OsARP6 Is Involved in Internode Elongation by Regulating Cell-Cycle-Related Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subcellular Localization
2.2. Yeast Two-Hybrid Assay (Y2H)
2.3. Bimolecular Fluorescence Complementation (BiFC)
2.4. Genotyping
2.5. Histological Analyses
2.6. Reverse Transcription and Quantitative PCR (RT-qPCR)
2.7. ChIP Assay
3. Results
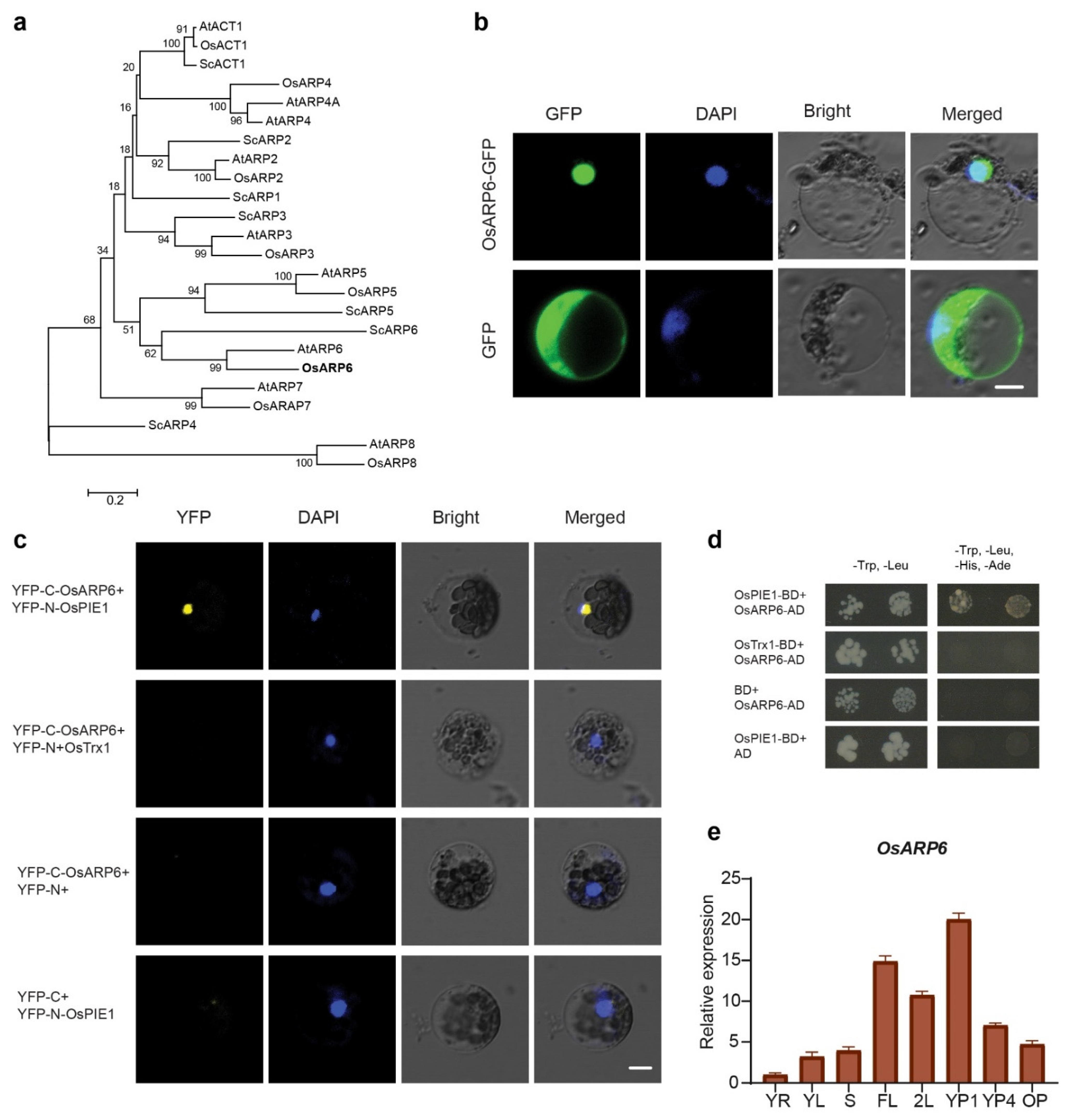
3.1. Phylogenetic Analysis and Subcellular Localization of OsARP6
3.2. OsARP6 Interacts with OsPIE1
3.3. Expression Pattern of OsARP6
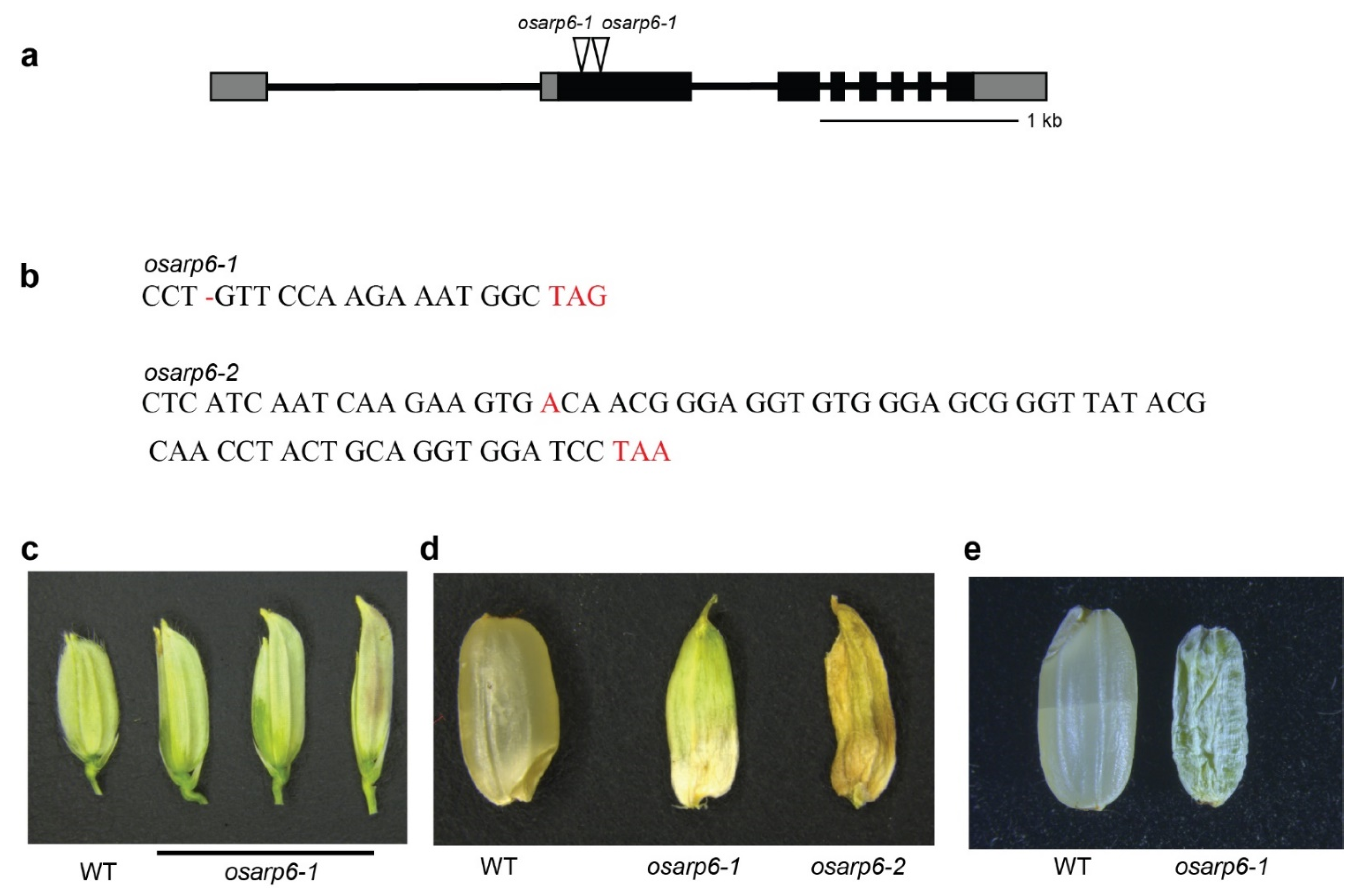
3.4. Generation of Rice osarp6 Mutants with CRISPR Cas9
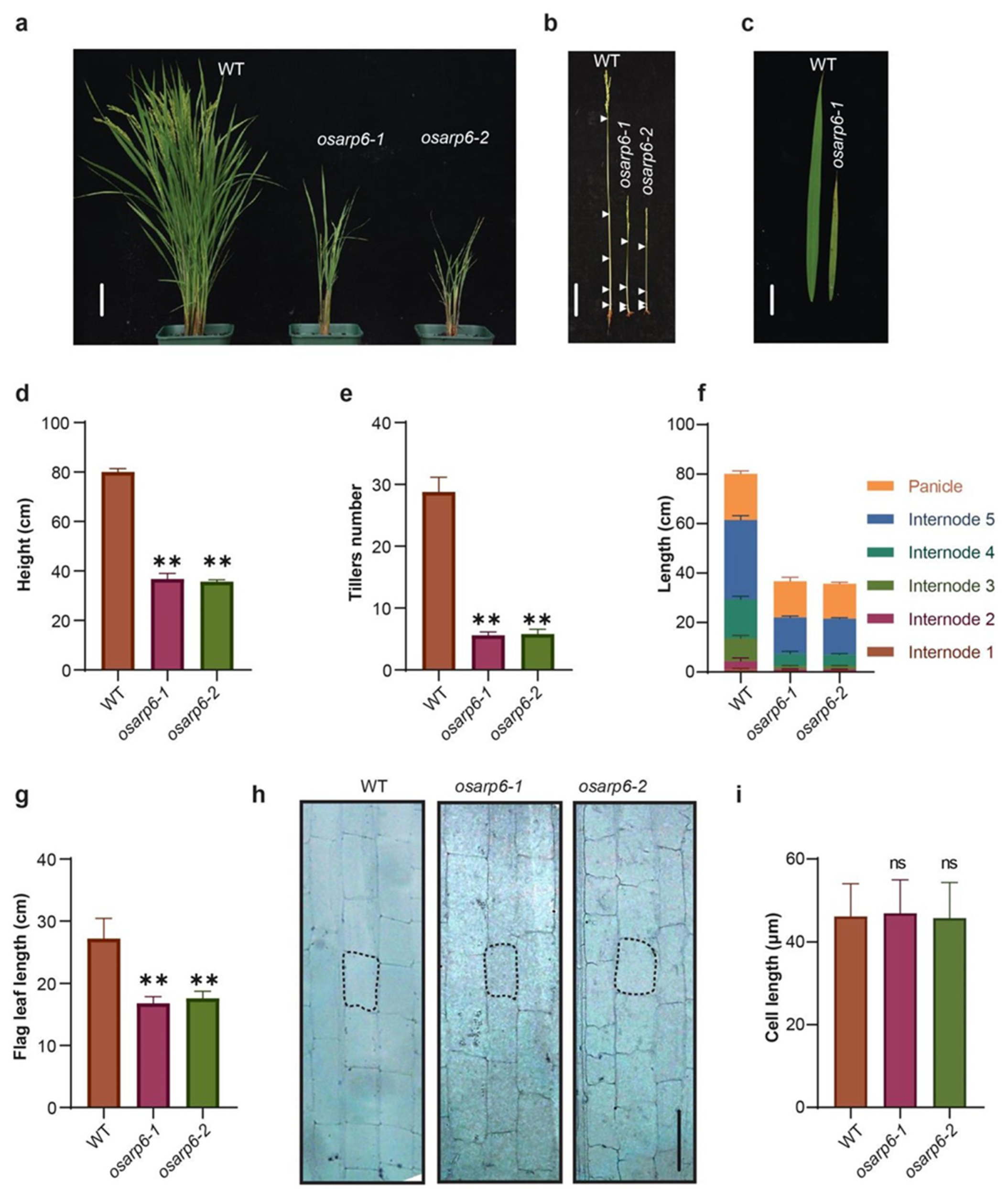
3.5. Loss of OsARP6 Function Resulted in Pleotropic Phenotypes
3.6. The Reduced Height of osarp6 Is Due to Defect in Cell Proliferation
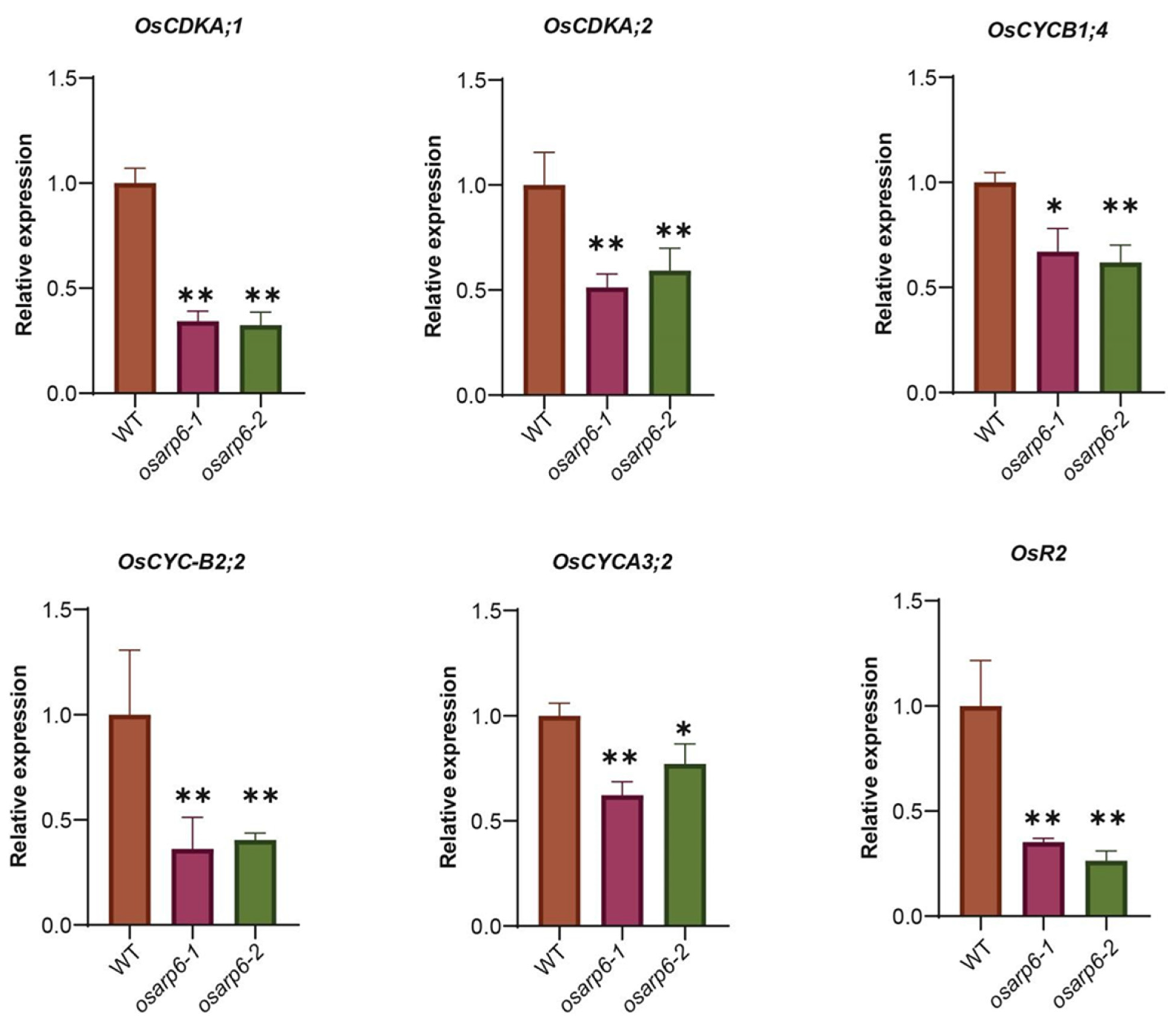
3.7. OsARP6 Is Involved Regulating the Expression of Various Cell-Cycle-Related Genes
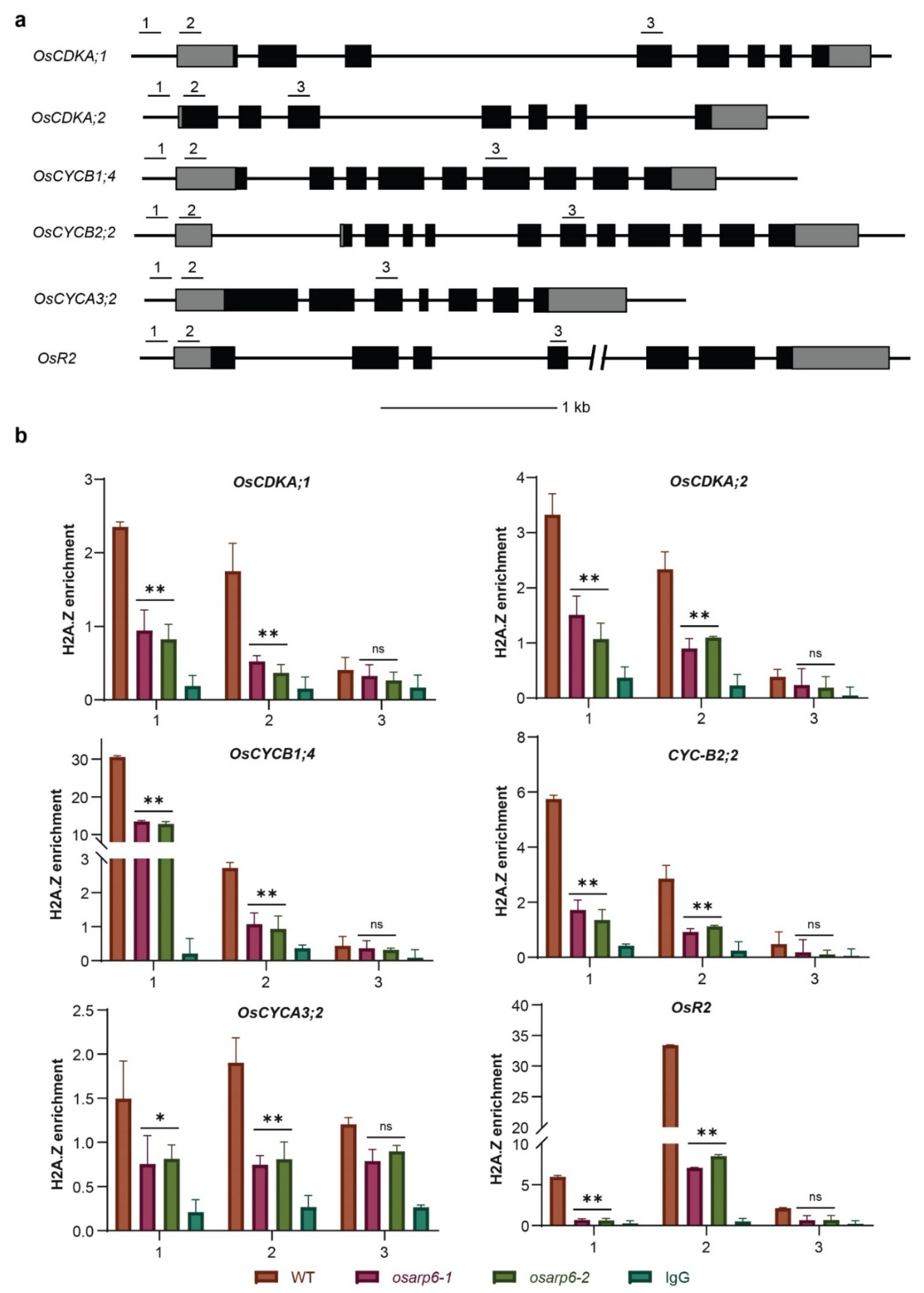
3.8. OsARP6 Is Required for H2A.Z Deposition at the Promoters and TSS of Cell-Cycle-Related Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, X.C.; Hwa, C.M. Genetic modification of plant architecture and variety improvement in rice. Heredity 2008, 101, 396–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khush, G.S. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–822. [Google Scholar] [CrossRef]
- Sakamoto, T.; Matsuoka, M. Generating high-yielding varieties by genetic manipulation of plant architecture. Curr. Opin. Biotechnol. 2004, 15, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, P.; Zhang, X.; Li, X.; Yan, X.; Fu, D.; Wu, G. The genetic and molecular basis of crop height based on a rice model. Planta 2018, 247, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Dechassa, M.L.; Tremethick, D.J. New insights into nucleosome and chromatin structure: An ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 2012, 13, 436–447. [Google Scholar] [CrossRef] [Green Version]
- Ho, L.; Crabtree, G.R. Chromatin remodelling during development. Nature 2010, 463, 474–484. [Google Scholar] [CrossRef]
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Krogan, N.J.; Keogh, M.-C.; Datta, N.; Sawa, C.; Ryan, O.W.; Ding, H.; Haw, R.; Pootoolal, J.; Tong, A.; Canadien, V.; et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 2003, 12, 1565–1576. [Google Scholar] [CrossRef] [Green Version]
- March-Díaz, R.; Reyes, J.C. The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant 2009, 2, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Rudnizky, S.; Bavly, A.; Malik, O.; Pnueli, L.; Melamed, P.; Kaplan, A. H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat. Commun. 2016, 7, 12958. [Google Scholar] [CrossRef]
- Thatcher, T.; Gorovsky, M.A. Phylogenetic analysis of the core histones H2A, H2B, H3, and H4. Nucleic Acids Res. 1994, 22, 174–179. [Google Scholar] [CrossRef]
- Mizuguchi, G.; Shen, X.; Landry, J.; Wu, W.-H.; Sen, S.; Wu, C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 2004, 303, 343–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meagher, R.B.; Kandasamy, M.K.; Deal, R.B.; McKinney, E.C. Actin-related proteins in chromatin-level control of the cell cycle and developmental transitions. Trends Cell Biol. 2007, 17, 325–332. [Google Scholar] [CrossRef]
- Jarillo, J.A.; Piñeiro, M. H2A.Z mediates different aspects of chromatin function and modulates flowering responses in Arabidopsis. Plant J. 2015, 83, 96–109. [Google Scholar] [CrossRef] [Green Version]
- Noh, Y.-S.; Amasino, R.M. PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 2003, 15, 1671–1682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, K.; Park, C.; Lee, J.; Oh, M.; Noh, B.; Lee, I. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 2007, 134, 1931–1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deal, R.B.; Topp, C.; McKinney, E.C.; Meagher, R.B. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 2007, 19, 74–83. [Google Scholar] [CrossRef] [Green Version]
- March-Díaz, R.; Garcia-Dominguez, M.; Florencio, F.J.; Reyes, J.C. SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 2007, 143, 893–901. [Google Scholar] [CrossRef] [Green Version]
- Kandasamy, M.K.; Deal, R.B.; McKinney, E.C.; Meagher, R.B. Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J. 2005, 41, 845–858. [Google Scholar] [CrossRef]
- Lázaro, A.; Gómez-Zambrano, Á.; López-González, L.; Piñeiro, M.; Jarillo, J.A. Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 2008, 59, 653–666. [Google Scholar] [CrossRef] [Green Version]
- Crevillén, P.; Gómez-Zambrano, Á.; López, J.A.; Vázquez, J.; Piñeiro, M.; Jarillo, J.A. Arabidopsis YAF9 histone readers modulate flowering time through NuA4-complex-dependent H4 and H2A.Z histone acetylation at FLC chromatin. New Phytol. 2019, 222, 1893–1908. [Google Scholar] [CrossRef]
- Gómez-Zambrano, Á.; Crevillén, P.; Franco-Zorrilla, J.M.; Lopez, J.A.; Moreno-Romero, J.; Roszak, P.; Santos-González, J.; Jurado, S.; Vázquez, J.; Köhler, C.; et al. Arabidopsis SWC4 binds DNA and recruits the SWR1 complex to modulate histone H2A.Z deposition at key regulatory genes. Mol. Plant 2018, 11, 815–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deal, R.B.; Kandasamy, M.K.; McKinney, E.C.; Meagher, R.B. The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 2005, 17, 2633–2646. [Google Scholar] [CrossRef] [Green Version]
- Meagher, R.B.; Deal, R.B.; Kandasamy, M.K.; McKinney, E.C. Nuclear actin-related proteins as epigenetic regulators of development. Plant Physiol. 2005, 139, 1576–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zahraeifard, S.; Foroozani, M.; Sepehri, A.; Oh, D.-H.; Wang, G.; Mangu, V.; Chen, B.; Baisakh, N.; Dassanayake, M.; Smith, A.P. Rice H2A.Z negatively regulates genes responsive to nutrient starvation but promotes expression of key housekeeping genes. J. Exp. Bot. 2018, 69, 4907–4919. [Google Scholar] [CrossRef] [Green Version]
- Lei, Y.; Lu, L.; Liu, H.-Y.; Li, S.; Xing, F.; Chen, L.-L. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 2014, 7, 1494–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, K.; Zhang, J.; Yang, Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR–Cas9-mediated genome editing in model plants and major crops. Mol. Plant 2014, 7, 923–926. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Wang, S.; Zheng, H.; Li, H.; Zhang, F.; Su, Y.; Xu, Z.; Lin, H.; Qian, Q.; Ding, Y. SIP1 participates in regulation of flowering time in rice by recruiting OsTrx1 to Ehd1. New Phytol. 2018, 219, 422–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, P.; Wang, S.; Jiang, H.; Cheng, B.; Wu, K.; Ding, Y. The COMPASS-like complex promotes flowering and panicle branching in rice. Plant Physiol. 2018, 176, 2761–2771. [Google Scholar] [CrossRef] [Green Version]
- Kabsch, W.; Mannherz, H.G.; Suck, D.; Pai, E.; Holmes, K. Atomic structure of the actin: DNase I complex. Nat. Cell Biol. 1990, 347, 37–44. [Google Scholar] [CrossRef]
- Lees-Miller, J.P.; Helfman, D.M.; Schroer, T.A. A vertebrate actin-related protein is a component of a multisubunit complex involved in microtubule-based vesicle motility. Nat. Cell Biol. 1992, 359, 244–246. [Google Scholar] [CrossRef]
- Pines, J. Cyclins and cyclin-dependent kinases: A biochemical view. Biochem. J. 1995, 308, 697–711. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E. Cyclin-dependent protein kinases: Key regulators of the eukaryotic cell cycle. BioEssays 1995, 17, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Norbury, C.; Nurse, P. Animal cell cycles and their control. Annu. Rev. Biochem. 1992, 61, 441–470. [Google Scholar] [CrossRef] [PubMed]
- Hemerly, A.; Bergounioux, C.; van Montagu, M.; Inzé, D.; Ferreira, P. Genes regulating the plant cell cycle: Isolation of a mitotic-like cyclin from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1992, 89, 3295–3299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabian-Marwedel, T.; Umeda, M.; Sauter, M. The rice cyclin-dependent kinase—Activating kinase R2 regulates S-phase progression. Plant Cell 2002, 14, 197–210. [Google Scholar] [CrossRef]
- Evans, T.; Rosenthal, E.T.; Youngblom, J.; Distel, D.; Hunt, T. Cyclin: A protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 1983, 33, 389–396. [Google Scholar] [CrossRef]
- Setiady, Y.Y.; Sekine, M.; Hariguchi, N.; Yamamoto, T.; Kouchi, H.; Shinmyo, A. Tobacco mitotic cyclins: Cloning, characterization, gene expression and functional assay. Plant J. 1995, 8, 949–957. [Google Scholar] [CrossRef]
- Fuerst, R.; Soni, R.; Murray, J.; Lindsey, K. Modulation of cyclin transcript levels in cultured cells of Arabidopsis thaliana. Plant Physiol. 1996, 112, 1023–1033. [Google Scholar] [CrossRef] [Green Version]
- Reichheld, J.-P.; Chaubet, N.; Shen, W.-H.; Renaudin, J.-P.; Gigot, C. Multiple A-type cyclins express sequentially during the cell cycle in Nicotiana tabacum BY2 cells. Proc. Natl. Acad. Sci. USA 1996, 93, 13819–13824. [Google Scholar] [CrossRef] [Green Version]
- Ito, M.; Marie-Claire, C.; Sakabe, M.; Ohno, T.; Hata, S.; Kouchi, H.; Hashimoto, J.; Fukuda, H.; Komamine, A.; Watanabe, A. Cell-cycle-regulated transcription of A- and B-type plant cyclin genes in synchronous cultures. Plant J. 1997, 11, 983–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genschik, P.; Criqui, M.C.; Parmentier, Y.; Derevier, A.; Fleck, J. Cell cycle-dependent proteolysis in plants: Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor MG132. Plant Cell 1998, 10, 2063. [Google Scholar]
- Peters, J.-M. SCF and APC: The yin and yang of cell cycle regulated proteolysis. Curr. Opin. Cell Biol. 1998, 10, 759–768. [Google Scholar] [CrossRef]
- Yoshida, T.; Shimada, K.; Oma, Y.; Kalck, V.; Akimura, K.; Taddei, A.; Iwahashi, H.; Kugou, K.; Ohta, K.; Gasser, S.M.; et al. Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores. PLoS Genet. 2010, 6, e1000910. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-H.; Alami, S.; Luk, E.; Wu, C.-H.; Sen, S.; Mizuguchi, G.; Wei, D.; Wu, C. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 2005, 12, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Kim, S.; Kim, S.Y.; Kim, M.; Hyun, Y.; Lee, H.; Choe, S.; Michaels, S.; Lee, I. SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell 2005, 17, 2647–2660. [Google Scholar] [CrossRef] [Green Version]
- Martin-Trillo, M.; Lázaro, A.; Poethig, R.S.; Gómez-Mena, C.; Piñeiro, M.; Zapater, J.M.M.; Jarillo, J.A. EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development 2006, 133, 1241–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gowans, G.J.; Schep, A.N.; Wong, K.M.; King, D.A.; Greenleaf, W.J.; Morrison, A.J. INO80 chromatin remodeling coordinates metabolic homeostasis with cell division. Cell Rep. 2018, 22, 611–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rust, K.; Tiwari, M.D.; Mishra, V.K.; Grawe, F.; Wodarz, A. Myc and the Tip60 chromatin remodeling complex control neuroblast maintenance and polarity in Drosophila. EMBO J. 2018, 37, e98659. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; McKinney, E.C.; Meagher, R.B. Cell cycle-dependent association of Arabidopsis actin-related proteins AtARP4 and AtARP7 with the nucleus. Plant J. 2003, 33, 939–948. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; Deal, R.B.; McKinney, E.C.; Meagher, R.B. Plant actin-related proteins. Trends Plant Sci. 2004, 9, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhao, L.; Wang, L.; Zhang, M.; Su, Z.; Cheng, Y.; Zhao, H.; Qin, Y. ERECTA signaling controls Arabidopsis inflorescence architecture through chromatin-mediated activation of PRE 1 expression. New Phytol. 2017, 214, 1579–1596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sura, W.; Kabza, M.; Karlowski, W.; Bieluszewski, T.; Kus-Slowinska, M.; Pawełoszek, Ł.; Sadowski, J.; Ziolkowski, P.A. Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. Plant Cell 2017, 29, 791–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Parents (Self-Pollinated) | Offsprings | |||||
|---|---|---|---|---|---|---|
| Total | WT | Heterozygous | Homozygous | Observed Ratio | Expected Ratio | |
| osarp6-1 (Heterozygous) | 119 | 32 | 60 | 27 | 1:1.88:0.84 | 1:2:1 |
| osarp6-2 (Heterozygous) | 134 | 37 | 68 | 29 | 1:1.83:0.78 | 1:2:1 |
| Total | 253 | 69 | 128 | 56 | 1:1.85:0.81 | 1:2:1 |
| Genotype | Internode Length (cm) | Panicle Length (cm) | ||||
|---|---|---|---|---|---|---|
| Fifth | Fourth | Third | Second | First | ||
| WT | 32.2 ± 1.8 | 15.7 ± 1.3 | 9.3 ± 1.2 | 3.3 ± 1.4 | 1.1 ± 0.4 | 18.6 ± 1.2 |
| osarp6-1 | 14.7 ± 0.6 | 5.0 ± 1.0 | 1.2 ± 0.2 | 0.8 ± 0.1 | 0.4 ± 0.1 | 14.7 ± 1.5 |
| osarp6-2 | 14.5 ± 0.5 | 4.6 ± 0.5 | 1.3 ± 0.1 | 0.7 ± 0.1 | 0.4 ± 0.0 | 14.2 ± 0.6 |
| osarp6-1 (heterozygous) | 24.7 ± 0.5 | 10.6 ± 1.5 | 5.1 ± 0.2 | 3.1 ± 0.4 | 0.6 ± 0.1 | 15.4 ± 0.2 |
| osarp6-2 (heterozygous) | 25.0 ± 0.7 | 10.0 ± 0.4 | 5.2 ± 0.5 | 3.1 ± 0.1 | 0.5 ± 0.1 | 15.4 ± 0.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikram, A.U.; Ding, Y.; Su, Y. OsARP6 Is Involved in Internode Elongation by Regulating Cell-Cycle-Related Genes. Biomolecules 2021, 11, 1100. https://doi.org/10.3390/biom11081100
Ikram AU, Ding Y, Su Y. OsARP6 Is Involved in Internode Elongation by Regulating Cell-Cycle-Related Genes. Biomolecules. 2021; 11(8):1100. https://doi.org/10.3390/biom11081100
Chicago/Turabian StyleIkram, Aziz Ul, Yong Ding, and Yanhua Su. 2021. "OsARP6 Is Involved in Internode Elongation by Regulating Cell-Cycle-Related Genes" Biomolecules 11, no. 8: 1100. https://doi.org/10.3390/biom11081100





