Lab-on-a-Chip Platforms as Tools for Drug Screening in Neuropathologies Associated with Blood–Brain Barrier Alterations
Abstract
:1. Introduction
2. LOC Materials and Manufacturing Advancements for Biomedical Research
2.1. PDMS LOCs
2.2. Glass LOCs
2.3. Biomedical Applications of LOCs
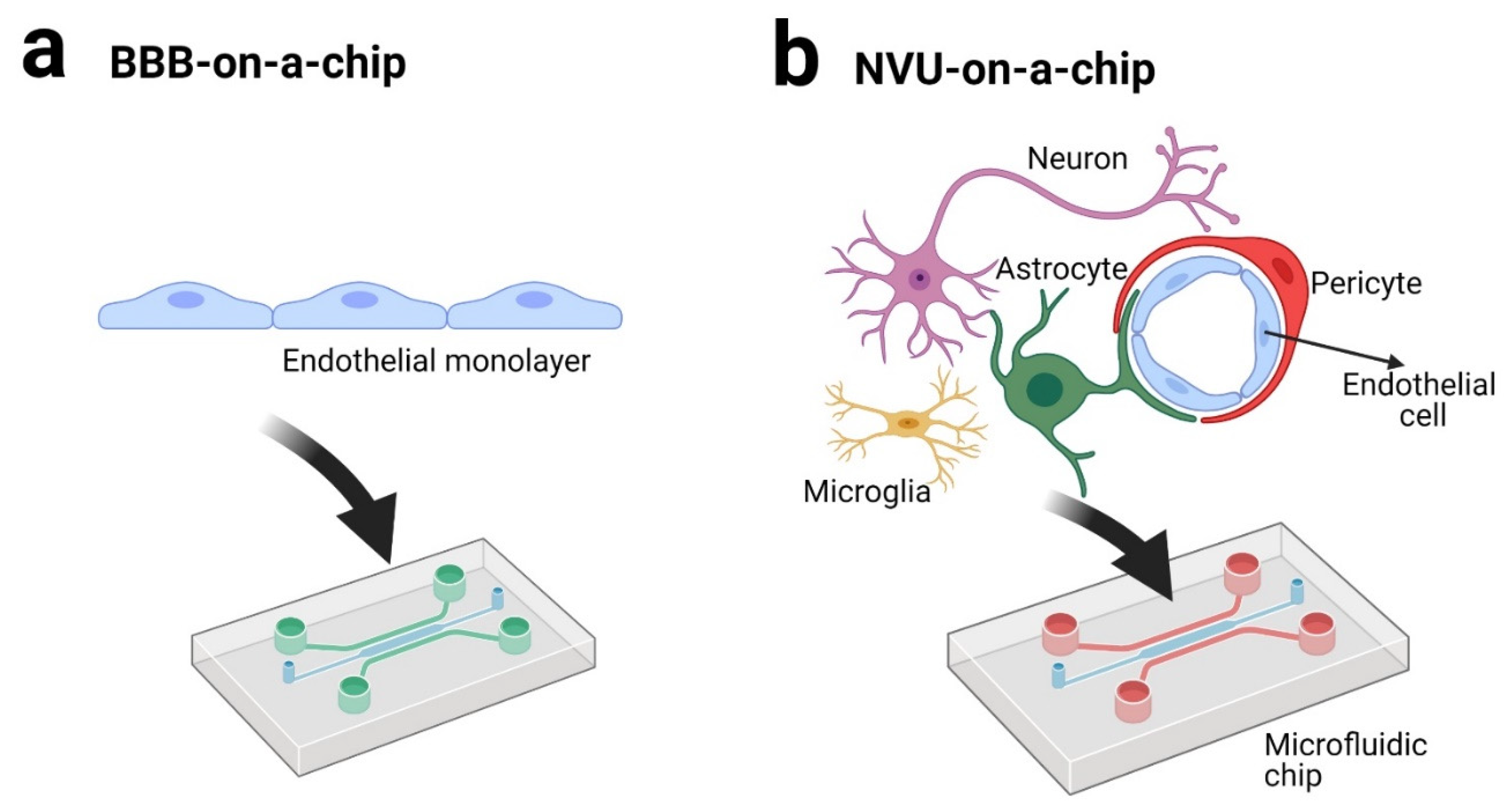
3. BBB andNVU on a Chip
4. Microdevices as a Pharmacological Screening Tool
5. Drug Screening in Neurodegenerative Disorders Using Microdevices
6. LOC Microdevices in Translational Medicine with Impact in Neurological Disorders
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Gupta, S.; Ramesh, K.; Ahmed, S.; Kakkar, V. Lab-on-chip technology: Are view on design trends and future scope in biomedical applications. Int. J. Bio-Sci. Bio-Technol. 2016, 8, 311–322. [Google Scholar] [CrossRef] [Green Version]
- Gardeniers, J.; Van den Berg, A. Lab-on-a-chip systems for biomedical and environmental monitoring. Anal. Bioanal. Chem. 2004, 378, 1700–1703. [Google Scholar] [CrossRef] [PubMed]
- Azizipour, N.; Avazpour, R.; Rosenzweig, D.H.; Sawan, M.; Ajji, A. Evolution of biochip technology: Are view fromlab-on-a-chip to organ-on-a-chip. Micromachines 2020, 11, 599. [Google Scholar] [CrossRef]
- Polini, A.; Prodanov, L.; Bhise, N.S.; Manoharan, V.; Dokmeci, M.R.; Khademhosseini, A. Organs-on-a-chip: A new tool fordrug discovery. Expert Opin. Drug Discov. 2014, 9, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Ghallab, Y.H.; Badawy, W. Lab-on-a-Chip: Techniques, Circuits, and Biomedical Applications; ArtechHouse: Norwood, MA, USA, 2010. [Google Scholar]
- Hou, X.; Zhang, Y.S.; Trujillo-de Santiago, G.; Alvarez, M.M.; Ribas, J.; Jonas, S.J.; Weiss, P.S.; Andrews, A.M.; Aizenberg, J.; Khademhosseini, A. Interplay between materials and microfluidics. Nat. Rev. Mater. 2017, 2, 1–15. [Google Scholar] [CrossRef]
- Dong, R.; Liu, Y.; Mou, L.; Deng, J.; Jiang, X. Microfluidics-based biomaterials and biodevices. Adv. Mater. 2019, 31, 1805033. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, S.; Zheng, J.; Li, Y.; Huang, H. Recent progress in microfluidic models of the blood-brain barrier. Micromachines 2019, 10, 375. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J.B.; Hanson, R.L.; Almughamsi, H.M.; Pang, C.; Fish, T.R.; Woolley, A.T. Microfluidics: Innovations in materials and their fabrication and functionalization. Anal. Chem. 2019, 92, 150–168. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Zhou, J.; Wu, H. Materials for microfluidic chip fabrication. Acc. Chem. Res. 2013, 46, 2396–2406. [Google Scholar] [CrossRef] [PubMed]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.J.; Fluri, K.; Seiler, K.; Fan, Z.; Effenhauser, C.S.; Manz, A. Micromachining a miniaturized capillary electrophoresis-based chemical analysis system on a chip. Science 1993, 261, 895–897. [Google Scholar] [CrossRef] [PubMed]
- Gale, B.K.; Jafek, A.R.; Lambert, C.J.; Goenner, B.L.; Moghimifam, H.; Nze, U.C.; Kamarapu, S.K. A review of current methods in microfluidic device fabrication and future commercialization prospects. Inventions 2018, 3, 60. [Google Scholar] [CrossRef] [Green Version]
- Whitesides, G.M.; Ostuni, E.; Takayama, S.; Jiang, X.; Ingber, D.E. Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 2001, 3, 335–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mata, A.; Fleischman, A.J.; Roy, S. Characterization of polydimethylsiloxane (PDMS) properties for biomedical micro/nanosystems. Biomed. Microdevices 2005, 7, 281–293. [Google Scholar] [CrossRef]
- Adriani, G.; Ma, D.; Pavesi, A.; Kamm, R.D.; Goh, E.L. A 3D neurovascular microfluidic model consisting of neurons, astrocytes and cerebral endothelial cells as a blood–brain barrier. Lab Chip 2017, 17, 448–459. [Google Scholar] [CrossRef]
- Maoz, B.M.; Herland, A.; FitzGerald, E.A.; Grevesse, T.; Vidoudez, C.; Pacheco, A.R.; Sheehy, S.P.; Park, T.-E.; Dauth, S.; Mannix, R. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat. Biotechnol. 2018, 36, 865–874. [Google Scholar] [CrossRef]
- Ning, R.; Zhuang, Q.; Lin, J.-M. Biomaterial-based microfluidics for cell culture and analysis. In Cell Analysis on Microfluidics; Springer: Singapore, 2018; pp. 181–224. [Google Scholar]
- Zilberzwige-Tal, S.; Gazit, E. Go with the flow—Microfluidics approaches for amyloid research. Chem. Asian J. 2018, 13, 3437–3447. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, W.; Jiang, X. Cell-based assays on microfluidics for drug screening. ACS Sens. 2019, 4, 1465–1475. [Google Scholar] [CrossRef]
- Oddo, A.; Peng, B.; Tong, Z.; Wei, Y.; Tong, W.Y.; Thissen, H.; Voelcker, N.H. Advances in microfluidicblood–brain barrier (BBB) models. Trends Biotechnol. 2019, 37, 1295–1314. [Google Scholar] [CrossRef]
- Park, T.-E.; Mustafaoglu, N.; Herland, A.; Hasselkus, R.; Mannix, R.; FitzGerald, E.A.; Prantil-Baun, R.; Watters, A.; Henry, O.; Benz, M. Hypoxia-enhanced blood-brain barrier chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Eddington, D.T.; Puccinelli, J.P.; Beebe, D.J. Thermal aging and reduced hydrophobic recovery of polydimethylsiloxane. Sens. Actuators B Chem. 2006, 114, 170–172. [Google Scholar] [CrossRef]
- Bodas, D.; Khan-Malek, C. Formation of more stable hydrophilic surfaces of PDMS by plasma and chemical treatments. Microelectron. Eng. 2006, 83, 1277–1279. [Google Scholar] [CrossRef]
- Hong, S.M.; Kim, S.H.; Kim, J.H.; Hwang, H.I. Hydrophilic surface modification of PDMS using atmospheric RF plasma. In Proceedings of the Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2006; p. 108. [Google Scholar]
- Tan, S.H.; Nguyen, N.-T.; Chua, Y.C.; Kang, T.G. Oxygen plasma treatment for reducing hydrophobicity of a sealed polydimethylsiloxane microchannel. Biomicrofluidics 2010, 4, 032204. [Google Scholar] [CrossRef] [Green Version]
- Hillborg, H.; Sandelin, M.; Gedde, U.W. Hydrophobic recovery of polydimethylsiloxane after exposure to partial discharges as a function of crosslink density. Polymer 2001, 42, 7349–7362. [Google Scholar] [CrossRef]
- Hoek, I.; Tho, F.; Arnold, W.M. Sodium hydroxide treatment of PDMS based microfluidic devices. Lab Chip 2010, 10, 2283–2285. [Google Scholar] [CrossRef]
- De Menezes Atayde, C.; Doi, I. Highly stable hydrophilic surfaces of PDMS thin layer obtained by UV radiation and oxygen plasma treatments. Phys. Status Solidi C 2010, 7, 189–192. [Google Scholar] [CrossRef]
- Lee, D.; Yang, S. Surface modification of PDMS by atmospheric-pressure plasma-enhanced chemical vapor deposition and analysis of long-lasting surface hydrophilicity. Sens. Actuators B Chem. 2012, 162, 425–434. [Google Scholar] [CrossRef]
- Hemmilä, S.; Cauich-Rodríguez, J.V.; Kreutzer, J.; Kallio, P. Rapid, simple, and cost-effective treatments to achieve long-term hydrophilic PDMS surfaces. Appl. Surf. Sci. 2012, 258, 9864–9875. [Google Scholar] [CrossRef]
- Gaš, B.; Zuska, J.; Coufal, P.; van de Goor, T. Optimization of the high-frequency contactless conductivity detector for capillary electrophoresis. Electrophoresis 2002, 23, 3520–3527. [Google Scholar] [CrossRef]
- Jahangiri, F.; Hakala, T.; Jokinen, V. Long-term hydrophilization of polydimethylsiloxane (PDMS) for capillary filling microfluidic chips. Microfluid. Nanofluidics 2020, 24, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Korolj, A.; Lai, B.F.L.; Radisic, M. Advancesinorgan-on-a-chipengineering. Nat. Rev. Mater. 2018, 3, 257–278. [Google Scholar] [CrossRef]
- Morarka, A.; Agrawal, S.; Kale, S.; Kale, A.; Ogale, S.; Paknikar, K.; Bodas, D. Quantum dot based immunosensor using 3D circular microchannels fabricated in PDMS. Biosens. Bioelectron. 2011, 26, 3050–3053. [Google Scholar] [CrossRef]
- Chan, H.N.; Chen, Y.; Shu, Y.; Chen, Y.; Tian, Q.; Wu, H. Direct, one-step molding of 3D-printed structures for convenient fabrication of truly 3D PDMS microfluidic chips. Microfluid. Nanofluidics 2015, 19, 9–18. [Google Scholar] [CrossRef]
- Sugioka, K.; Cheng, Y. Ultra fast lasers—Reliable tools for advanced materials processing. Light Sci. Appl. 2014, 3, e149. [Google Scholar] [CrossRef]
- Takano, A.; Ogawa, T.; Tanaka, M.; Futai, N. On-chip incubation system for long-term microfluidic cell culture. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 8404–8407. [Google Scholar]
- Jo, B.-H.; VanLerberghe, L.M.; Motsegood, K.M.; Beebe, D.J. Three-dimensional micro-channel fabrication in polydimethylsiloxane (PDMS) elastomer. J. Microelectromech. Syst. 2000, 9, 76–81. [Google Scholar] [CrossRef]
- Fleger, M.; Neyer, A. PDMS microfluidic chip with integrated wave guides for optical detection. Microelectron. Eng. 2006, 83, 1291–1293. [Google Scholar] [CrossRef]
- Park, J.; Lee, B.K.; Jeong, G.S.; Hyun, J.K.; Lee, C.J.; Lee, S.-H. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer’s disease. Lab Chip 2015, 15, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Cho, Y.H.; Park, M.S.; Kim, B.H. Microchannel fabrication on glass materials for microfluidic devices. Int. J. Precis. Eng. Manuf. 2019, 20, 479–495. [Google Scholar] [CrossRef]
- Matsuo, S.; Sumi, H.; Kiyama, S.; Tomita, T.; Hashimoto, S. Femtosecond laser-assisted etching of Pyrex glass with aqueoussolution of KOH. Appl. Surf. Sci. 2009, 255, 9758–9760. [Google Scholar] [CrossRef]
- Chang, T.-L.; Chen, Z.-C.; Lee, Y.-W.; Li, Y.-H.; Wang, C.-P. Ultrafast laser ablation of soda-lime glass for fabricating microfluidic pillar array channels. Microelectron. Eng. 2016, 158, 95–101. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Datta, A.; Berg, J.M.; Gangopadhyay, S. Studies on surface wettability of poly (dimethyl) siloxane (PDMS) and glass under oxygen-plasma treatment and correlation with bond strength. J. Microelectromech. Syst. 2005, 14, 590–597. [Google Scholar] [CrossRef]
- Plecis, A.; Chen, Y. Fabrication of microfluidic devices based on glass—PDMS—Glass technology. Microelectron. Eng. 2007, 84, 1265–1269. [Google Scholar] [CrossRef]
- Haubert, K.; Drier, T.; Beebe, D. PDMS bonding by means of a portable, low-cost corona system. Lab Chip 2006, 6, 1548–1549. [Google Scholar] [CrossRef] [PubMed]
- Aran, K.; Sasso, L.A.; Kamdar, N.; Zahn, J.D. Irreversible, direct bonding of nanoporous polymer membranes to PDMS or glass microdevices. Lab Chip 2010, 10, 548–552. [Google Scholar] [CrossRef] [Green Version]
- Sima, F.; Xu, J.; Wu, D.; Sugioka, K. Ultrafast laser fabrication of functional biochips: New avenues for exploring 3D micro-and nano-environments. Micromachines 2017, 8, 40. [Google Scholar] [CrossRef] [Green Version]
- Sugioka, K.; Cheng, Y. Femtosecond laser processing for optofluidic fabrication. Lab Chip 2012, 12, 3576–3589. [Google Scholar] [CrossRef]
- Jipa, F.; Iosub, S.; Calin, B.; Axente, E.; Sima, F.; Sugioka, K. High repetition rate UV versus VIS picosecond laser fabricationof 3D microfluidic channels embedded in photosensitive glass. Nanomaterials 2018, 8, 583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanada, Y.; Sugioka, K.; Shihira-Ishikawa, I.; Kawano, H.; Miyawaki, A.; Midorikawa, K. 3D microfluidic chips with integrated functional microelements fabricated by a femtosecond laser for studying the gliding mechanism of cyanobacteria. Lab Chip 2011, 11, 2109–2115. [Google Scholar] [CrossRef] [PubMed]
- Sima, F.; Sugioka, K.; Vázquez, R.M.; Osellame, R.; Kelemen, L.; Ormos, P. Three-dimensional femtosecond laser processing for lab-on-a-chip applications. Nanophotonics 2018, 7, 613–634. [Google Scholar] [CrossRef]
- Jipa, F.; Orobeti, S.; Butnaru, C.; Zamfirescu, M.; Axente, E.; Sima, F.; Sugioka, K. Picosecond Laser Processing of Photosensitive Glass for Generation of Biologically Relevant Microenvironments. Appl. Sci. 2020, 10, 8947. [Google Scholar] [CrossRef]
- Sima, F.; Kawano, H.; Hirano, M.; Miyawaki, A.; Obata, K.; Serien, D.; Sugioka, K. Mimicking intravasation–extravasation with a3D glass nanofluidic model for the chemotaxis-free migration of cancer cells in confined spaces. Adv. Mater. Technol. 2020, 5, 2000484. [Google Scholar] [CrossRef]
- Sima, F.; Kawano, H.; Miyawaki, A.; Kelemen, L.; Ormos, P.; Wu, D.; Xu, J.; Midorikawa, K.; Sugioka, K. 3D biomimetic chips for cancer cell migration in nanometer-sized spaces using “Ship-in-a-Bottle” femtosecond laser processing. ACS Appl. Biomater. 2018, 1, 1667–1676. [Google Scholar] [CrossRef]
- Wu, D.; Wu, S.Z.; Xu, J.; Niu, L.G.; Midorikawa, K.; Sugioka, K. Hybrid femtosecond laser microfabrication to achieve true 3D glass/polymer composite biochips with multiscale features and high performance: The concept of ship-in-a-bottle biochip. Laser Photonics Rev. 2014, 8, 458–467. [Google Scholar] [CrossRef]
- Ramadan, Q.; Zourob, M. Organ-on-a-chip engineering: Toward bridging the gap between lab and industry. Biomicrofluidics 2020, 14, 041501. [Google Scholar] [CrossRef]
- Jain, A.; Barrile, R.; van der Meer, A.D.; Mammoto, A.; Mammoto, T.; DeCeunynck, K.; Aisiku, O.; Otieno, M.A.; Louden, C.S.; Hamilton, G.A. Primary human lung alveolus-on-a-chip model of intravascular thrombosis for assessment of therapeutics. Clin. Pharmacol. Ther. 2018, 103, 332–340. [Google Scholar] [CrossRef]
- Torisawa, Y.-S.; Spina, C.S.; Mammoto, T.; Mammoto, A.; Weaver, J.C.; Tat, T.; Collins, J.J.; Ingber, D.E. Bone marrow–on–a–chip replicates hematopoietic niche physiology in vitro. Nat. Methods 2014, 11, 663–669. [Google Scholar] [CrossRef] [Green Version]
- Kimura, H.; Yamamoto, T.; Sakai, H.; Sakai, Y.; Fujii, T. An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab Chip 2008, 8, 741–746. [Google Scholar] [CrossRef]
- Agarwal, A.; Goss, J.A.; Cho, A.; McCain, M.L.; Parker, K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013, 13, 3599–3608. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.S.; Arneri, A.; Bersini, S.; Shin, S.-R.; Zhu, K.; Goli-Malekabadi, Z.; Aleman, J.; Colosi, C.; Busignani, F.; Dell’Erba, V. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016, 110, 45–59. [Google Scholar] [CrossRef] [Green Version]
- Stucki, A.O.; Stucki, J.D.; Hall, S.R.; Felder, M.; Mermoud, Y.; Schmid, R.A.; Geiser, T.; Guenat, O.T. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip 2015, 15, 1302–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mun, K.S.; Arora, K.; Huang, Y.; Yang, F.; Yarlagadda, S.; Ramananda, Y.; Abu-El-Haija, M.; Palermo, J.J.; Appakalai, B.N.; Nathan, J.D. Patient-derived pancreas-on-a-chip to model cystic fibrosis-related disorders. Nat. Commun. 2019, 10, 1–12. [Google Scholar]
- Wufuer, M.; Lee, G.; Hur, W.; Jeon, B.; Kim, B.J.; Choi, T.H.; Lee, S. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Skardal, A.; Murphy, S.V.; Devarasetty, M.; Mead, I.; Kang, H.-W.; Seol, Y.-J.; Zhang, Y.S.; Shin, S.-R.; Zhao, L.; Aleman, J. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Ramme, A.P.; Koenig, L.; Hasenberg, T.; Schwenk, C.; Magauer, C.; Faust, D.; Lorenz, A.K.; Krebs, A.-C.; Drewell, C.; Schirrmann, K. Autologous induced pluripotent stem cell-derived four-organ-chip. Future Sci. OA 2019, 5, FSO413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, K.-J.; Suh, K.-Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010, 10, 36–42. [Google Scholar] [CrossRef]
- Maschmeyer, I.; Lorenz, A.; Ramme, A.; Hasenberg, T.; Schimek, K.; Hübner, J.; Lauster, R.; Marx, U. A microfluidic four-organ-chip for interconnected long-termco-culture of human intestine, liver, skin and kidney equivalents. Toxicol. Lett. 2015, 2, S176. [Google Scholar] [CrossRef]
- Junaid, A.; Mashaghi, A.; Hankemeier, T.; Vulto, P. An end-user perspective on Organ-on-a-Chip: Assays and usability aspects. Curr. Opin. Biomed. Eng. 2017, 1, 15–22. [Google Scholar] [CrossRef]
- Miccoli, B.; Braeken, D.; Li, Y.-C.E. Brain-on-a-chip devices for drug screening and disease modeling applications. Curr. Pharm. Des. 2018, 24, 5419–5436. [Google Scholar] [CrossRef]
- Jahromi, M.A.M.; Abdoli, A.; Rahmanian, M.; Bardania, H.; Bayandori, M.; Basri, S.M.M.; Kalbasi, A.; Aref, A.R.; Karimi, M.; Hamblin, M.R. Microfluidic brain-on-a-chip: Perspectives for mimicking neural system disorders. Mol. Neurobiol. 2019, 56, 8489–8512. [Google Scholar] [CrossRef]
- Ndyabawe, K.; Kisaalita, W.S. Engineering Microsystems to recapitulate brain physiology on a chip. Drug Discov. Today 2019, 24, 1725–1730. [Google Scholar] [CrossRef]
- Zhao, Y.; Demirci, U.; Chen, Y.; Chen, P. Multi scale brain research on a microfluidic chip. Lab Chip 2020, 20, 1531–1543. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Zhu, Y.; Qin, J. Human brain organoid-on-a-chip to model prenatal nicotine exposure. Lab Chip 2018, 18, 851–860. [Google Scholar] [CrossRef]
- Jorfi, M.; D’Avanzo, C.; Tanzi, R.E.; Kim, D.Y.; Irimia, D. Human neurospheroid arrays for in vitro studies of Alzheimer’s disease. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato-Negishi, M.; Tsuda, Y.; Onoe, H.; Takeuchi, S. A neurospheroid network-stamping method for neural transplantation to the brain. Biomaterials 2010, 31, 8939–8945. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Lim, J.; Park, J.; Lee, W.; Yoon, D.S.; Kim, S.H.; Kim, M.-K.; Lee, S.-H.; Kim, D.-H. Construction of neurospheroids via surface modified concave microwells. J. Ind. Eng. Chem. 2018, 62, 341–351. [Google Scholar] [CrossRef]
- Iadecola, C. The neurovascular unit coming of age: A journey through neurovascular coupling in health and disease. Neuron 2017, 96, 17–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamieson, J.J.; Gerecht, S. Chipping away at blood-brain-barrier modeling. Cell Stem Cell 2019, 24, 831–832. [Google Scholar] [CrossRef]
- Van der Helm, M.; vanderMeer, A.; Eijkel, J.; vandenBerg, A.; Segerink, L. Microfluidic organ-on-chip technology for blood-brain barrier research. Tissue Barriers 2016, 4, e1142493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkins, B.T.; Davis, T.P. The blood-brain barrier/neurovascular unit in health and disease. Pharmacol. Rev. 2005, 57, 173–185. [Google Scholar] [CrossRef]
- Bertini, G.; Bramanti, P.; Constantin, G.; Pellitteri, M.; Radu, B.M.; Radu, M.; Fabene, P.F. New players in the neurovascular unit: Insights from experimental and clinical epilepsy. Neurochem. Int. 2013, 63, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Muoio, V.; Persson, P.; Sendeski, M. The neurovascular unit—Concept review. Acta Physiol. 2014, 210, 790–798. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.J. Astrocyte–endothelial interactions and blood–brain barrier permeability. J. Anat. 2002, 200, 523–534. [Google Scholar] [CrossRef]
- Bélanger, M.; Allaman, I.; Magistretti, P.J. Brain energy metabolism: Focus on astrocyte-neuronmetabolic cooperation. Cell Metab. 2011, 14, 724–738. [Google Scholar] [CrossRef] [Green Version]
- Savtchouk, I.; Volterra, A. Gliotransmission: Beyond black-and-white. J. Neurosci. 2018, 38, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Kettenmann, H.; Kirchhoff, F.; Verkhratsky, A. Microglia: New roles for the synaptic stripper. Neuron 2013, 77, 10–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BioRender Home Page. Available online: https://biorender.com (accessed on 18 May 2021).
- DeFelice, F.G.; Munoz, D.P. Opportunities and challenges in developing relevant animal models for Alzheimer’s disease. Ageing Res. Rev. 2016, 26, 112–114. [Google Scholar] [CrossRef]
- Khandaker, G.M.; Cousins, L.; Deakin, J.; Lennox, B.R.; Yolken, R.; Jones, P.B. Inflammation and immunity in schizophrenia: Implications for pathophysiology and treatment. Lancet Psychiatry 2015, 2, 258–270. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.A.; Codreanu, S.G.; Shi, M.; Sherrod, S.D.; Markov, D.A.; Neely, M.D.; Britt, C.M.; Hoilett, O.S.; Reiserer, R.S.; Samson, P.C. Metabolic consequences of inflammatory disruption of the blood-brain barrier in an organ-on-chip model of the human neurovascular unit. J. Neuroinflamm. 2016, 13, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Herland, A.; van derMeer, A.D.; FitzGerald, E.A.; Park, T.-E.; Sleeboom, J.J.; Ingber, D.E. Distinct contributions of astrocytes and pericytes to neuroinflammation identified in a 3D human blood-brain barrier on a chip. PLoS ONE 2016, 11, e0150360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.D.; Khafagy, E.-S.; Khanafer, K.; Takayama, S.; ElSayed, M.E. Organization of endothelialcells, pericytes, and astrocytes into a 3D microfluidic in vitro model of the blood–brain barrier. Mol. Pharm. 2016, 13, 895–906. [Google Scholar] [CrossRef]
- Griep, L.M.; Wolbers, F.; de Wagenaar, B.; ter Braak, P.M.; Weksler, B.; Romero, I.A.; Couraud, P.O.; Vermes, I.; van der Meer, A.D.; van den Berg, A. BBB on chip: Microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed. Microdevices 2013, 15, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Wevers, N.R.; Kasi, D.G.; Gray, T.; Wilschut, K.J.; Smith, B.; VanVught, R.; Shimizu, F.; Sano, Y.; Kanda, T.; Marsh, G. A perfused human blood–brain barrier on-a-chip for high-throughput assessment of barrier function and antibody transport. Fluids Barriers CNS 2018, 15, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Li, Z.; Yu, Y.; Sizdahkhani, S.; Ho, W.S.; Yin, F.; Wang, L.; Zhu, G.; Zhang, M.; Jiang, L. A dynamic in vivo-like organotypic blood-brain barrier model to probe metastatic brain tumors. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Booth, R.; Kim, H. Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB). Lab Chip 2012, 12, 1784–1792. [Google Scholar] [CrossRef] [PubMed]
- Motallebnejad, P.; Thomas, A.; Swisher, S.L.; Azarin, S.M. An isogenic hiPSC-derived BBB-on-a-chip. Biomicrofluidics 2019, 13, 064119. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.I.; Abaci, H.E.; Shuler, M.L. Microfluidicblood–brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol. Bioeng. 2017, 114, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Kim, S.; Buonocore, J.; Park, J.; Welsh, C.J.; Li, J.; Han, A. A three-dimensional arrayed microfluidic blood–brain barrier model with integrated electrical sensor array. IEEE Trans. Biomed. Eng. 2017, 65, 431–439. [Google Scholar] [CrossRef]
- Sellgren, K.L.; Hawkins, B.T.; Grego, S. An optically transparent membrane supports shear stress studies in a three-dimensional microfluidic neurovascular unit model. Biomicrofluidics 2015, 9, 061102. [Google Scholar] [CrossRef] [Green Version]
- Falanga, A.P.; Pitingolo, G.; Celentano, M.; Cosentino, A.; Melone, P.; Vecchione, R.; Guarnieri, D.; Netti, P.A. Shuttle-mediated nanoparticle transport across an in vitro brain endothelium under flow conditions. Biotechnol. Bioeng. 2017, 114, 1087–1095. [Google Scholar] [CrossRef]
- Moya, M.L.; Triplett, M.; Simon, M.; Alvarado, J.; Booth, R.; Osburn, J.; Soscia, D.; Qian, F.; Fischer, N.O.; Kulp, K. A reconfigurable in vitro model for studying the blood–brain barrier. Ann. Biomed. Eng. 2020, 48, 780–793. [Google Scholar] [CrossRef]
- Ahn, S.I.; Sei, Y.J.; Park, H.-J.; Kim, J.; Ryu, Y.; Choi, J.J.; Sung, H.-J.; MacDonald, T.J.; Levey, A.I.; Kim, Y. Microengineered human blood–brain barrier platform for understanding nanoparticle transport mechanisms. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Lee, S.; Chung, M.; Lee, S.R.; Jeon, N.L. 3D brain angiogenesis model to reconstitute functional human blood–brain barrier in vitro. Biotechnol. Bioeng. 2020, 117, 748–762. [Google Scholar] [CrossRef]
- Nguyen, P.Q.H.; Duong, D.D.; Kwun, J.D.; Lee, N.Y. Hybrid elastomer–plastic microfluidic device as a convenient model for mimicking the blood–brain barrier in vitro. Biomed. Microdevices 2019, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Vatine, G.D.; Barrile, R.; Workman, M.J.; Sances, S.; Barriga, B.K.; Rahnama, M.; Barthakur, S.; Kasendra, M.; Lucchesi, C.; Kerns, J. Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell 2019, 24, 995–1005.e1006. [Google Scholar] [CrossRef]
- Kilic, O.; Pamies, D.; Lavell, E.; Schiapparelli, P.; Feng, Y.; Hartung, T.; Bal-Price, A.; Hogberg, H.T.; Quinones-Hinojosa, A.; Guerrero-Cazares, H. Brain-on-a-chip model enables analysis of human neuronal differentiation and chemotaxis. Lab Chip 2016, 16, 4152–4162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabhakarpandian, B.; Shen, M.-C.; Nichols, J.B.; Mills, I.R.; Sidoryk-Wegrzynowicz, M.; Aschner, M.; Pant, K. SyM-BBB: A microfluidic blood brain barrier model. Lab Chip 2013, 13, 1093–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deosarkar, S.P.; Prabhakarpandian, B.; Wang, B.; Sheffield, J.B.; Krynska, B.; Kiani, M.F. A novel dynamic neonatal blood-brain barrier on a chip. PLoS ONE 2015, 10, e0142725. [Google Scholar] [CrossRef] [Green Version]
- Stoica, R.; Rusu, C.M.; Staicu, C.E.; Burlacu, A.E.; Radu, M.; Radu, B.M. Ca2+ homeostasis in brain microvascular endothelial cells. Int. Rev. Cell Mol. Biol. 2021; 362, part A, in press. [Google Scholar]
- Alcendor, D.J.; Block, F.E., III; Cliffel, D.E.; Daniels, J.S.; Ellacott, K.L.; Goodwin, C.R.; Hofmeister, L.H.; Li, D.; Markov, D.A.; May, J.C. Neurovascular unit on a chip: Implications for translational applications. Stem Cell Res. Ther. 2013, 4, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Huh, D.; Torisawa, Y.-S.; Hamilton, G.A.; Kim, H.J.; Ingber, D.E. Microengineered physiological biomimicry: Organs-on-chips. Lab Chip 2012, 12, 2156–2164. [Google Scholar] [CrossRef]
- Gumbleton, M.; Audus, K.L. Progress and limitations in the use of in vitro cell cultures to serve as a permeability screen for the blood-brain barrier. J. Pharm. Sci. 2001, 90, 1681–1698. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, N.; Kingshott, P.; Sumer, H.; Sharma, C.S.; Rath, S.N. On-chip anticancer drug screening–recent progress in microfluidic platforms to address challenges in chemotherapy. Biosens. Bioelectron. 2019, 137, 236–254. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S. Organs-on-a-chip: Current applications and consideration points for in vitro ADME-Toxstudies. Drug Metab. Pharmacokinet. 2018, 33, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Musteata, F.M. Pharmacokinetic applications of microdevices and microsampling techniques. Bioanalysis 2009, 1, 171–185. [Google Scholar] [CrossRef]
- Mitxelena-Iribarren, O.; Zabalo, J.; Arana, S.; Mujika, M. Improved microfluidic platform for simultaneous multipledrug screening towards personalized treatment. Biosens. Bioelectron. 2019, 123, 237–243. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.; Shao, Z.; Luo, G.; Ding, M.; Liang, Q. Simultaneous assay of oxygen-dependent cytotoxicity and genotoxicity of anticancer drugs on an integrated microchip. Anal. Chem. 2018, 90, 11899–11907. [Google Scholar] [CrossRef]
- Benam, K.H.; Villenave, R.; Lucchesi, C.; Varone, A.; Hubeau, C.; Lee, H.-H.; Alves, S.E.; Salmon, M.; Ferrante, T.C.; Weaver, J.C. Smallair way-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 2016, 13, 151–157. [Google Scholar] [CrossRef]
- Ewart, L.; Fabre, K.; Chakilam, A.; Dragan, Y.; Duignan, D.B.; Eswaraka, J.; Gan, J.; Guzzie-Peck, P.; Otieno, M.; Jeong, C.G. Navigating tissue chips from development to dissemination: A pharmaceutical industry perspective. Exp. Biol. Med. 2017, 242, 1579–1585. [Google Scholar] [CrossRef] [Green Version]
- Low, L.; Tagle, D. Tissue chips–innovative tools for drug development and disease modeling. Lab Chip 2017, 17, 3026–3036. [Google Scholar] [CrossRef]
- Mastrangeli, M.; Millet, S.; van den Eijnden-van Raaij, J. Organ-on-chip in development: Towards a roadmap for organs-on-chip. ALTEX Altern. Anim. Exp. 2019, 36, 650–668. [Google Scholar] [CrossRef]
- Mastrangeli, M.; Millet, S.; Mummery, C.; Loskill, P.; Braeken, D.; Eberle, W.; Cipriano, M.; Fernandez, L.; Graef, M.; Gidrol, X. Building blocks for a European organ-on-chip road map. ALTEX Altern. Anim. Exp. 2019, 36, 481–492. [Google Scholar]
- Moraes, C.; Mehta, G.; Lesher-Perez, S.C.; Takayama, S. Organs-on-a-chip: A focus on compartmentalized microdevices. Ann. Biomed. Eng. 2012, 40, 1211–1227. [Google Scholar] [CrossRef] [PubMed]
- Ugolini, G.S.; Cruz-Moreira, D.; Visone, R.; Redaelli, A.; Rasponi, M. Microfabricated physiological models for in vitro drug screening applications. Micromachines 2016, 7, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chirra, H.D.; Shao, L.; Ciaccio, N.; Fox, C.B.; Wade, J.M.; Ma, A.; Desai, T.A. Planar microdevices for enhanced in vivo retention and oral bioavailability of poorly permeable drugs. Adv. Healthc. Mater. 2014, 3, 1648–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontana, F.; Ferreira, M.P.; Correia, A.; Hirvonen, J.; Santos, H.A. Microfluidics as a cutting-edge technique for drug delivery applications. J. Drug Deliv. Sci. Technol. 2016, 34, 76–87. [Google Scholar] [CrossRef]
- Cavero, I.; Guillon, J.-M.; Holzgrefe, H.H. Human organotypic bioconstructs from organ-on-chip devices for human-predictive biological insights on drug candidates. Expert Opin. Drug Saf. 2019, 18, 651–677. [Google Scholar] [CrossRef] [PubMed]
- Orbach, S.M.; Less, R.R.; Kothari, A.; Rajagopalan, P. In vitro intestinal and liver models for toxicity testing. ACS Biomater. Sci. Eng. 2017, 3, 1898–1910. [Google Scholar] [CrossRef]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic organ-on-a-chip models of human intestine. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 659–668. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; LesherPerez, S.C.; Yamanishi, C.; Labuz, J.M.; Leung, B.; Takayama, S. Pharmacokinetic profile that reduces nephrotoxicity of gentamicin in a perfused kidney-on-a-chip. Biofabrication 2016, 8, 015021. [Google Scholar] [CrossRef] [PubMed]
- Lind, J.U.; Busbee, T.A.; Valentine, A.D.; Pasqualini, F.S.; Yuan, H.; Yadid, M.; Park, S.-J.; Kotikian, A.; Nesmith, A.P.; Campbell, P.H. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater. 2017, 16, 303–308. [Google Scholar] [CrossRef]
- Phan, D.T.; Wang, X.; Craver, B.M.; Sobrino, A.; Zhao, D.; Chen, J.C.; Lee, L.Y.; George, S.C.; Lee, A.P.; Hughes, C.C. A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab Chip 2017, 17, 511–520. [Google Scholar] [CrossRef]
- Oleaga, C.; Riu, A.; Rothemund, S.; Lavado, A.; McAleer, C.W.; Long, C.J.; Persaud, K.; Narasimhan, N.S.; Tran, M.; Roles, J. Investigation of the effect of hepatic metabolism on off-target cardiotoxicity in a multi-organ human-on-a-chip system. Biomaterials 2018, 182, 176–190. [Google Scholar] [CrossRef]
- Theobald, J.; Ghanem, A.; Wallisch, P.; Banaeiyan, A.A.; Andrade-Navarro, M.A.; Taškova, K.; Haltmeier, M.; Kurtz, A.; Becker, H.; Reuter, S. Liver-kidney-on-chip to study toxicity of drug metabolites. ACS Biomater. Sci. Eng. 2018, 4, 78–89. [Google Scholar] [CrossRef]
- Koo, Y.; Hawkins, B.T.; Yun, Y. Three-dimensional (3D) tetra-culture brain on chip platform for organophosphate toxicity screening. Sci. Rep. 2018, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Isoherranen, N.; Madabushi, R.; Huang, S.M. Emerging role of organ-on-a-chip technologies in quantitative clinical pharmacology evaluation. Clin. Transl. Sci. 2019, 12, 113–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J. Application of organ-on-chip in drug discovery. J. Biosci. Med. 2020, 8, 119–134. [Google Scholar] [CrossRef] [Green Version]
- Ai, Y.; Zhang, F.; Wang, C.; Xie, R.; Liang, Q. Recent progress inlab-on-a-chip for pharmaceutical analysis and pharmacological/toxicological test. TrAC Trends Anal. Chem. 2019, 117, 215–230. [Google Scholar] [CrossRef]
- Weinhart, M.; Hocke, A.; Hippenstiel, S.; Kurreck, J.; Hedtrich, S. 3D organ models—Revolution in pharmacological research? Pharmacol. Res. 2019, 139, 446–451. [Google Scholar] [CrossRef]
- Aldewachi, H.; Al-Zidan, R.N.; Conner, M.T.; Salman, M.M. High-throughput screening platforms in the discovery of novel drugs for neurodegenerative diseases. Bioengineering 2021, 8, 30. [Google Scholar] [CrossRef]
- Palop, J.J.; Mucke, L. Amyloid-β–induced neuronal dysfunction in Alzheimer’s disease: From synapses toward neural networks. Nat. Neurosci. 2010, 13, 812. [Google Scholar] [CrossRef]
- Sui, Y.-T.; Bullock, K.M.; Erickson, M.A.; Zhang, J.; Banks, W. Alpha synuclein is transported into and out of the brain by the blood–brain barrier. Peptides 2014, 62, 197–202. [Google Scholar] [CrossRef] [Green Version]
- Bicker, J.; Alves, G.; Fortuna, A.; Falcão, A. Blood–brain barrier models and their relevance for a successful development of CNS drug delivery systems: Areview. Eur. J. Pharm. Biopharm. 2014, 87, 409–432. [Google Scholar] [CrossRef]
- Terrell-Hall, T.B.; Ammer, A.G.; Griffith, J.I.; Lockman, P.R. Permeability across a novel microfluidic blood-tumor barrier model. Fluids Barriers CNS 2017, 14, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bang, S.; Lee, S.-R.; Ko, J.; Son, K.; Tahk, D.; Ahn, J.; Im, C.; Jeon, N.L. A low permeability microfluidicblood-brain barrier platform with direct contact between perfusable vascular network and astrocytes. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Song, H.L.; Shim, S.; Kim, D.H.; Won, S.H.; Joo, S.; Kim, S.; Jeon, N.L.; Yoon, S.Y. β-Amyloid is transmitted via neuronal connections along axonal membranes. Ann. Neurol. 2014, 75, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Pediaditakis, I.; Kodella, K.R.; Manatakis, D.V.; Hinojosa, C.D.; Manolakos, E.S.; Rubin, L.L.; Hamilton, G.A.; Karalis, K. Modeling Alpha-Synuclein Pathology in a HumanBrain-Chip to Assess Blood-Brain Barrier Disruptionin Parkinson’s Disease. bioRxiv 2020. [Google Scholar] [CrossRef]
- He, Y.; Yao, Y.; Tsirka, S.E.; Cao, Y. Cell-culture models of the blood–brain barrier. Stroke 2014, 45, 2514–2526. [Google Scholar] [CrossRef] [Green Version]
- Cohrs, R.J.; Martin, T.; Ghahramani, P.; Bidaut, L.; Higgins, P.J.; Shahzad, A. Translational medicine definition by the European society for translational medicine. In New Horizons in Translational Medicine; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Ioannidis, J.P. Materializing research promises: Opportunities, priorities and conflicts in translational medicine. J. Transl. Med. 2004, 2, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Garcia, A.; Oliva-Ramirez, J.; Bautista-Flores, C.; Hosseini, S. 3D In vitro human organ mimicry devices for drug discovery, development, and assessment. Adv. Polym. Technol. 2020, 2020, 41. [Google Scholar] [CrossRef]
- Wehling, M. Translational medicine: Science or wishful thinking? J. Transl. Med. 2008, 6, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Abgrall, P.; Gue, A. Lab-on-chip technologies: Making a microfluidic network and coupling it into a complete microsystem—A review. J. Micromechanics Microengineering 2007, 17, R15. [Google Scholar] [CrossRef]
- Radu, B.M.; Osculati, A.M.M.; Suku, E.; Banciu, A.; Tsenov, G.; Merigo, F.; DiChio, M.; Banciu, D.D.; Tognoli, C.; Kacer, P. All muscarinic acetylcholine receptors (M1-M5) are expressed in murine brain microvascular endothelium. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Q.Y.; Haqqani, A.S.; Leclerc, S.; Liu, Z.; Fauteux, F.; Baumann, E.; Delaney, C.E.; Ly, D.; Star, A.T. Differential expression of receptors mediating receptor-mediated transcytosis (RMT) in brain microvessels, brain parenchyma and peripheral tissues of the mouse and the human. Fluids Barriers CNS 2020, 17, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Cameron, T.; Bennet, T.; Rowe, E.M.; Anwer, M.; Wellington, C.L.; Cheung, K.C. Review of design considerations for brain-on-a-chip models. Micromachines 2021, 12, 441. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Ye, M.; Levy, A.; Rothstein, J.; Bergles, D.; Searson, P.C. The blood-brain barrier: An engineering perspective. Front. Neuroeng. 2013, 6, 7. [Google Scholar] [CrossRef] [Green Version]
- Soldner, F.; Hockemeyer, D.; Beard, C.; Gao, Q.; Bell, G.W.; Cook, E.G.; Hargus, G.; Blak, A.; Cooper, O.; Mitalipova, M. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 2009, 136, 964–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheridan, S.D.; Theriault, K.M.; Reis, S.A.; Zhou, F.; Madison, J.M.; Daheron, L.; Loring, J.F.; Haggarty, S.J. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS ONE 2011, 6, e26203. [Google Scholar] [CrossRef] [Green Version]
- Brennand, K.J.; Simone, A.; Jou, J.; Gelboin-Burkhart, C.; Tran, N.; Sangar, S.; Li, Y.; Mu, Y.; Chen, G.; Yu, D. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011, 473, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lopez-Santiago, L.F.; Yuan, Y.; Jones, J.M.; Zhang, H.; O’Malley, H.A.; Patino, G.A.; O’Brien, J.E.; Rusconi, R.; Gupta, A. Dravet syndrome patient-derived neurons suggest a novel epilepsy mechanism. Ann. Neurol. 2013, 74, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Jiao, J.; Yang, Y.; Shi, Y.; Chen, J.; Gao, R.; Fan, Y.; Yao, H.; Liao, W.; Sun, X.-F.; Gao, S. Modeling Dravetsyndrome using induced pluripotent stem cells (iPSCs) and directly converted neurons. Hum. Mol. Genet. 2013, 22, 4241–4252. [Google Scholar] [CrossRef]
- Lim, R.G.; Quan, C.; Reyes-Ortiz, A.M.; Lutz, S.E.; Kedaigle, A.J.; Gipson, T.A.; Wu, J.; Vatine, G.D.; Stocksdale, J.; Casale, M.S. Huntington’s disease iPSC-derived brain microvascular endothelial cells reveal WNT-mediated angiogenic and blood-brain barrier deficits. Cell Rep. 2017, 19, 1365–1377. [Google Scholar] [CrossRef] [Green Version]
- Vatine, G.D.; Al-Ahmad, A.; Barriga, B.K.; Svendsen, S.; Salim, A.; Garcia, L.; Garcia, V.J.; Ho, R.; Yucer, N.; Qian, T. Modeling psychomotor retardation using iPSCs from MCT8-deficient patients indicates a prominent role for the blood-brain barrier. Cell Stem Cell 2017, 20, 831–843.e835. [Google Scholar] [CrossRef] [Green Version]
- Ochalek, A.; Mihalik, B.; Avci, H.X.; Chandrasekaran, A.; Téglási, A.; Bock, I.; Giudice, M.L.; Táncos, Z.; Molnár, K.; László, L. Neurons derived from sporadic Alzheimer’s disease iPSCs revealel evated TAU hyperphosphorylation, increased amyloid levels, and GSK3B activation. Alzheimer’s Res. Ther. 2017, 9, 1–19. [Google Scholar]
- Ananiev, G.; Williams, E.C.; Li, H.; Chang, Q. Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model. PLoS ONE 2011, 6, e25255. [Google Scholar] [CrossRef]
- Dajani, R.; Koo, S.E.; Sullivan, G.J.; Park, I.H. Investigation of Rett syndrome using pluripotent stem cells. J. Cell. Biochem. 2013, 114, 2446–2453. [Google Scholar] [CrossRef] [Green Version]
- Yi, F.; Qu, J.; Li, M.; Suzuki, K.; Kim, N.Y.; Liu, G.-H.; Belmonte, J.C.I. Establishment of hepatic and neural differentiation platforms of Wilson’s disease specific induced pluripotent stem cells. Protein Cell 2012, 3, 855–863. [Google Scholar] [CrossRef] [Green Version]
- Paşca, S.P.; Portmann, T.; Voineagu, I.; Yazawa, M.; Shcheglovitov, A.; Paşca, A.M.; Cord, B.; Palmer, T.D.; Chikahisa, S.; Nishino, S. Using iPSC-derived neurons to uncover cellular phenotype sassociated with Timothy syndrome. Nat. Med. 2011, 17, 1657. [Google Scholar] [CrossRef] [PubMed]
- Krey, J.F.; Paşca, S.P.; Shcheglovitov, A.; Yazawa, M.; Schwemberger, R.; Rasmusson, R.; Dolmetsch, R.E. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat. Neurosci. 2013, 16, 201–209. [Google Scholar] [CrossRef] [Green Version]
- U.S. Food and Drug Administration Home Page. Available online: https://www.fda.gov/medical-devices/medical-device-regulatory-science-research-programs-conducted-osel/microfluidics-program-research-microfluidics-based-medical-devices (accessed on 18 May 2021).
- Bors, L.A.; Erdő, F. Over coming the blood–brain barrier. Challenges and tricks for CNS drug delivery. Sci. Pharm. 2019, 87, 6. [Google Scholar] [CrossRef] [Green Version]
- Mann, S.A.; Heide, J.; Knott, T.; Airini, R.; Epureanu, F.B.; Deftu, A.-F.; Deftu, A.-T.; Radu, B.M.; Amuzescu, B. Recording of multiple ion current components and action potentials in human induced pluripotent stem cell-derived cardiomyocytes via automated patch-clamp. J. Pharmacol. Toxicol. Methods 2019, 100, 106599. [Google Scholar] [CrossRef]
- Amuzescu, B.; Radu, B.M.; Mihailescu, D.F.; Alexander, S. Method for inVitro Detection of the Proarrhythmogenic Risk of a Drug Candidate on Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes (hiPSC-CM). European Patent EP3546935B1, 23 December 2020. [Google Scholar]
- Reyes, D.R.; van Heeren, H.; Guha, S.; Herbertson, L.; Tzannis, A.P.; Ducrée, J.; Bissig, H.; Becker, H. Accelerating innovation and commercialization through standardization of microfluidic-based medical devices. Lab. Chip 2021, 21, 9–21. [Google Scholar] [CrossRef] [PubMed]

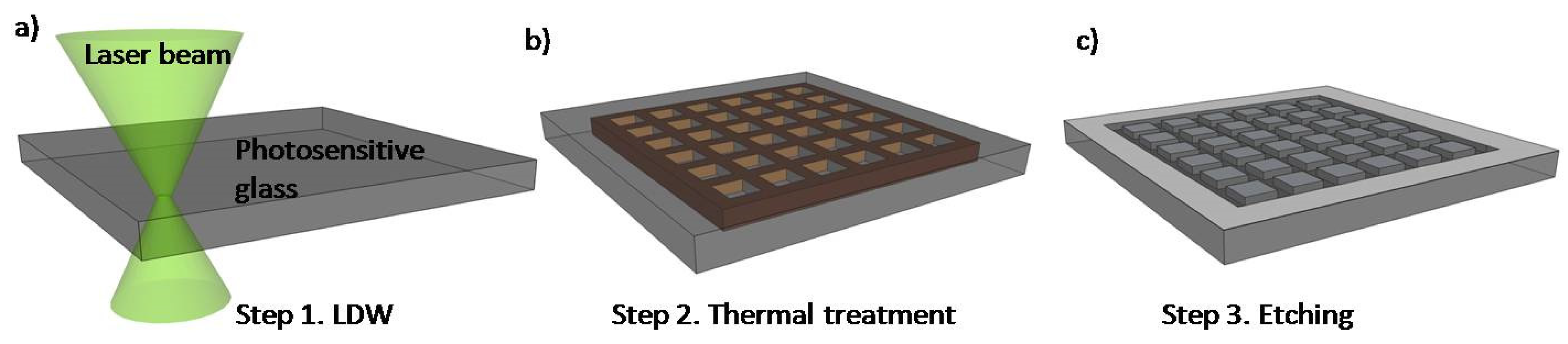

| Material/Property | Silicon/Glass | Elastomers | Thermosets | Thermoplastics | Hydrogel | Paper |
|---|---|---|---|---|---|---|
| optical transparency | no/high | high | high | medium to high | low to medium | low |
| hydrophobicity | hydrophilic | hydrophobic | hydrophobic | hydrophobic | hydrophilic | amphiphilic |
| thermostability | very high | medium | high | medium to high | low | medium |
| resistance to oxidizer | excellent | moderate | good | moderate to good | low | low |
| solvent compatibility | very high | low | high | medium to high | low | medium |
| permeability to oxygen (Barrer a) | <0.01 | ≈500 | 0.03–1 | 0.05–5 | >1 | >1 |
| surface charge | very stable | not stable | stable | stable | N/A | N/A |
| common technique for microfabrication/features | photolithography, laser-assisted etching | casting | casting, photopolymerization | thermo-molding | casting, photopolymerization, 3D bioprinting | photolithography, printing |
| smallest channel dimension | <100 nm | <1 μm | <100 nm | ≈100 nm | ≈10 μm | ≈200 μm |
| channel profile | limited 3D/3D | 3D | arbitrary 3D | 3D | 3D | 2D |
| multilayer channels | hard/easy | easy | easy | easy | Medium | easy |
| throughput | medium to high | high | high | high | low to medium | high |
| Organ/Tissue Type | Chip Material | Membrane Material | Application | Reference |
|---|---|---|---|---|
| Alveolus-on-a-chip | PDMS | PDMS | Interface alveolar epithelium/endothelium for the study of inflammation-induced thrombosis | [60] |
| Bone marrow-on-a-chip | PDMS | PDMS | Analysis of the cellular response to drugs and radiation | [61] |
| Gut-on-a-chip | PDMS | Polyester | Development of a platform for drug screening and substance toxicity testing | [62] |
| Heart-on-a-chip | PDMS | No membrane | Testing the inotropic effect of isoproterenol on cardiac contractility | [63] |
| PMMA and PDMS | No membrane | Evaluation of cardiovascular toxicity of some pharmaceutical products | [64] | |
| Intestine–liver–brain–kidney-on-a-chip | PDMS | PDMS | Production and testing of an autologous iPSC derived four-organ-on-a-chip in long-term cocultivation conditions (i.e., 14 days) | [69] |
| Kidney-on-a-chip | PDMS | Polyester | Analysis in conditions close to the physiological ones of renal tubule cells | [70] |
| Lung-on-a-chip | PDMS | PDMS | Mimicking and analyzing the long alveolar barrier | [65] |
| Lung–liver–heart-on-a-chip | PMMA and PDMS | Polyester | Assessment of the importance of interactions between organs in response to drugs | [68] |
| Pancreas-on-a-chip | PDMS | Polyester | Investigating the role of CFTR (Cystic Fibrosis Transmembrane Conductance Regulator) in insulin production | [66] |
| Skin-on-a-chip | PDMS | Polyester | Mimicking edema and inflammation of the skin and testing dexamethasone effects | [67] |
| Organ/Tissue Type | Type of Cells | Application | Reference |
|---|---|---|---|
| Brain organoid-on-a-chip | 3D brain organoids derived from human-induced pluripotent stem cells (hiPSCs) | Modeling the neurodevelopmental disorders under environmental exposure (e.g., nicotine) | [77] |
| 3D brain-on-a-chip | Neurospheroids obtained from prenatal E16 rat cortical neurons | In vitro brain model for neurodegenerative disease (e.g., Alzheimers’ disease) and high-throughput drug screening | [42] |
| Brain-on-a-chip | Neurospheroids obtained from human neural progenitor and human iPSC-derived neural progenitor cells | Investigating the development of Alzheimer’s disease and testing drugs against this neuropathology | [78] |
| Neurospheroid network-on-a-chip | Neurospheroids obtained from primary culture obtained from the cerebral cortex of Wistar rats | Studying neural transplantation therapy for treating severe degenerative brain disease | [79] |
| 3D brain-on-a-chip | Neurospheroids obtained from prenatal rat (E18) cortical neurons | Modulation of cell–ECM interactions at the neuronal level by analyzing neurospheroids and their study in pathological conditions | [80] |
| Model | Chip Material | Membrane Material | Culture Type | Cells | Application | Reference |
|---|---|---|---|---|---|---|
| BBB | PDMS and glass | Polycarbonate | Co-culture | Endothelial cells (b.End3) and astrocytes (C8D1A) | BBB permeability | [100] |
| BBB | PDMS | Polyethylene terephthalate | Co-culture | Endothelial cells (BMEC from hiPCS) and astrocytes (from IMR90-4 iPSCs) | BBB permeability due to TNF-α in liver failure/melanoma | [101] |
| BBB | OrganoPlate | No membrane | Tri-culture | Endothelial cells (TY10), astrocytes (hAst) and pericytes (hBPCT) | BBB permeability for different types of molecules (antibodies) | [98] |
| BBB | Objet Vero Clear, silicone, and PDMS | Polycarbonate | Co-culture | Endothelial cells (BMEC from iPSC) and astrocytes (Rat primary culture) | BBB permeability for drugs | [102] |
| BBB | PDMS | Polycarbonate | Co-culture | Primary mouse brain microvascular endothelial cells and primary mouse astrocytes | Cellular interactions in the BBB under physiological or shear stress conditions | [103] |
| BBB | PDMS | Polyester and polytetrafluoroethylene | Co-culture | Endothelial cells (b.End3) and astrocytes (C8D1A) | Analysis of cell cultures on porous membranes | [104] |
| BBB | PMMA | Polyester | Monoculture | Endothelial cells (b.End3) | Transport of nanoparticles across the BBB | [105] |
| BBB | PDMS and polyvinylidene fluoride (PVDF) | Polyvinylidene fluoride (PVDF) | Co-culture | Human cerebral microvascular endothelial cells (hCMEC/D3) and normal human astrocytes | Reproducible platform for the BBB study under static or continuous flow conditions | [106] |
| BBB | PDMS | Polycarbonate | Tri-culture | Human cerebral microvascular endothelial cells (HBMEC), pericytes, and astrocytes | BBB model for the investigation of neuroinflammation | [107] |
| BBB | PDMS | No membrane | Multi-Culture | Endothelial cells (HBMEC and HUVEC), pericytes (HhPC-PL), astrocytes (NHA), and primary normal human lung fibroblasts (LF) | In vitro reproduction of angiogenesis in the central nervous system | [108] |
| BBB | PDMS, PMMA, and PC | N/A | Co-culture | Endothelial cells (HUVEC) and human astrocytes | Testing the biocompatibility of the APTES-coated PDMS surface, on which different types of coating were applied | [109] |
| NVU | PDMS | No membrane | Tri-culture | Human iPSC-derived blood–brain barrier cells Human primary astrocytes Human primary pericytes | Complex platform for the study of neurological diseases | [110] |
| NVU | PDMS | PDMS | Co-culture (×2) | Human teratocarcinoma NTERA-2 cl. D1 (hNT2) cells and human endothelial cells (hBMEC) Human teratocarcinoma NTERA-2 cl. D1 (hNT2) cells and Human fetal neural progenitor cells (hNPCs) | Differentiation of cells on the chip and analysis of the importance of cell interactions in neurodevelopment | [111] |
| NVU | PDMS | No membrane | Multi-Culture | Endothelial cells (HUVEC and hCMEC/D3), neurons (primary culture), and astrocytes (primary culture) | Neurovascular unit development | [17] |
| NVU | PDMS andpolycarbonate | Polyethylene terephthalate andpolycarbonate | Multi-Culture | Human hippocampal neural stem cells HIP-009 cells, cortical human brain microvascular endothelial cells (hBMVECs), human astrocytes, and human brain pericytes of cortical origin | Effect of intravascular administration of methamphetamine | [18] |
| Type of Analysis | Comparison | Type of Cells | References |
|---|---|---|---|
| TEER ZO1 immunostaining | Slightly higher resistance values upon 7 days in culture for BBB-on-a-chip compared to the transwell system Similar ZO1 immunostaining | human endothelial cells hCMEC/D3 | [97] |
| TEER ZO1 immunostaining | Astrocyte conditioned medium improves the resistance values of BBB-on-a-chip BBB-on-a-chip has higher resistance values than the transwell model | Rat brain endothelial cells (RBEC) isolated from neonatal rats neonatal rat astrocytes | [113] |
| TEER | μBBB had significantly higher (10-fold) resistance values than the transwell model for co-cultures | b.End3 endothelial cells, with and without co-cultured C8-D1A astrocytes | [100] |
| Barrier permeability and cytokine release profile | Similar permeability of the human 3D BBB-on-a-chip compared to the non-human cells BBB models or to the inflammatory stimulated models (depending on the presence of astrocytes or pericytes) Significantly higher permeability of the human 3D BBB-on-a-chip compared to co-cultures in static transwell plates | Co-culture of human brain microvascular endothelial cells, human brain pericytes, human astrocytes (from cortex) | [95] |
| P-glycoprotein (P-gp) permeability | BBB-on-a-chip model, but not the transwell model, enable the study of P-gp efflux pump permeability and its pharmacological blockade (e.g., verapamil) | Human iPS cell line IMR90-4 | [23] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Staicu, C.E.; Jipa, F.; Axente, E.; Radu, M.; Radu, B.M.; Sima, F. Lab-on-a-Chip Platforms as Tools for Drug Screening in Neuropathologies Associated with Blood–Brain Barrier Alterations. Biomolecules 2021, 11, 916. https://doi.org/10.3390/biom11060916
Staicu CE, Jipa F, Axente E, Radu M, Radu BM, Sima F. Lab-on-a-Chip Platforms as Tools for Drug Screening in Neuropathologies Associated with Blood–Brain Barrier Alterations. Biomolecules. 2021; 11(6):916. https://doi.org/10.3390/biom11060916
Chicago/Turabian StyleStaicu, Cristina Elena, Florin Jipa, Emanuel Axente, Mihai Radu, Beatrice Mihaela Radu, and Felix Sima. 2021. "Lab-on-a-Chip Platforms as Tools for Drug Screening in Neuropathologies Associated with Blood–Brain Barrier Alterations" Biomolecules 11, no. 6: 916. https://doi.org/10.3390/biom11060916
APA StyleStaicu, C. E., Jipa, F., Axente, E., Radu, M., Radu, B. M., & Sima, F. (2021). Lab-on-a-Chip Platforms as Tools for Drug Screening in Neuropathologies Associated with Blood–Brain Barrier Alterations. Biomolecules, 11(6), 916. https://doi.org/10.3390/biom11060916










