Sub-Lethal 5-Fluorouracil Dose Challenges Planarian Stem Cells Promoting Transcriptional Profile Changes in the Pluripotent Sigma-Class Neoblasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals, 5FU Treatment, and Regeneration Experiments
2.2. Whole Mount In Situ Hybridization and Post-Hybridization Immunofluorescence
2.3. Bromodeoxyuridine Labeling and Immunofluorescence on Tissue Sections
2.4. Cytofluorimetric Analysis of DNA Content
2.5. TUNEL Assay
2.6. Statistical Analysis
3. Results
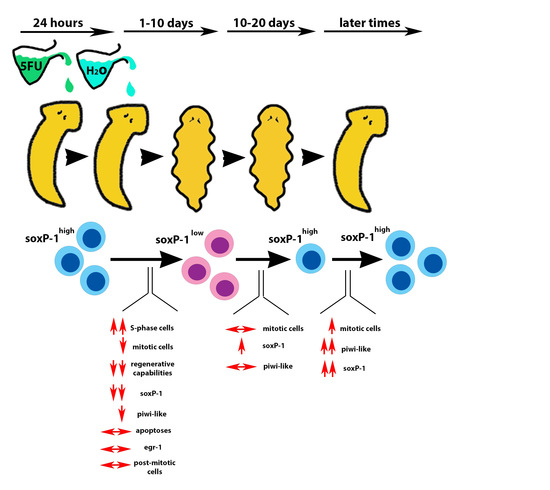
3.1. Analysis of Stem Cell Markers Following SDT-600 Treatment
3.2. Analysis of Cell Cycle Progression and Apoptosis Following SDT-600 Treatment
3.3. Correlation between the Transcriptional Change in the DjsoxP-1 Level and the Expression of Lineage Differentiation Markers
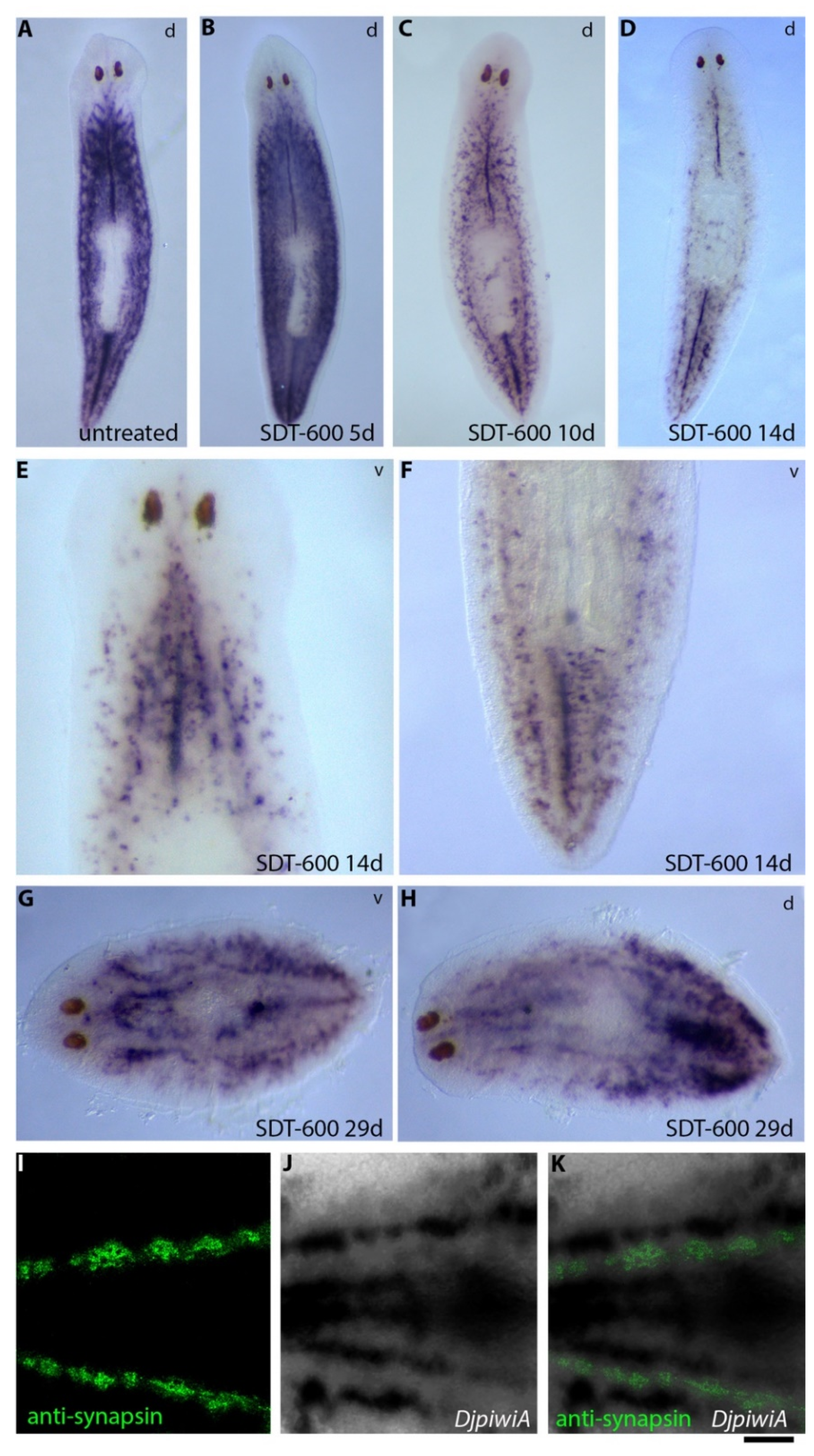
3.4. Analysis of Regenerative Performances and DjsoxP-1 Reactivation Following SDT-600 Treatment
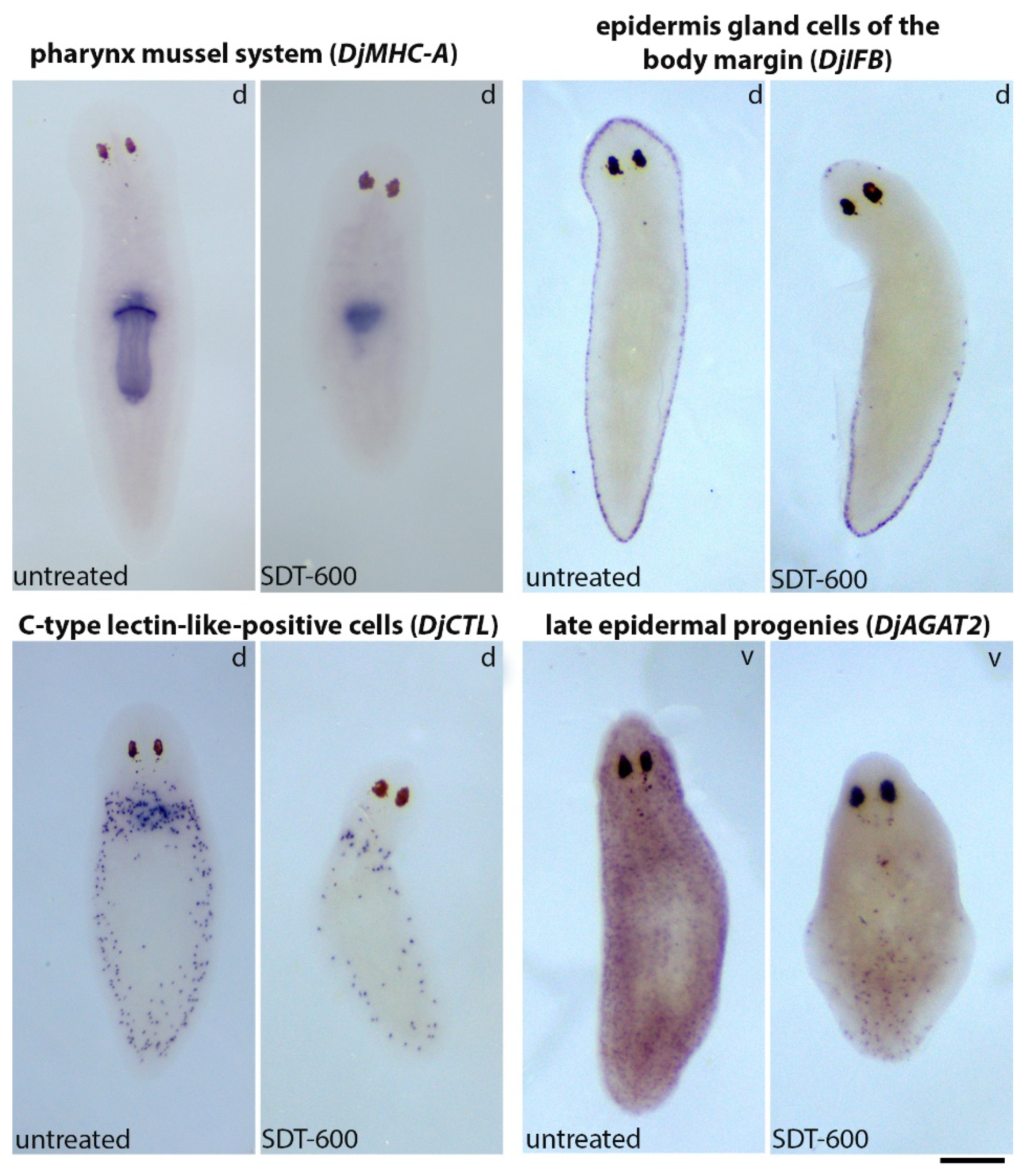
3.5. Analysis of Differentiated Tissues at Late Times after SDT-600 Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Alessandra, S.; Rossi, L. Planarian stem cell heterogeneity. Stem Cells Heterog. Nov. Concepts 2019, 1123, 39–54. [Google Scholar]
- Rossi, L.; Salvetti, A.; Batistoni, R.; Deri, P.; Gremigni, V. Planarians, a tale of stem cells. Cell Mol. Life Sci. 2008, 65, 16–23. [Google Scholar] [CrossRef]
- Reddien, P.W.; Oviedo, N.J.; Jennings, J.R.; Jenkin, J.C.; Alvarado, A.S. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 2005, 310, 1327–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibata, N.; Rouhana, L.; Agata, K. Cellular and molecular dissection of pluripotent adult somatic stem cells in planarians. Dev. Growth Differ. 2010, 52, 27–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Wolfswinkel, J.C.; Wagner, D.E.; Reddien, P.W. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 2014, 15, 326–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, A.G.; Kosaka, N.; Abnave, P.; Sahu, S.; Aboobaker, A.A. The abrogation of condensin function provides independent evidence for defining the self-renewing population of pluripotent stem cells. Dev. Biol. 2018, 433, 218–226. [Google Scholar] [CrossRef]
- Gambino, G.; Ippolito, C.; Modeo, L.; Salvetti, A.; Rossi, L. 5-Fluorouracil-treated planarians, a versatile model system for studying stem cell heterogeneity and tissue aging. Biol. Cell 2020, 112, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Orii, H.; Agata, K.; Watanabe, K. POU-domain genes in planarian Dugesia japonica: The structure and expression. Biochem. Biophys. Res. Commun. 1993, 192, 1395–1402. [Google Scholar] [CrossRef]
- Cassella, L.; Salvetti, A.; Iacopetti, P.; Ippolito, C.; Ghezzani, C.; Gimenez, G.; Ghigo, E.; Rossi, L. Putrescine independent wound response phenotype is produced by ODC-like RNAi in planarians. Sci. Rep. 2017, 7, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Gambino, G.; Falleni, A.; Nigro, M.; Salvetti, A.; Cecchettini, A.; Ippolito, C.; Guidi, P.; Rossi, L. Dynamics of interaction and effects of microplastics on planarian tissue regeneration and cellular homeostasis. Aquat. Toxicol. 2020, 218, 105354. [Google Scholar] [CrossRef]
- Salvetti, A.; Rossi, L.; Deri, P.; Batistoni, R. An MCM2-related gene is expressed in proliferating cells of intact and regenerating planarians. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2000, 218, 603–614. [Google Scholar]
- Rossi, L.; Bonuccelli, L.; Iacopetti, P.; Evangelista, M.; Ghezzani, C.; Tana, L.; Salvetti, A. Prohibitin 2 regulates cell proliferation and mitochondrial cristae morphogenesis in planarian stem cells. Stem Cell Rev. Rep. 2014, 10, 871–887. [Google Scholar] [CrossRef]
- Rossi, L.; Salvetti, A.; Lena, A.; Batistoni, R.; Deri, P.; Pugliesi, C.; Loreti, E.; Gremigni, V. DjPiwi-1, a member of the PAZ-Piwi gene family, defines a subpopulation of planarian stem cells. Dev. Genes Evol. 2006, 216, 335. [Google Scholar] [CrossRef]
- Rossi, L.; Salvetti, A.; Marincola, F.M.; Lena, A.; Deri, P.; Mannini, L.; Batistoni, R.; Wang, E.; Gremigni, V. Deciphering the molecular machinery of stem cells: A look at the neoblast gene expression profile. Genome Biol. 2007, 8, 1–17. [Google Scholar] [CrossRef]
- Salvetti, A.; Rossi, L.; Bonuccelli, L.; Lena, A.; Pugliesi, C.; Rainaldi, G.; Evangelista, M.; Gremigni, V. Adult stem cell plasticity: Neoblast repopulation in non-lethally irradiated planarians. Dev. Biol. 2009, 328, 305–314. [Google Scholar] [CrossRef]
- Rossi, L.; Cassella, L.; Iacopetti, P.; Ghezzani, C.; Tana, L.; Gimenez, G.; Ghigo, E.; Salvetti, A. Insight into stem cell regulation from sub-lethally irradiated worms. Gene 2018, 662, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- King, R.S.; Newmark, P.A. In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev. Biol. 2013, 13, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ermakov, A.M.; Ermakova, O.N.; Kudravtsev, A.A.; Kreshchenko, N.D. Study of planarian stem cell proliferation by means of flow cytometry. Mol. Biol. Rep. 2012, 39, 3073–3080. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.E.; Ho, J.J.; Reddien, P.W. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell 2012, 10, 299–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, K.; Shibata, N.; Orii, H.; Amikura, R.; Sakurai, T.; Agata, K.; Kobayashi, S.; Watanabe, K. Identification and origin of the germline stem cells as revealed by the expression of nanos-related gene in planarians. Dev. Growth Differ. 2006, 48, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Eisenhoffer, G.T.; Kang, H.; Sánchez Alvarado, A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 2008, 3, 327–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, K.C.; Cheng, L.C.; Vu, H.T.; Lange, J.J.; McKinney, S.A.; Seidel, C.W.; Alvarado, A. SEgr-5 is a post-mitotic regulator of planarian epidermal differentiation. Elife 2015, 4, e10501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearson, B.J.; Sánchez Alvarado, A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development 2010, 137, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Flores, N.M.; Oviedo, N.J.; Sage, J. Essential role for the planarian intestinal GATA transcription factor in stem cells and regeneration. Dev. Biol. 2016, 418, 179–188. [Google Scholar] [CrossRef] [Green Version]
- Nogi, T.; Levin, M. Characterization of innexin gene expression and functional roles of gap-junctional communication in planarian regeneration. Dev. Biol. 2005, 287, 314–335. [Google Scholar] [CrossRef] [Green Version]
- Tazaki, A.; Kato, K.; Orii, H.; Agata, K.; Watanabe, K. The body margin of the planarian Dugesia japonica: Characterization by the expression of an intermediate filament gene. Dev. Genes Evol. 2002, 212, 365–373. [Google Scholar] [CrossRef]
- Tazaki, A.; Gaudieri, S.; Ikeo, K.; Gojobori, T.; Watanabe, K.; Agata, K. Neural network in planarian revealed by an antibody against planarian synaptotagmin homologue. Biochem. Biophys. Res. Commun. 1999, 260, 426–432. [Google Scholar] [CrossRef]
- Sakai, T.; Kato, K.; Watanabe, K.; Orii, H. Planarian pharynx regeneration revealed by the expression of myosin heavy chain-A. Int. J. Dev. Biol. 2002, 46, 329–332. [Google Scholar]
- Gao, L.; Han, Y.; Deng, H.; Hu, W.; Zhen, H.; Li, N.; Qin, N.; Yan, M.; Wu, W.; Liu, B.; et al. The role of a novel C-type lectin-like protein from planarian in innate immunity and regeneration. Dev. Comp. Immunol. 2017, 67, 413–426. [Google Scholar] [CrossRef]
- Bunz, F. Thymidylate synthase and 5-fluorouracil: A cautionary tale. Cancer Biol. Ther. 2008, 7, 995–996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, E.; Allegra, C.J. The role of thymidylate synthase in cellular regulation. Adv. Enzym. Regul. 1996, 36, 143–163. [Google Scholar] [CrossRef]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Moro, Y.; Kogashiwa, Y.; Sakurai, H.; Takahashi, R.; Kimura, T.; Hirasaki, M.; Matsumoto, Y.; Sugasawa, M.; Kohno, N. In vitro study of the anti-cancer effect of alternate-day 5-fluorouracil in head and neck cancer cells. Anticancer Res. 2019, 39, 6041–6047. [Google Scholar] [CrossRef] [PubMed]
- Ovejero, S.; Bueno, A.; Sacristán, M.P. Working on genomic stability: From the S-phase to mitosis. Genes 2020, 11, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raz, A.A.; Wurtzel, O.; Reddien, P.W. Planarian stem cells specify fate yet retain potency during the cell cycle. Cell Stem Cell 2021, S1934-5909, 00154–00155. [Google Scholar]
- Molinaro, A.M.; Lindsay-Mosher, N.; Pearson, B.J. Identification of TOR-responsive slow-cycling neoblasts in planarians. EMBO Rep. 2021, 22, e50292. [Google Scholar] [CrossRef]
- Wurtzel, O.; Cote, L.E.; Poirier, A.; Satija, R.; Regev, A.; Reddien, P.W. A generic and cell-type-specific wound response precedes regeneration in planarians. Dev. Cell 2015, 35, 632–645. [Google Scholar] [CrossRef] [Green Version]
- Owlarn, S.; Klenner, F.; Schmidt, D.; Rabert, F.; Tomasso, A.; Reuter, H.; Mulaw, M.A.; Moritz, S.; Gentile, L.; Weidinger, G.; et al. Generic wound signals initiate regeneration in missing-tissue contexts. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Wang, Y.; Zayas, R.M.; Guo, T.; Newmark, P.A. Nanos function is essential for development and regeneration of planarian germ cells. Proc. Natl. Acad. Sci. USA 2007, 104, 5901–5906. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gambino, G.; Ippolito, C.; Evangelista, M.; Salvetti, A.; Rossi, L. Sub-Lethal 5-Fluorouracil Dose Challenges Planarian Stem Cells Promoting Transcriptional Profile Changes in the Pluripotent Sigma-Class Neoblasts. Biomolecules 2021, 11, 949. https://doi.org/10.3390/biom11070949
Gambino G, Ippolito C, Evangelista M, Salvetti A, Rossi L. Sub-Lethal 5-Fluorouracil Dose Challenges Planarian Stem Cells Promoting Transcriptional Profile Changes in the Pluripotent Sigma-Class Neoblasts. Biomolecules. 2021; 11(7):949. https://doi.org/10.3390/biom11070949
Chicago/Turabian StyleGambino, Gaetana, Chiara Ippolito, Monica Evangelista, Alessandra Salvetti, and Leonardo Rossi. 2021. "Sub-Lethal 5-Fluorouracil Dose Challenges Planarian Stem Cells Promoting Transcriptional Profile Changes in the Pluripotent Sigma-Class Neoblasts" Biomolecules 11, no. 7: 949. https://doi.org/10.3390/biom11070949
APA StyleGambino, G., Ippolito, C., Evangelista, M., Salvetti, A., & Rossi, L. (2021). Sub-Lethal 5-Fluorouracil Dose Challenges Planarian Stem Cells Promoting Transcriptional Profile Changes in the Pluripotent Sigma-Class Neoblasts. Biomolecules, 11(7), 949. https://doi.org/10.3390/biom11070949