Biomed. Pharmacother 2020, 128, 110255. [Google Scholar] [CrossRef]
- Ma, C.; Wang, L.; Xie, X.-Q. Ligand Classifier of Adaptively Boosting Ensemble Decision Stumps (LiCABEDS) and its application on modeling ligand functionality for 5HT-subtype GPCR families. J. Chem. Inf. Model. 2011, 51, 521–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Wang, L.; Yang, P.; Myint, K.Z.; Xie, X.-Q. LiCABEDS II. Modeling of ligand selectivity for G-protein-coupled cannabinoid receptors. J. Chem. Inf. Model. 2013, 53, 11–26. [Google Scholar] [CrossRef] [Green Version]
- Reda, C.; Kaufmann, E.; Delahaye-Duriez, A. Machine learning applications in drug development. Comput. Struct. Biotechnol. J. 2020, 18, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Tsou, L.K.; Yeh, S.H.; Ueng, S.H.; Chang, C.P.; Song, J.S.; Wu, M.H.; Chang, H.F.; Chen, S.R.; Shih, C.; Chen, C.T.; et al. Comparative study between deep learning and QSAR classifications for TNBC inhibitors and novel GPCR agonist discovery. Sci. Rep. 2020, 10, 16771. [Google Scholar] [CrossRef]
- Bian, Y.; Xie, X.-Q. Generative chemistry: Drug discovery with deep learning generative models. J. Mol. Model. 2021, 27, 71. [Google Scholar] [CrossRef]
- Kumar, R.; Sharma, A.; Siddiqui, M.H.; Tiwari, R.K. Prediction of Human Intestinal Absorption of Compounds Using Artificial Intelligence Techniques. Curr. Drug Discov. Technol. 2017, 14, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Jacob, L.; Vert, J.P. Protein-ligand interaction prediction: An improved chemogenomics approach. Bioinformatics 2008, 24, 2149–2156. [Google Scholar] [CrossRef] [Green Version]
- AlQuraishi, M. AlphaFold at CASP13. Bioinformatics 2019, 35, 4862–4865. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Zidek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Shen, Q.; Wang, G.; Li, S.; Liu, X.; Lu, S.; Chen, Z.; Song, K.; Yan, J.; Geng, L.; Huang, Z.; et al. ASD v3.0: Unraveling allosteric regulation with structural mechanisms and biological networks. Nucleic Acids Res. 2016, 44, D527–D535. [Google Scholar] [CrossRef] [Green Version]
- Irwin, J.J.; Shoichet, B.K. ZINC--a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raymond, E.; Carhart, D.H.S.; Venkataraghavan, R. Atom pairs as molecular features in structure-activity studies: Definition and applications. J. Chem. Inf. Comput. Sci. 1985, 25, 64–73. [Google Scholar] [CrossRef]
- Durant, J.L.; Leland, B.A.; Henry, D.R.; Nourse, J.G. Reoptimization of MDL keys for use in drug discovery. J. Chem. Inf. Comput. Sci. 2002, 42, 1273–1280. [Google Scholar] [CrossRef] [Green Version]
- Rogers, D.; Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- Steinbeck, C.; Hoppe, C.; Kuhn, S.; Floris, M.; Guha, R.; Willighagen, E.L. Recent developments of the chemistry development kit (CDK)—An open-source java library for chemo- and bioinformatics. Curr. Pharm. Des. 2006, 12, 2111–2120. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Isa, N.A.M.; Mamat, W.M.F.W. Clustered-hybrid multilayer perceptron network for pattern recognition application. Appl. Soft Comput. 2011, 11, 1457–1466. [Google Scholar]
- Safavian, S.R.; Landgrebe, D. A survey of decision tree classifier methodology. IEEE Trans. Syst. Man Cybern. 1991, 21, 660–674. [Google Scholar] [CrossRef] [Green Version]
- Plewczynski, D.; von Grotthuss, M.; Rychlewski, L.; Ginalski, K. Virtual high throughput screening using combined random forest and flexible docking. Comb. Chem. High Throughput Screen. 2009, 12, 484–489. [Google Scholar] [CrossRef]
- Friedman, N.; Geiger, D.; Goldszmidt, M. Bayesian Network Classifiers. Mach. Learn. 1997, 29, 131–163. [Google Scholar] [CrossRef] [Green Version]
- Tolles, J.; Meurer, W.J. Logistic Regression: Relating Patient Characteristics to Outcomes. JAMA 2016, 316, 533–534. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Zhou, B.; Jin, W. Visualization of Single Cell RNA-Seq Data Using t-SNE in R. Methods Mol. Biol 2020, 2117, 159–167. [Google Scholar] [CrossRef]
- Carletta, J. Assessing Agreement on Classification Tasks: The Kappa Statistic. Comput. Linguist. 1996, 22, 249–254. [Google Scholar]
- Matthews, B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozymeBiochim. Biophys. Acta Protein Struct. 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Korkmaz, S. Deep Learning-Based Imbalanced Data Classification for Drug Discovery. J. Chem. Inf. Model. 2020, 60, 4180–4190. [Google Scholar] [CrossRef] [PubMed]

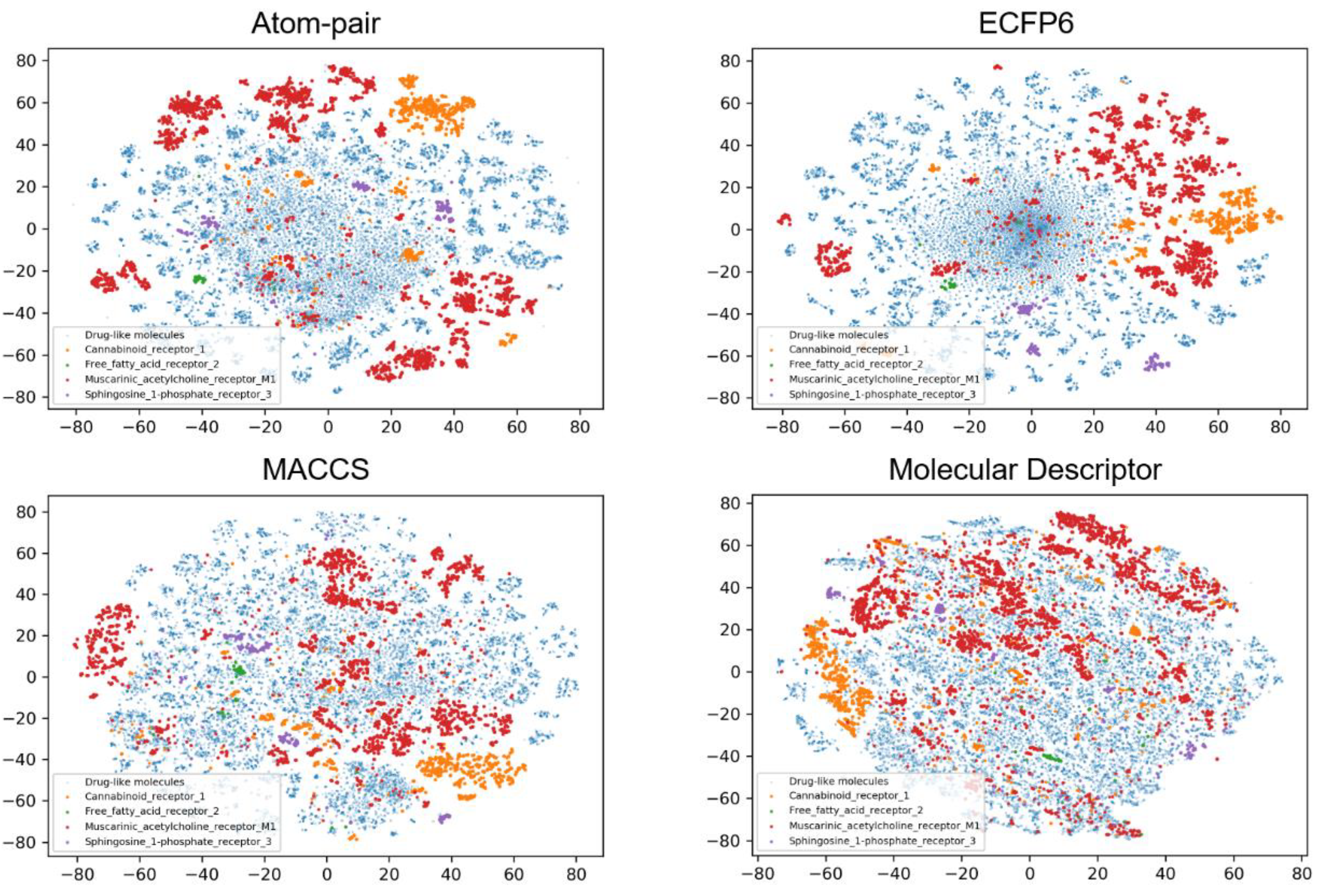
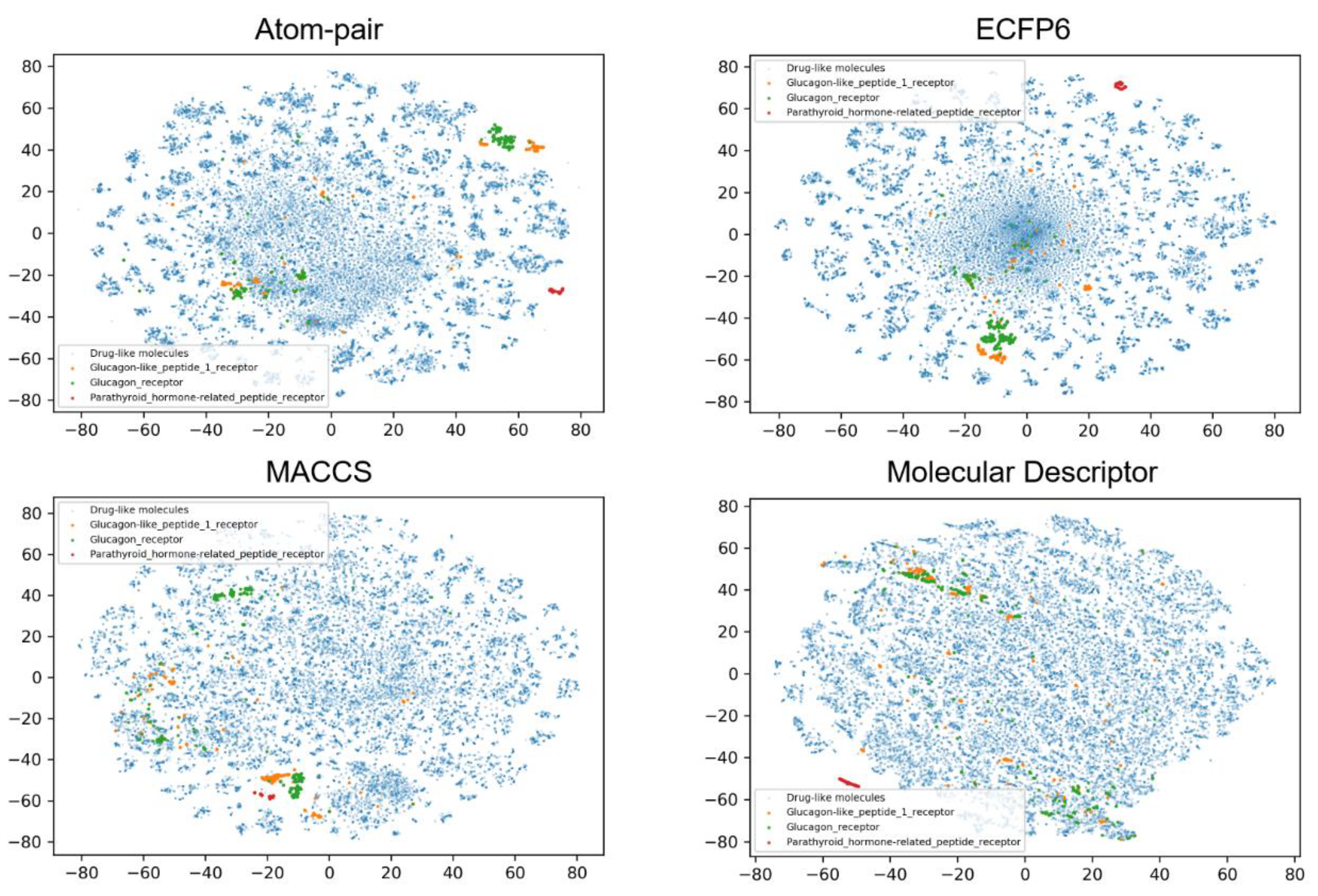
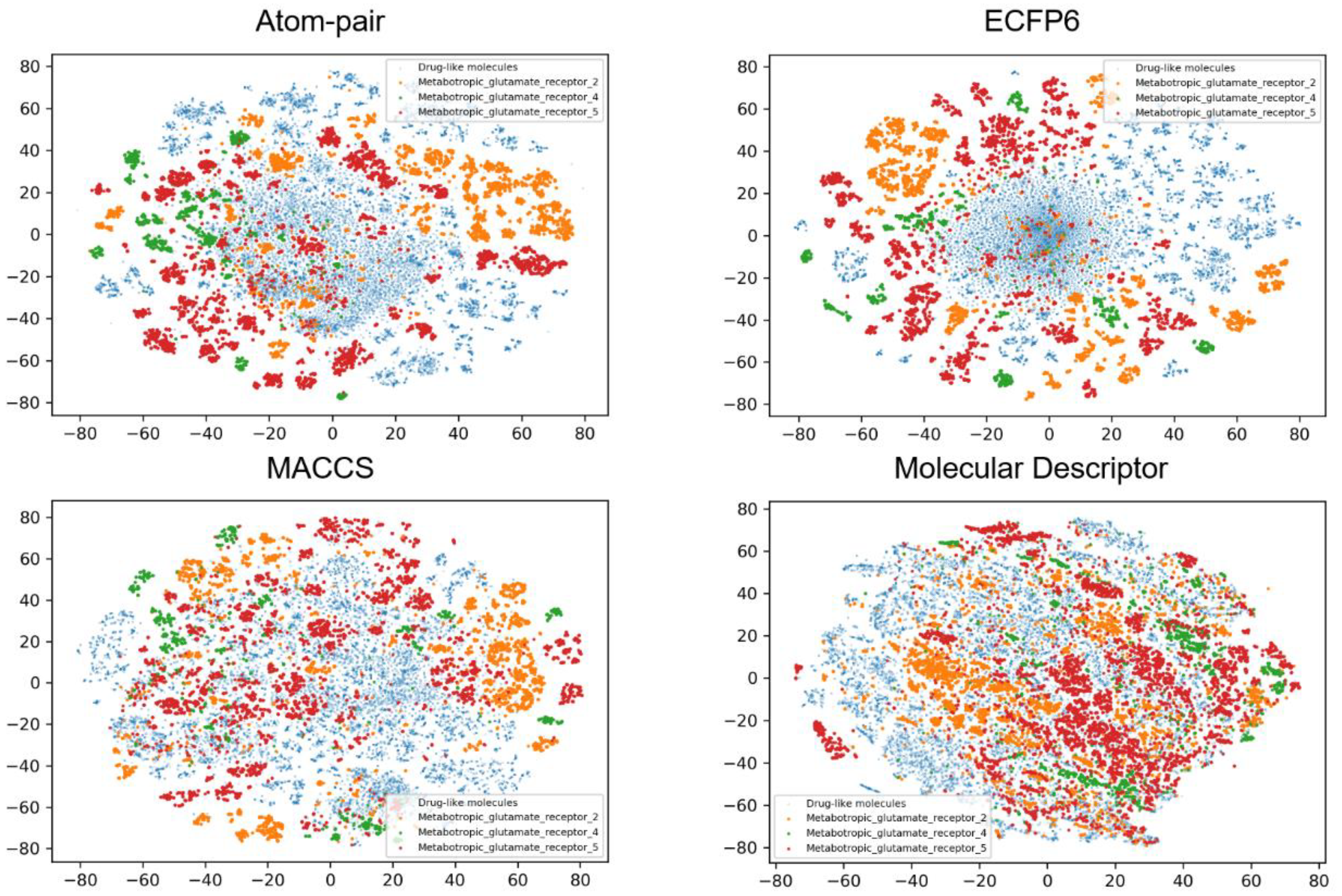
| Dataset | Training Set | Validation Set | Test Set | Total |
|---|---|---|---|---|
| Drug-like compounds | 5691 | 1423 | 1779 | 8893 |
| CB1—Cannabinoid receptor 1 (Class A) | 1242 | 311 | 383 | 1936 |
| FFA2—Free fatty acid receptor 2 (Class A) | 60 | 15 | 14 | 89 |
| mAchR M1—Muscarinic acetylcholine receptor M1 (Class A) | 4019 | 1005 | 1225 | 6249 |
| S1PR3—Sphingosine 1-phosphate receptor 3 (Class A) | 323 | 81 | 114 | 518 |
| GLP1-R—Glucagon-like peptide 1 receptor (Class B) | 202 | 51 | 61 | 314 |
| GCGR—Glucagon receptor (Class B) | 285 | 72 | 65 | 422 |
| PTHrP—Parathyroid hormone/parathyroid hormone-related peptide receptor (Class B) | 56 | 15 | 17 | 88 |
| mGlu2—Metabotropic glutamate receptor 2 (Class C) | 3519 | 880 | 1075 | 5474 |
| mGlu4—Metabotropic glutamate receptor 4 (Class C) | 1342 | 336 | 410 | 2088 |
| mGlu5—Metabotropic glutamate receptor 5 (Class C) | 5295 | 1324 | 1744 | 8363 |
| Datasets | Model | AUC | ACC | Bal_ACC | f1-Score | CK | MCC | Precision | Recall |
|---|---|---|---|---|---|---|---|---|---|
| Atom-pair | SVM | 0.966 | 0.972 | 0.936 | 0.949 | 0.965 | 0.965 | 0.965 | 0.936 |
| NB | 0.820 | 0.627 | 0.679 | 0.625 | 0.550 | 0.559 | 0.644 | 0.679 | |
| MLP | 0.960 | 0.954 | 0.925 | 0.933 | 0.943 | 0.943 | 0.944 | 0.925 | |
| LR | 0.942 | 0.912 | 0.895 | 0.907 | 0.891 | 0.891 | 0.922 | 0.895 | |
| RF | 0.942 | 0.956 | 0.890 | 0.925 | 0.946 | 0.946 | 0.971 | 0.890 | |
| DT | 0.861 | 0.770 | 0.748 | 0.743 | 0.715 | 0.716 | 0.743 | 0.748 | |
| ECFP6 | SVM | 0.974 | 0.976 | 0.950 | 0.963 | 0.971 | 0.971 | 0.978 | 0.950 |
| NB | 0.916 | 0.868 | 0.847 | 0.872 | 0.835 | 0.839 | 0.911 | 0.847 | |
| MLP | 0.957 | 0.958 | 0.919 | 0.931 | 0.948 | 0.948 | 0.948 | 0.919 | |
| LR | 0.955 | 0.940 | 0.916 | 0.932 | 0.925 | 0.925 | 0.954 | 0.916 | |
| RF | 0.946 | 0.964 | 0.897 | 0.931 | 0.956 | 0.956 | 0.980 | 0.897 | |
| DT | 0.896 | 0.822 | 0.812 | 0.789 | 0.780 | 0.780 | 0.771 | 0.812 | |
| MACCS | SVM | 0.961 | 0.963 | 0.926 | 0.934 | 0.954 | 0.954 | 0.944 | 0.926 |
| NB | 0.822 | 0.629 | 0.684 | 0.588 | 0.551 | 0.555 | 0.562 | 0.684 | |
| MLP | 0.954 | 0.940 | 0.914 | 0.915 | 0.925 | 0.925 | 0.918 | 0.914 | |
| LR | 0.898 | 0.839 | 0.815 | 0.834 | 0.800 | 0.800 | 0.857 | 0.815 | |
| RF | 0.945 | 0.961 | 0.895 | 0.924 | 0.951 | 0.951 | 0.962 | 0.895 | |
| DT | 0.890 | 0.854 | 0.796 | 0.791 | 0.820 | 0.820 | 0.790 | 0.796 | |
| Molecular Descriptors | SVM | 0.809 | 0.788 | 0.644 | 0.682 | 0.734 | 0.736 | 0.751 | 0.644 |
| NB | 0.824 | 0.618 | 0.690 | 0.576 | 0.538 | 0.541 | 0.531 | 0.690 | |
| MLP | 0.943 | 0.936 | 0.893 | 0.893 | 0.920 | 0.920 | 0.902 | 0.893 | |
| LR | 0.887 | 0.840 | 0.793 | 0.810 | 0.801 | 0.801 | 0.834 | 0.793 | |
| RF | 0.941 | 0.953 | 0.887 | 0.919 | 0.941 | 0.941 | 0.966 | 0.887 | |
| DT | 0.880 | 0.828 | 0.779 | 0.767 | 0.787 | 0.788 | 0.758 | 0.779 |
| Datasets | Model | AUC | ACC | Bal_ACC | f1-Score | CK | MCC | Precision | Recall |
|---|---|---|---|---|---|---|---|---|---|
| Atom-pair and Molecular Descriptors | SVM | 0.825 | 0.818 | 0.672 | 0.715 | 0.771 | 0.773 | 0.791 | 0.672 |
| NB | 0.834 | 0.660 | 0.704 | 0.667 | 0.588 | 0.597 | 0.688 | 0.704 | |
| MLP | 0.959 | 0.945 | 0.925 | 0.918 | 0.932 | 0.932 | 0.914 | 0.925 | |
| LR | 0.959 | 0.947 | 0.923 | 0.931 | 0.934 | 0.934 | 0.941 | 0.923 | |
| RF | 0.944 | 0.962 | 0.894 | 0.927 | 0.952 | 0.953 | 0.975 | 0.894 | |
| DT | 0.875 | 0.827 | 0.770 | 0.765 | 0.785 | 0.785 | 0.762 | 0.770 | |
| ECFP6 and Molecular Descriptors | SVM | 0.819 | 0.809 | 0.661 | 0.703 | 0.759 | 0.761 | 0.776 | 0.661 |
| NB | 0.922 | 0.871 | 0.859 | 0.875 | 0.838 | 0.841 | 0.901 | 0.859 | |
| MLP | 0.973 | 0.960 | 0.951 | 0.945 | 0.950 | 0.950 | 0.939 | 0.951 | |
| LR | 0.960 | 0.961 | 0.925 | 0.939 | 0.952 | 0.952 | 0.957 | 0.925 | |
| RF | 0.949 | 0.968 | 0.902 | 0.932 | 0.960 | 0.960 | 0.978 | 0.902 | |
| DT | 0.876 | 0.814 | 0.773 | 0.759 | 0.770 | 0.770 | 0.747 | 0.773 | |
| MACCS and Molecular Descriptors | SVM | 0.812 | 0.793 | 0.649 | 0.688 | 0.739 | 0.741 | 0.760 | 0.649 |
| NB | 0.853 | 0.659 | 0.743 | 0.621 | 0.588 | 0.592 | 0.579 | 0.743 | |
| MLP | 0.943 | 0.951 | 0.891 | 0.913 | 0.939 | 0.939 | 0.950 | 0.891 | |
| LR | 0.922 | 0.897 | 0.856 | 0.871 | 0.872 | 0.872 | 0.896 | 0.856 | |
| RF | 0.945 | 0.961 | 0.895 | 0.926 | 0.951 | 0.951 | 0.971 | 0.895 | |
| DT | 0.894 | 0.837 | 0.806 | 0.785 | 0.799 | 0.799 | 0.767 | 0.806 |
| Datasets | Model | AUC | ACC | Bal_ACC | f1-Score | CK | MCC | Precision | Recall |
|---|---|---|---|---|---|---|---|---|---|
| Atom-pair and ECFP6 and MACCS and Molecular Descriptors | SVM | 0.837 | 0.841 | 0.693 | 0.735 | 0.800 | 0.802 | 0.807 | 0.693 |
| NB | 0.884 | 0.776 | 0.792 | 0.778 | 0.725 | 0.729 | 0.800 | 0.792 | |
| MLP | 0.962 | 0.967 | 0.941 | 0.941 | 0.960 | 0.960 | 0.958 | 0.928 | |
| LR | 0.968 | 0.967 | 0.940 | 0.950 | 0.959 | 0.959 | 0.962 | 0.940 | |
| RF | 0.950 | 0.970 | 0.904 | 0.933 | 0.962 | 0.962 | 0.976 | 0.904 | |
| DT | 0.883 | 0.823 | 0.787 | 0.774 | 0.781 | 0.781 | 0.765 | 0.787 |
| Datasets | AUC | ACC | Bal_ACC | f1-Score | CK | MCC | Precision | Recall |
|---|---|---|---|---|---|---|---|---|
| Atom-pair | 0.915 | 0.865 | 0.846 | 0.847 | 0.835 | 0.837 | 0.865 | 0.846 |
| ECFP6 | 0.941 | 0.921 | 0.890 | 0.903 | 0.903 | 0.903 | 0.924 | 0.890 |
| MACCS | 0.912 | 0.864 | 0.838 | 0.831 | 0.834 | 0.834 | 0.839 | 0.838 |
| Molecular Descriptors | 0.881 | 0.827 | 0.781 | 0.775 | 0.787 | 0.788 | 0.790 | 0.781 |
| Atom-pair and Molecular Descriptors | 0.899 | 0.860 | 0.815 | 0.821 | 0.827 | 0.829 | 0.845 | 0.815 |
| ECFP6 and Molecular Descriptors | 0.917 | 0.897 | 0.845 | 0.859 | 0.872 | 0.872 | 0.883 | 0.845 |
| MACCS and Molecular Descriptors | 0.895 | 0.850 | 0.807 | 0.801 | 0.815 | 0.816 | 0.821 | 0.807 |
| Atom-pair and ECFP6 and MACCS and Molecular Descriptors | 0.914 | 0.891 | 0.843 | 0.852 | 0.865 | 0.866 | 0.878 | 0.841 |
| Datasets | Model | Drug-Like | CB1 | FFA2 | mAchR M1 | S1P3 | GLP1-R | GCGR | PTHrP | mGlu2 | mGlu4 | mGlu5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Atom-pair | SVM | 0.954 | 0.987 | 0.889 | 0.983 | 0.991 | 0.822 | 0.870 | 1.000 | 0.979 | 0.979 | 0.981 |
| NB | 0.609 | 0.683 | 0.373 | 0.715 | 0.720 | 0.456 | 0.673 | 0.919 | 0.676 | 0.387 | 0.661 | |
| MLP | 0.928 | 0.967 | 0.923 | 0.971 | 0.974 | 0.789 | 0.838 | 1.000 | 0.956 | 0.940 | 0.975 | |
| LR | 0.869 | 0.952 | 0.880 | 0.934 | 0.978 | 0.765 | 0.840 | 1.000 | 0.911 | 0.913 | 0.936 | |
| RF | 0.925 | 0.970 | 0.783 | 0.972 | 0.950 | 0.792 | 0.869 | 1.000 | 0.966 | 0.966 | 0.978 | |
| DT | 0.698 | 0.802 | 0.690 | 0.824 | 0.778 | 0.558 | 0.528 | 1.000 | 0.743 | 0.711 | 0.840 | |
| ECFP6 | SVM | 0.960 | 0.989 | 0.923 | 0.981 | 0.991 | 0.903 | 0.902 | 1.000 | 0.986 | 0.973 | 0.987 |
| NB | 0.813 | 0.812 | 0.800 | 0.901 | 0.944 | 0.785 | 0.871 | 1.000 | 0.892 | 0.868 | 0.906 | |
| MLP | 0.934 | 0.979 | 0.750 | 0.968 | 0.987 | 0.885 | 0.847 | 1.000 | 0.973 | 0.944 | 0.971 | |
| LR | 0.902 | 0.980 | 0.833 | 0.955 | 0.978 | 0.875 | 0.885 | 1.000 | 0.954 | 0.942 | 0.952 | |
| RF | 0.938 | 0.973 | 0.783 | 0.976 | 0.982 | 0.784 | 0.878 | 1.000 | 0.979 | 0.967 | 0.982 | |
| DT | 0.757 | 0.847 | 0.625 | 0.848 | 0.861 | 0.618 | 0.645 | 1.000 | 0.836 | 0.754 | 0.885 | |
| MACCS | SVM | 0.943 | 0.978 | 0.815 | 0.975 | 0.973 | 0.847 | 0.826 | 1.000 | 0.978 | 0.967 | 0.972 |
| NB | 0.612 | 0.648 | 0.275 | 0.665 | 0.719 | 0.317 | 0.438 | 1.000 | 0.695 | 0.428 | 0.670 | |
| MLP | 0.909 | 0.965 | 0.929 | 0.956 | 0.964 | 0.714 | 0.789 | 1.000 | 0.952 | 0.930 | 0.960 | |
| LR | 0.799 | 0.882 | 0.720 | 0.849 | 0.942 | 0.729 | 0.775 | 1.000 | 0.834 | 0.754 | 0.885 | |
| RF | 0.936 | 0.976 | 0.696 | 0.976 | 0.964 | 0.841 | 0.866 | 1.000 | 0.972 | 0.959 | 0.974 | |
| DT | 0.791 | 0.867 | 0.500 | 0.876 | 0.793 | 0.643 | 0.653 | 0.971 | 0.875 | 0.824 | 0.913 | |
| Molecular Descriptors | SVM | 0.759 | 0.812 | 0.000 | 0.836 | 0.865 | 0.465 | 0.566 | 1.000 | 0.787 | 0.568 | 0.839 |
| NB | 0.677 | 0.570 | 0.329 | 0.671 | 0.752 | 0.470 | 0.406 | 0.829 | 0.576 | 0.402 | 0.652 | |
| MLP | 0.909 | 0.938 | 0.733 | 0.960 | 0.965 | 0.752 | 0.759 | 1.000 | 0.949 | 0.908 | 0.955 | |
| LR | 0.828 | 0.880 | 0.583 | 0.881 | 0.900 | 0.766 | 0.705 | 1.000 | 0.838 | 0.669 | 0.860 | |
| RF | 0.925 | 0.969 | 0.727 | 0.971 | 0.959 | 0.811 | 0.870 | 1.000 | 0.966 | 0.943 | 0.969 | |
| DT | 0.784 | 0.846 | 0.483 | 0.856 | 0.787 | 0.626 | 0.653 | 0.941 | 0.823 | 0.753 | 0.886 |
| Datasets | Model | Drug-Like | CB1 | FFA2 | mAchR M1 | S1P3 | GLP1-R | GCGR | PTHrP | mGlu2 | mGlu4 | mGlu5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Atom-pair and Molecular Descriptors | SVM | 0.782 | 0.832 | 0.000 | 0.864 | 0.870 | 0.578 | 0.608 | 1.000 | 0.832 | 0.638 | 0.860 |
| NB | 0.648 | 0.699 | 0.550 | 0.742 | 0.755 | 0.510 | 0.706 | 0.919 | 0.725 | 0.393 | 0.693 | |
| MLP | 0.922 | 0.944 | 0.889 | 0.965 | 0.911 | 0.828 | 0.837 | 0.971 | 0.953 | 0.912 | 0.967 | |
| LR | 0.926 | 0.964 | 0.846 | 0.965 | 0.974 | 0.870 | 0.859 | 1.000 | 0.952 | 0.928 | 0.957 | |
| RF | 0.936 | 0.972 | 0.727 | 0.975 | 0.968 | 0.811 | 0.898 | 1.000 | 0.975 | 0.952 | 0.979 | |
| DT | 0.763 | 0.843 | 0.500 | 0.861 | 0.773 | 0.627 | 0.580 | 0.941 | 0.825 | 0.817 | 0.887 | |
| ECFP6 and Molecular Descriptors | SVM | 0.773 | 0.828 | 0.000 | 0.858 | 0.865 | 0.523 | 0.596 | 1.000 | 0.818 | 0.613 | 0.856 |
| NB | 0.827 | 0.829 | 0.800 | 0.904 | 0.964 | 0.796 | 0.894 | 1.000 | 0.885 | 0.822 | 0.905 | |
| MLP | 0.941 | 0.979 | 0.963 | 0.975 | 0.961 | 0.828 | 0.866 | 1.000 | 0.960 | 0.942 | 0.975 | |
| LR | 0.942 | 0.981 | 0.833 | 0.974 | 0.965 | 0.862 | 0.901 | 0.971 | 0.968 | 0.956 | 0.971 | |
| RF | 0.944 | 0.977 | 0.727 | 0.978 | 0.987 | 0.808 | 0.899 | 1.000 | 0.979 | 0.973 | 0.983 | |
| DT | 0.753 | 0.882 | 0.364 | 0.832 | 0.850 | 0.592 | 0.609 | 1.000 | 0.818 | 0.776 | 0.869 | |
| MACCS and Molecular Descriptors | SVM | 0.761 | 0.813 | 0.000 | 0.839 | 0.865 | 0.488 | 0.585 | 1.000 | 0.795 | 0.581 | 0.842 |
| NB | 0.673 | 0.645 | 0.371 | 0.718 | 0.786 | 0.377 | 0.480 | 0.971 | 0.694 | 0.428 | 0.690 | |
| MLP | 0.930 | 0.958 | 0.727 | 0.968 | 0.954 | 0.800 | 0.837 | 1.000 | 0.967 | 0.933 | 0.964 | |
| LR | 0.878 | 0.923 | 0.696 | 0.929 | 0.922 | 0.815 | 0.806 | 1.000 | 0.894 | 0.804 | 0.916 | |
| RF | 0.937 | 0.971 | 0.727 | 0.973 | 0.968 | 0.833 | 0.875 | 1.000 | 0.973 | 0.954 | 0.977 | |
| DT | 0.780 | 0.848 | 0.500 | 0.873 | 0.827 | 0.591 | 0.696 | 1.000 | 0.846 | 0.786 | 0.888 |
| Datasets | Model | Drug-Like | CB1 | FFA2 | mAchR M1 | S1P3 | GLP1-R | GCGR | PTHrP | mGlu2 | mGlu4 | mGlu5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Atom-pair and ECFP6 and MACCS and Molecular Descriptors | SVM | 0.804 | 0.852 | 0.000 | 0.889 | 0.900 | 0.578 | 0.621 | 1.000 | 0.860 | 0.702 | 0.877 |
| NB | 0.739 | 0.803 | 0.632 | 0.848 | 0.891 | 0.678 | 0.821 | 0.971 | 0.837 | 0.512 | 0.823 | |
| MLP | 0.953 | 0.981 | 0.800 | 0.982 | 0.991 | 0.865 | 0.870 | 1.000 | 0.970 | 0.964 | 0.975 | |
| LR | 0.953 | 0.979 | 0.880 | 0.981 | 0.974 | 0.881 | 0.894 | 1.000 | 0.972 | 0.961 | 0.973 | |
| RF | 0.948 | 0.979 | 0.727 | 0.979 | 0.982 | 0.833 | 0.882 | 1.000 | 0.981 | 0.973 | 0.985 | |
| DT | 0.753 | 0.869 | 0.500 | 0.849 | 0.830 | 0.596 | 0.617 | 1.000 | 0.841 | 0.775 | 0.882 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, T.; Bian, Y.; McGuire, T.; Xie, X.-Q. Integrated Multi-Class Classification and Prediction of GPCR Allosteric Modulators by Machine Learning Intelligence. Biomolecules 2021, 11, 870. https://doi.org/10.3390/biom11060870
Hou T, Bian Y, McGuire T, Xie X-Q. Integrated Multi-Class Classification and Prediction of GPCR Allosteric Modulators by Machine Learning Intelligence. Biomolecules. 2021; 11(6):870. https://doi.org/10.3390/biom11060870
Chicago/Turabian StyleHou, Tianling, Yuemin Bian, Terence McGuire, and Xiang-Qun Xie. 2021. "Integrated Multi-Class Classification and Prediction of GPCR Allosteric Modulators by Machine Learning Intelligence" Biomolecules 11, no. 6: 870. https://doi.org/10.3390/biom11060870
APA StyleHou, T., Bian, Y., McGuire, T., & Xie, X.-Q. (2021). Integrated Multi-Class Classification and Prediction of GPCR Allosteric Modulators by Machine Learning Intelligence. Biomolecules, 11(6), 870. https://doi.org/10.3390/biom11060870






