Effects of the Clock Modulator Nobiletin on Circadian Rhythms and Pathophysiology in Female Mice of an Alzheimer’s Disease Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Circadian Activity and Period Measurement
2.3. Noninvasive Piezoelectric Transducer Sleep/Wake Recording
2.4. Metabolic Chamber Analysis
2.5. Glucose Tolerance and Cold Tolerance Tests
2.6. Real-Time PCR Analysis
2.7. Western Blotting
2.8. Immunohistochemistry
2.9. Quantifications and Statistical Analysis
3. Results
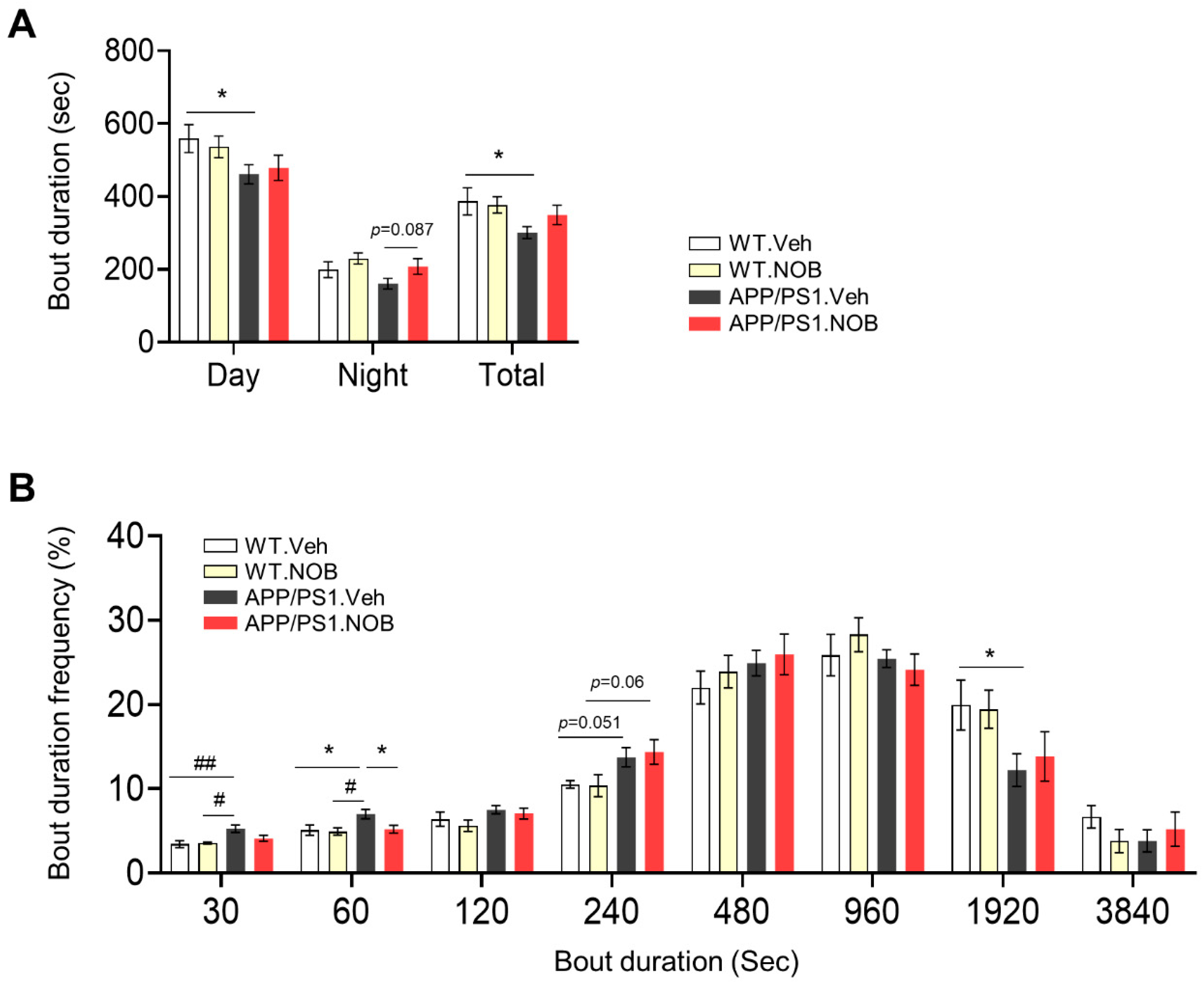
3.1. Sleep Behavior and Circadian Free-Running Rhythmin Female APP/PS1 Mice and NOB Effects
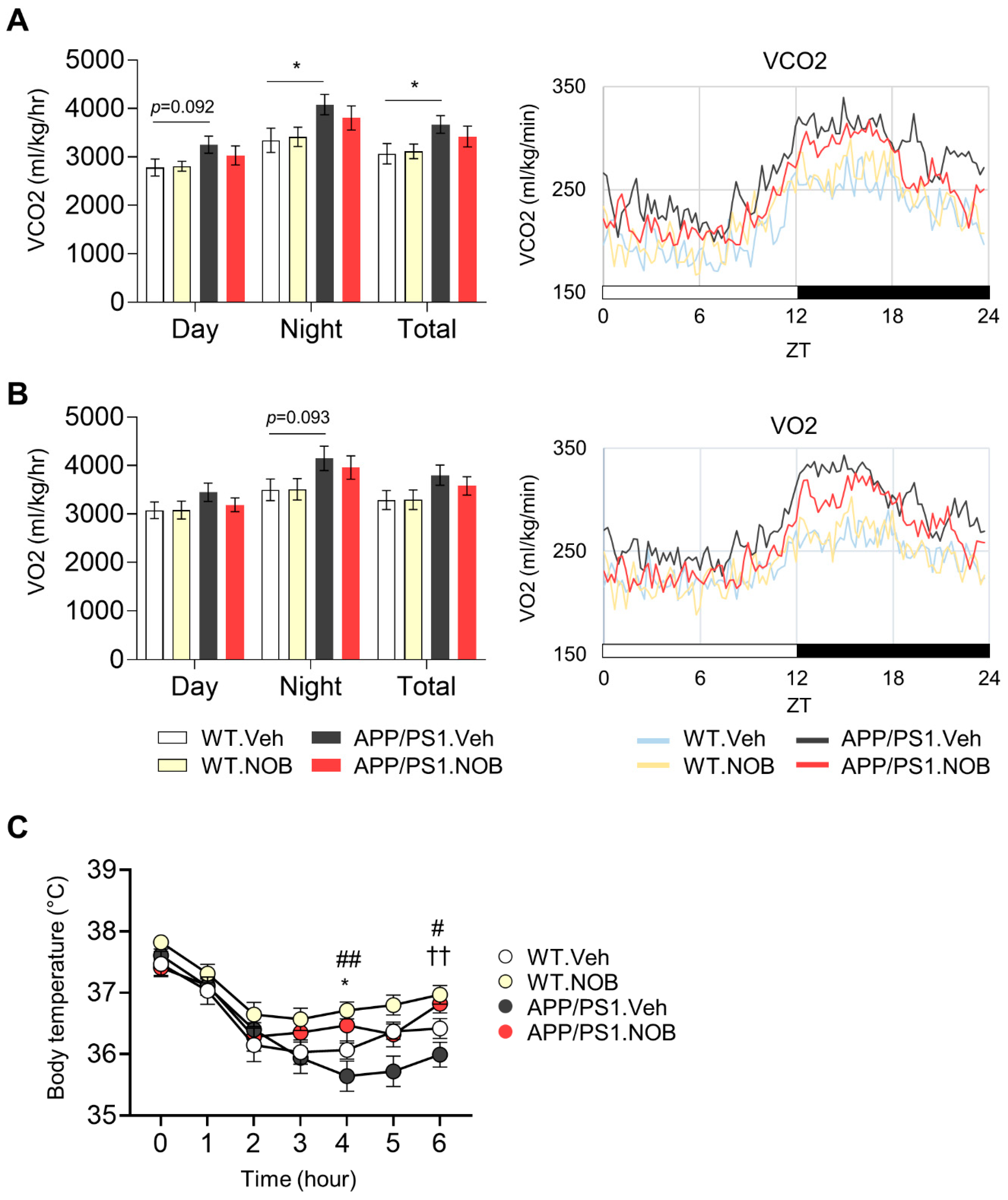
3.2. Effects of NOB on Systemic Metabolism in APP/PS1 Mice
3.3. NOB Regulates Circadian Gene Expression in the Cortex
3.4. NOB Remodels Clock-Controlled Gene Expression in the Cortex
3.5. NOB Attenuates Amyloid Beta (Aβ) Plaque Deposition in APP/PS1 Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scheltens, P. Alzheimer’s disease. Lancet 2016, 388, 505–517. [Google Scholar] [CrossRef]
- Videnovic, A.; Lazar, A.; Barker, R.A.; Overeem, S. ‘The clocks that time us’—Circadian rhythms in neurodegenerative disorders. Nat. Rev. Neurol. 2014, 10, 683–693. [Google Scholar] [CrossRef]
- Musiek, E.S.; Holtzman, D.M. Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science 2016, 354, 1004–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coogan, A.; Schutová, B.; Husung, S.; Furczyk, K.; Baune, B.T.; Kropp, P.; Häßler, F.; Thome, J. The Circadian System in Alzheimer’s Disease: Disturbances, Mechanisms, and Opportunities. Biol. Psychiatry 2013, 74, 333–339. [Google Scholar] [CrossRef]
- Kondratova, A.A.; Kondratov, R.V. The circadian clock and pathology of the ageing brain. Nat. Rev. Neurosci. 2012, 13, 325–335. [Google Scholar] [CrossRef]
- Kang, J.E. Amyloid-beta dynamics are regulated by orexin and the sleep-wake cycle. Science 2009, 326, 1005–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holth, J.K.; Fritschi, S.K.; Wang, C.; Pedersen, N.P.; Cirrito, J.R.; Mahan, T.E.; Finn, M.B.; Manis, M.; Geerling, J.C.; Fuller, P.M.; et al. The sleep-wake cycle regulates brain interstitial fluid tau in mice and CSF tau in humans. Science 2019, 363, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Jiang, W.; Zhang, E.E. Orexin signaling regulates both the hippocampal clock and the circadian oscillation of Alzheimer’s disease-risk genes. Sci. Rep. 2016, 6, 36035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roh, J.H.; Jiang, H.; Finn, M.B.; Stewart, F.R.; Mahan, T.; Cirrito, J.R.; Heda, A.; Snider, B.J.; Li, M.; Yanagisawa, M.; et al. Potential role of orexin and sleep modulation in the pathogenesis of Alzheimer’s disease. J. Exp. Med. 2014, 211, 2487–2496. [Google Scholar] [CrossRef]
- Xie, L.; Kang, H.; Xu, Q.; Chen, M.J.; Liao, Y.; Thiyagarajan, M.; O’Donnell, J.; Christensen, D.J.; Nicholson, C.; Iliff, J.J.; et al. Sleep Drives Metabolite Clearance from the Adult Brain. Science 2013, 342, 373–377. [Google Scholar] [CrossRef] [Green Version]
- Tranah, G.J.; Ma, T.B.; Stone, K.L.; Ancoli-Israel, S.; Paudel, M.L.; Ensrud, K.; Cauley, J.A.; Redline, S.; Hillier, T.A.; Cummings, S.R.; et al. Circadian activity rhythms and risk of incident dementia and mild cognitive impairment in older women. Ann. Neurol. 2011, 70, 722–732. [Google Scholar] [CrossRef] [Green Version]
- Lim, A.S. Modification of the relationship of the apolipoprotein E epsilon4 allele to the risk of Alzheimer disease and neurofibrillary tangle density by sleep. JAMA Neurol. 2013, 70, 1544–1551. [Google Scholar] [CrossRef] [Green Version]
- Ashbrook, L.H.; Krystal, A.D.; Fu, Y.-H.; Ptáček, L.J. Genetics of the human circadian clock and sleep homeostat. Neuropsychopharmacology 2020, 45, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Mattis, J.; Sehgal, A. Circadian Rhythms, Sleep, and Disorders of Aging. Trends Endocrinol. Metab. 2016, 27, 192–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, A.C.; Welsh, D.K.; Ko, C.H.; Tran, H.G.; Zhang, E.; Priest, A.A.; Buhr, E.D.; Singer, O.; Meeker, K.; Verma, I.M.; et al. Intercellular Coupling Confers Robustness against Mutations in the SCN Circadian Clock Network. Cell 2007, 129, 605–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; HogenEsch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. In Proceedings of the National Academy of Sciences, Dallas, TX, USA, 19 September 2014; Volume 111, pp. 16219–16224. [Google Scholar]
- Kress, G.J. Regulation of amyloid-beta dynamics and pathology by the circadian clock. J. Exp. Med. 2018, 215, 1059–1068. [Google Scholar] [CrossRef]
- Cermakian, N.; Lamont, E.W. Circadian clock gene expression in brain regions of Alzheimer’s disease patients and control subjects. J. Biol. Rhythm. 2011, 26, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Pasinetti, G.M.; Eberstein, J.A. Metabolic syndrome and the role of dietary lifestyles in Alzheimer’s disease. J. Neurochem. 2008, 106, 1503–1514. [Google Scholar] [CrossRef] [Green Version]
- Ho, L.; Qin, W.; Pompl, P.N. Diet-induced insulin resistance promotes amyloidosis in a transgenic mouse model of Alzheimer’s disease. FASEB J. 2004, 18, 902–904. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Sato, N. Diabetes-accelerated memory dysfunction via cerebrovascular inflammation and Abeta deposition in an Alzheimer mouse model with diabetes. Proc. Natl. Acad. Sci. USA 2010, 107, 7036–7041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, J.M.; Dixit, S.; Saulsberry, A.C. Reversal of high fat diet-induced obesity improves glucose tolerance, inflammatory response, beta-amyloid accumulation and cognitive decline in the APP/PSEN1 mouse model of Alzheimer’s disease. Neurobiol. Dis. 2017, 100, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Loehfelm, A.; Boucsein, A.; Pretz, D.; Tups, A. Timing Matters: Circadian Effects on Energy Homeostasis and Alzheimer’s Disease. Trends Endocrinol. Metab. 2019, 30, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Arnold, S.E.; Arvanitakis, Z.; Macauley, S.; Koenig, A.M.; Wang, H.-Y.; Ahima, R.S.; Craft, S.; Gandy, S.; Buettner, C.; Stoeckel, L.E.; et al. Brain insulin resistance in type 2 diabetes and Alzheimer disease: Concepts and conundrums. Nat. Rev. Neurol. 2018, 14, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.; Cai, W.; Konishi, M.; Kahn, C.R. Insulin signaling in the hippocampus and amygdala regulates metabolism and neurobehavior. Proc. Natl. Acad. Sci. USA 2019, 116, 6379–6384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steen, E.; Terry, B.M.; Enrique, J.R. Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer’s disease--is this type 3 diabetes? J. Alzheimers Dis. 2005, 7, 63–80. [Google Scholar] [CrossRef] [Green Version]
- Talbot, K.; Wang, H.-Y.; Kazi, H.; Han, L.-Y.; Bakshi, K.P.; Stucky, A.; Fuino, R.L.; Kawaguchi, K.R.; Samoyedny, A.J.; Wilson, R.S.; et al. Demonstrated brain insulin resistance in Alzheimer’s disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J. Clin. Investig. 2012, 122, 1316–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, K.F.; Befroy, D.; Dufour, S.; Dziura, J.; Ariyan, C.; Rothman, D.L.; DiPietro, L.; Cline, G.W.; Shulman, G.I. Mitochondrial Dysfunction in the Elderly: Possible Role in Insulin Resistance. Science 2003, 300, 1140–1142. [Google Scholar] [CrossRef] [Green Version]
- Sergi, D.; Naumovski, N.N.; Heilbronn, L.H.K.; Abeywardena, M.; O’Callaghan, N.; Lionetti, L.; Luscombe-Marsh, N.L.-M. Mitochondrial (Dys)function and Insulin Resistance: From Pathophysiological Molecular Mechanisms to the Impact of Diet. Front. Physiol. 2019, 10, 532. [Google Scholar] [CrossRef]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef]
- Mattson, M.P.; Arumugam, T.V. Hallmarks of Brain Aging: Adaptive and Pathological Modification by Metabolic States. Cell Metab. 2018, 27, 1176–1199. [Google Scholar] [CrossRef] [Green Version]
- Bass, J.; Lazar, M.A. Circadian time signatures of fitness and disease. Science 2016, 354, 994–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sulli, G.; Manoogian, E.N.; Taub, P.R.; Panda, S. Training the Circadian Clock, Clocking the Drugs, and Drugging the Clock to Prevent, Manage, and Treat Chronic Diseases. Trends Pharmacol. Sci. 2018, 39, 812–827. [Google Scholar] [CrossRef] [PubMed]
- Hatori, M.; Vollmers, C.; Zarrinpar, A.; DiTacchio, L.; Bushong, E.A.; Gill, S.; Leblanc, M.; Chaix, A.; Joens, M.; Fitzpatrick, J.A.; et al. Time-Restricted Feeding without Reducing Caloric Intake Prevents Metabolic Diseases in Mice Fed a High-Fat Diet. Cell Metab. 2012, 15, 848–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acosta-Rodriguez, V.A.; de Groot, M.H.M.; Rijo-Ferreira, F. Mice under Caloric Restriction Self-Impose a Temporal Restriction of Food Intake as Revealed by an Automated Feeder System. Cell Metab. 2017, 26, 267–277. [Google Scholar] [CrossRef] [Green Version]
- Makwana, K.; Gosai, N.; Poe, A.; Kondratov, R.V. Calorie restriction reprograms diurnal rhythms in protein translation to regulate metabolism. FASEB J. 2019, 33, 4473–4489. [Google Scholar] [CrossRef]
- Kojetin, D.J.; Burris, T.P. REV-ERB and ROR nuclear receptors as drug targets. Nat. Rev. Drug Discov. 2014, 13, 197–216. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Yoo, S.-H.; Takahashi, J.S. Development and Therapeutic Potential of Small-Molecule Modulators of Circadian Systems. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 231–252. [Google Scholar] [CrossRef]
- Hirota, T.; Kay, S.A. Identification of Small-Molecule Modulators of the Circadian Clock. Methods Enzymol. 2015, 551, 267–282. [Google Scholar] [CrossRef]
- Nohara, K.; Mallampalli, V.; Nemkov, T.; Wirianto, M.; Yang, J.; Ye, Y.; Sun, Y.; Han, L.; Esser, K.A.; Mileykovskaya, E.; et al. Nobiletin fortifies mitochondrial respiration in skeletal muscle to promote healthy aging against metabolic challenge. Nat. Commun. 2019, 10, 3923. [Google Scholar] [CrossRef] [Green Version]
- He, B.; Nohara, K.; Park, N.; Park, Y.S.; Guillory, B.; Zhao, Z.; Garcia, J.M.; Koike, N.; Lee, C.C.; Takahashi, J.S.; et al. The Small Molecule Nobiletin Targets the Molecular Oscillator to Enhance Circadian Rhythms and Protect against Metabolic Syndrome. Cell Metab. 2016, 23, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Nohara, K.; Shin, Y.; Park, N.; Jeong, K.; He, B.; Koike, N.; Yoo, S.-H.; Chen, Z. Ammonia-lowering activities and carbamoyl phosphate synthetase 1 (Cps1) induction mechanism of a natural flavonoid. Nutr. Metab. 2015, 12, 23. [Google Scholar] [CrossRef] [Green Version]
- Nohara, K.; Kim, E.; Wirianto, M.; Mileykovskaya, E.; Dowhan, W.; Chen, Z.; Yoo, S.-H. Cardiolipin Synthesis in Skeletal Muscle Is Rhythmic and Modifiable by Age and Diet. Oxid. Med. Cell. Longev. 2020, 2020, 1–12. [Google Scholar] [CrossRef]
- Evans, M.; Sharma, P.; Guthrie, N. Bioavailability of Citrus Polymethoxylated Flavones and Their Biological Role in Metabolic Syndrome and Hyperlipidemia. In Readings Advanced Pharmacokinetics—Theory, Methods and Applications; InTech: London, UK, 2012; pp. 1–19. [Google Scholar]
- Huang, H.; Li, L.; Shi, W. The Multifunctional Effects of Nobiletin and Its Metabolites In Vivo and In Vitro. Evid. Based Complementary Alternat. Med. 2016, 2016, 2918796. [Google Scholar]
- Mulvihill, E.E.; Burke, A.C.; Huff, M.W. Citrus Flavonoids as Regulators of Lipoprotein Metabolism and Atherosclerosis. Annu. Rev. Nutr. 2016, 36, 275–299. [Google Scholar] [CrossRef]
- Walle, T. Methoxylated flavones, a superior cancer chemopreventive flavonoid subclass? Semin. Cancer Biol. 2007, 17, 354–362. [Google Scholar] [CrossRef] [Green Version]
- Mileykovskaya, E.; Yoo, S.-H.; Dowhan, W.; Chen, Z. Nobiletin: Targeting the Circadian Network to Promote Bioenergetics and Healthy Aging. Biochemistry 2020, 85, 1554–1559. [Google Scholar] [CrossRef]
- Nakajima, A.; Ohizumi, Y. Potential Benefits of Nobiletin, A Citrus Flavonoid, against Alzheimer’s Disease and Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 3380. [Google Scholar] [CrossRef] [Green Version]
- Cirmi, S.; Ferlazzo, N.; Lombardo, G.E.; Ventura-Spagnolo, E.; Gangemi, S.; Calapai, G.; Navarra, M. Neurodegenerative Diseases: Might Citrus Flavonoids Play a Protective Role? Molecules 2016, 21, 1312. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Wirianto, M.; Kim, E.; Burish, M.; Yoo, S.-H.; Chen, Z. Clock-Modulating Activities of the Anti-Arrhythmic Drug Moricizine. Clocks Sleep 2021, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Lee, S.-Y.; Koike, N.; Kim, E.; Wirianto, M.; Burish, M.J.; Yagita, K.; Lee, H.K.; Chen, Z.; Chung, J.M.; et al. Circadian regulation of chemotherapy-induced peripheral neuropathic pain and the underlying transcriptomic landscape. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wirianto, M.; Yang, J.; Kim, E. The GSK-3beta-FBXL21 Axis Contributes to Circadian TCAP Degradation and Skeletal Muscle Function. Cell Rep. 2020, 32, 108140. [Google Scholar] [CrossRef] [PubMed]
- Duran-Aniotz, C.; Moreno-Gonzalez, I.; Gamez, N.; Perez-Urrutia, N.; Vegas-Gomez, L.; Soto, C.; Morales, R. Amyloid pathology arrangements in Alzheimer’s disease brains modulate in vivo seeding capability. Acta Neuropathol. Commun. 2021, 9, 1–13. [Google Scholar] [CrossRef]
- Morales, R.; Bravo-Alegria, J.; Moreno-Gonzalez, I. Transmission of cerebral amyloid pathology by peripheral administration of misfolded Abeta aggregates. Mol. Psychiatry 2021. [Google Scholar] [CrossRef]
- Jankowsky, J.L.; Fadale, D.J.; Anderson, J. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: Evidence for augmentation of a 42-specific gamma secretase. Hum. Mol. Genet. 2004, 13, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Jetten, A.M. Retinoid-Related Orphan Receptors (RORs): Critical Roles in Development, Immunity, Circadian Rhythm, and Cellular Metabolism. Nucl. Recept. Signal. 2009, 7, e003. [Google Scholar] [CrossRef] [Green Version]
- Marwarha, G.; Schommer, J.; Lude, J. Palmitate-induced C/EBP homologous protein activation leads to NF-kappaB-mediated increase in BACE1 activity and amyloid beta genesis. J. Neurochem. 2018, 144, 761–779. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Song, R.; Lu, W.; Liu, Z.; Wang, L.; Zhu, X.; Liu, Y.; Sun, Z.; Li, J.; Li, X. YXQN Reduces Alzheimer’s Disease-Like Pathology and Cognitive Decline in APPswePS1dE9 Transgenic Mice. Front. Aging Neurosci. 2017, 9, 157. [Google Scholar] [CrossRef]
- Vandal, M.; White, P.J.; Thornissac, M. Impaired thermoregulation and beneficial effects of thermoneutrality in the 3xTg-AD model of Alzheimer’s disease. Neurobiol. Aging 2016, 43, 47–57. [Google Scholar] [CrossRef]
- Nohara, K.; Nemkov, T.; D’Alessandro, A.; Yoo, S.-H.; Chen, Z. Coordinate Regulation of Cholesterol and Bile Acid Metabolism by the Clock Modifier Nobiletin in Metabolically Challenged Old Mice. Int. J. Mol. Sci. 2019, 20, 4281. [Google Scholar] [CrossRef] [Green Version]
- Hooijmans, C.R.; Graven, C.; Dederen, P.J.; Tanila, H.; van Groen, T.; Kiliaan, A.J. Amyloid beta deposition is related to decreased glucose transporter-1 levels and hippocampal atrophy in brains of aged APP/PS1 mice. Brain Res. 2007, 1181, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Das, T.K.; Chakrabarti, S.K.; Zulkipli, I.N.; Hamid, M.R.A. Curcumin Ameliorates the Impaired Insulin Signaling Involved in the Pathogenesis of Alzheimer’s Disease in Rats. J. Alzheimer’s Dis. Rep. 2019, 3, 59–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Yang, X.; Jiao, M.; Anoopkumar-Dukie, S.; Zeng, Y.; Mei, H. Housefly (Musca domestica) larvae powder, preventing oxidative stress injury via regulation of UCP4 and CyclinD1 and modulation of JNK and P38 signaling in APP/PS1 mice. Food Funct. 2018, 10, 235–243. [Google Scholar] [CrossRef]
- Perluigi, M.; Di Domenico, F.; Barone, E.; Butterfield, D. mTOR in Alzheimer disease and its earlier stages: Links to oxidative damage in the progression of this dementing disorder. Free Radic. Biol. Med. 2021, 169, 382–396. [Google Scholar] [CrossRef]
- Burillo, J.; Marqués, P.; Jiménez, B.; González-Blanco, C.; Benito, M.; Guillén, C. Insulin Resistance and Diabetes Mellitus in Alzheimer’s Disease. Cells 2021, 10, 1236. [Google Scholar] [CrossRef]
- Uddin, M.S.; Kabir, M.; Niaz, K.; Jeandet, P.; Clément, C. Molecular Insight into the Therapeutic Promise of Flavonoids against Alzheimer’s Disease. Molecules 2020, 25, 1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, A.R.; Stanton, D.; Roman, D.; Liu, A.C. Systems Level Understanding of Circadian Integration with Cell Physiology. J. Mol. Biol. 2020, 432, 3547–3564. [Google Scholar] [CrossRef]
- Petrenko, V.; Gandasi, N.R.; Sage, D.; Tengholm, A.; Barg, S.; Dibner, C. In pancreatic islets from type 2 diabetes patients, the dampened circadian oscillators lead to reduced insulin and glucagon exocytosis. Proc. Natl. Acad. Sci. USA 2020, 117, 2484–2495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Y.F.; Guo, Z.; Zheng, T.; Jiang, Y.; Yan, Y.; Yin, X. Intranasal insulin alleviates cognitive deficits and amyloid pathology in young adult APPswe/PS1dE9 mice. Aging Cell 2016, 15, 893–902. [Google Scholar] [CrossRef] [Green Version]
- Nakajima, A.; Aoyama, Y.; Shin, E.J.; Nam, Y. Nobiletin, a citrus flavonoid, improves cognitive impairment and reduces soluble ABeta levels in a triple transgenic mouse model of Alzheimer’s disease. Behav. Brain Res. 2015, 289, 69–77. [Google Scholar] [CrossRef]
- Seki, T.; Kamiya, T.; Furukawa, K.; Azumi, M. Nobiletin-rich Citrus reticulata peels, a kampo medicine for Alzheimer’s disease: A case series. Geriatr. Gerontol. Int. 2013, 13, 236–238. [Google Scholar] [CrossRef]
- Sundaram, S.; Nagaraj, S.; Mahoney, H.; Portugues, A. Inhibition of casein kinase 1delta/epsilon improves cognitive-affective behavior and reduces amyloid load in the APP-PS1 mouse model of Alzheimer’s disease. Sci. Rep. 2019, 9, 13743. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Kim, D.E.; Griffin, P.; Sheehan, P.; Kim, D.; Musiek, E.S.; Yoon, S. Inhibition of REV-ERBs stimulates microglial amyloid-beta clearance and reduces amyloid plaque deposition in the 5XFAD mouse model of Alzheimer’s disease. Aging Cell 2019, 19, e13078. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.; Wu, Z.; Meng, J.; Saito, T.; Saido, T.C.; Qing, H.; Nakanishi, H. An impaired intrinsic microglial clock system induces neuroinflammatory alterations in the early stage of amyloid precursor protein knock-in mouse brain. J. Neuroinflamm. 2019, 16, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Roby, D.A.; Ruiz, F.; Kermath, B.A.; Voorhees, J.R. Pharmacological activation of the nuclear receptor REV-ERB reverses cognitive deficits and reduces amyloid-beta burden in a mouse model of Alzheimer’s disease. PLoS ONE 2019, 14, e0215004. [Google Scholar] [CrossRef] [PubMed]
- Waters, A.N.; Society for Women’s Health Research Alzheimer’s Disease; Laitner, M.H. Biological sex differences in Alzheimer’s preclinical research: A call to action. Alzheimers Dement. 2021, 7, e12111. [Google Scholar]
- Toro, C.A.; Zhang, L.; Cao, J.; Cao, D. Sex differences in Alzheimer’s disease: Understanding the molecular impact. Brain Res. 2019, 1719, 194–207. [Google Scholar] [CrossRef]



| Forward (5′–3′) | Reverse (5′–3′) | |
|---|---|---|
| Clock | CCTTCAGCAGTCAGTCCATAAAC | AGACATCGCTGGCTGTGTTAA |
| Bmal1 | CCACCTCAGAGCCATTGATACA | GAGCAGGTTTAGTTCCACTTTGTCT |
| Per1 | TTCGTGGACTTGACACCTCTT | GGGAACGCTTTGCTTTAGAT |
| Per2 | ATGCTCGCCATCCACAAGA | GCGGAATCGAATGGGAGAAT |
| Cry1 | CTGGCGTGGAAGTCATCGT | CTGTCCGCCATTGAGTTCTATG |
| Npas2 | CAACAGACGGCAGCATCATCT | TTCTGATCCATGACATCCGC |
| Rora | GCACCTGACCGAAGACGAAA | GAGCGATCCGCTGACATCA |
| Rorb | GACCCACACCTACGAGGAAA | GTGATCTGGATGGCACACTG |
| Nr1d1 | CATGGTGCTACTGTGTAAGGTGTGT | CACAGGCGTGCACTCCATAG |
| Dbp | CTGGCCCGAGTCTTTTTGC | CCAGGTCCACGTATTCCACG |
| App | AGCACCGAGAGAGAATGTCC | GCAAGTTCTTGGCTTGACG |
| Bace1 | ACATTGCTGCCATCACTGAA | GCCTGGCAATCTCAGCATAG |
| Bace2 | TGAGGACCTTGTCACCATCCCAAA | TGGCCAAAGCAGCATAAGCAAGTC |
| ApoE | ATTGCGAAGATGAAGGCTCT | CCACTCGAGCTGATCTGTCA |
| Scna | TGACAGCAGTCGCTCAGA | CATGTCTTCCAGGATTCCTTC |
| Scnb | GGAGGAGCTGTGTTCTCTGG | TCCTCTGGCTTCAGGTCTGT |
| Lrp1 | ATTGAGGGCAAGATGACACA | CCAGTCTGTCCAGTACATCCAC |
| Igf1 | TGGTGACCGGCTACGTGAAG | CAAAGTACATCTTTCCGGACC |
| Igf1r | ATCGCGATTTCTGCGCCAACA | TTCTTCTCTTCATCGCCGCAGACT |
| Glut1 | AGCCCTGCTACAGTGTAT | AGGTCTCGGGTCACATC |
| Glut3 | TAAACCAGCTGGGCATCGTTGTTG | AATGATGGTTAAGCCAAGGAGCCC |
| Insr | GACAGCCACCACACTCACACTTC | GTGCAGCTCCTCATCACCATATCG |
| Irs1 | CCAGCCTGGCTATTTAGCTG | TTCTCTAGGAGCTGGGTGGA |
| Irs2 | TCTTTCACGACTGTGGCTTCCTT | CACTGGAGCTTTGCCCTCTGC |
| Lepr | GTGTGAGGAGGTACGTGGTGAAG | CCGAGGGAATTGACAGCCAGAAC |
| Ucp2 | ATGGTTGGTTTCAAGGCCACA | CGGTATCCAGAGGGAAAGTGAT |
| Ucp4 | TCGAGACAAACAAGGAAGGGG | GACCAAGGGGTCATTCTCAGC |
| Cpt1c | GCAGGAGATCTCACCGACAT | CCCTGGAATCCGTGTAGTGT |
| Gapdh | CAAGGTCATCCATGACAACTTTG | GGCCATCCACAGTCTTCTGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.; Nohara, K.; Wirianto, M.; Escobedo, G., Jr.; Lim, J.Y.; Morales, R.; Yoo, S.-H.; Chen, Z. Effects of the Clock Modulator Nobiletin on Circadian Rhythms and Pathophysiology in Female Mice of an Alzheimer’s Disease Model. Biomolecules 2021, 11, 1004. https://doi.org/10.3390/biom11071004
Kim E, Nohara K, Wirianto M, Escobedo G Jr., Lim JY, Morales R, Yoo S-H, Chen Z. Effects of the Clock Modulator Nobiletin on Circadian Rhythms and Pathophysiology in Female Mice of an Alzheimer’s Disease Model. Biomolecules. 2021; 11(7):1004. https://doi.org/10.3390/biom11071004
Chicago/Turabian StyleKim, Eunju, Kazunari Nohara, Marvin Wirianto, Gabriel Escobedo, Jr., Ji Ye Lim, Rodrigo Morales, Seung-Hee Yoo, and Zheng Chen. 2021. "Effects of the Clock Modulator Nobiletin on Circadian Rhythms and Pathophysiology in Female Mice of an Alzheimer’s Disease Model" Biomolecules 11, no. 7: 1004. https://doi.org/10.3390/biom11071004
APA StyleKim, E., Nohara, K., Wirianto, M., Escobedo, G., Jr., Lim, J. Y., Morales, R., Yoo, S. -H., & Chen, Z. (2021). Effects of the Clock Modulator Nobiletin on Circadian Rhythms and Pathophysiology in Female Mice of an Alzheimer’s Disease Model. Biomolecules, 11(7), 1004. https://doi.org/10.3390/biom11071004






