Understanding and Exploiting Post-Translational Modifications for Plant Disease Resistance
Abstract
:1. Introduction
2. The Framework of Plant Defence
3. Post-Translational Modifications Have a Critical Role in Defence
3.1. Phosphorylation
3.2. Ubiquitination
3.3. SUMOylation
3.4. Interaction between PTMs
4. Effectors Disrupt Host PTMs
5. Growth–Defence Trade-Offs
6. Exploiting PTMs to Produce Disease-Resistant Crops
7. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oerke, E.-C. Crop Losses to Pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- FAO. The Future of Food and Agriculture: Trends and Challenges; Food and Agriculture Organization of the United Nations, Ed.; Food and Agriculture Organization of the United Nations: Rome, Italy, 2017; ISBN 978-92-5-109551-5. [Google Scholar]
- IFPRI. Global Nutrition Report 2015: Actions and Accountability to Advance Nutrition and Sustainable Development; International Food Policy Research Institute: Washington, DC, USA, 2015. [Google Scholar]
- Bass, C.; Puinean, A.M.; Zimmer, C.T.; Denholm, I.; Field, L.M.; Foster, S.P.; Gutbrod, O.; Nauen, R.; Slater, R.; Williamson, M.S. The evolution of insecticide resistance in the peach potato aphid, Myzus persicae. Insect Biochem. Mol. Biol. 2014, 51, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Zayan, S.A. Impact of climate change on plant diseases and IPM strategies. In Plant Disease—Current Threats and Management Trends; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef] [Green Version]
- Panstruga, R.; Moscou, M.J. What is the molecular basis of nonhost resistance? Mol. Plant Microbe Interact. 2020, 33, 1253–1264. [Google Scholar] [CrossRef]
- Das, G.; Patra, J.K.; Baek, K.-H. Insight into MAS: A molecular tool for development of stress resistant and quality of rice through gene stacking. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.-G.; Zhao, K. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Zhu, H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rampitsch, C.; Bykova, N.V. Proteomics and plant disease: Advances in combating a major threat to the global food supply. Proteomics 2012, 12, 673–690. [Google Scholar] [CrossRef] [PubMed]
- Castro-Moretti, F.R.; Gentzel, I.N.; Mackey, D.; Alonso, A.P. Metabolomics as an emerging tool for the study of plant–pathogen interactions. Metabolites 2020, 10, 52. [Google Scholar] [CrossRef] [Green Version]
- Han, G.-Z. Origin and evolution of the plant immune system. New Phytol. 2019, 222, 70–83. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Naveed, Z.A.; Wei, X.; Chen, J.; Mubeen, H.; Ali, G.S. The PTI to ETI continuum in phytophthora-plant interactions. Front. Plant Sci. 2020, 11, 593905. [Google Scholar] [CrossRef]
- Pritchard, L.; Birch, P.R.J. The Zigzag model of plant–microbe interactions: Is it time to move on? Mol. Plant Pathol. 2014, 15, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Stotz, H.U.; Mitrousia, G.K.; de Wit, P.J.G.M.; Fitt, B.D.L. Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 2014, 19, 491–500. [Google Scholar] [CrossRef] [Green Version]
- Balmer, D.; Planchamp, C.; Mauch-Mani, B. On the move: Induced resistance in monocots. J. Exp. Bot. 2013, 64, 1249–1261. [Google Scholar] [CrossRef]
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, L.; Boller, T. FLS2: An LRR Receptor–like kinase involved in the perception of the bacterial elicitor flagellin in arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Kunze, G.; Zipfel, C.; Robatzek, S.; Niehaus, K.; Boller, T.; Felix, G. The N terminus of bacterial elongation factor Tu elicits innate immunity in arabidopsis plants. Plant Cell 2004, 16, 3496–3507. [Google Scholar] [CrossRef] [Green Version]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.G.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Zhang, X.-C.; Neece, D.; Ramonell, K.M.; Clough, S.; Kim, S.; Stacey, M.G.; Stacey, G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in arabidopsis. Plant Cell 2008, 20, 471–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef] [Green Version]
- Baggs, E.L.; Monroe, J.G.; Thanki, A.S.; O’Grady, R.; Schudoma, C.; Haerty, W.; Krasileva, K.V. Convergent loss of an EDS1/PAD4 Signaling pathway in several plant lineages reveals coevolved components of plant immunity and drought response. Plant Cell 2020, 32, 2158–2177. [Google Scholar] [CrossRef] [PubMed]
- Van der Hoorn, R.A.L.; Kamoun, S. From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 2008, 20, 2009–2017. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.G.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354. [Google Scholar] [CrossRef] [Green Version]
- Bernoux, M.; Ve, T.; Williams, S.; Warren, C.; Hatters, D.; Valkov, E.; Zhang, X.; Ellis, J.G.; Kobe, B.; Dodds, P.N. Structural and Functional Analysis of a Plant Resistance Protein TIR Domain Reveals Interfaces for Self-Association, signaling, and autoregulation. Cell Host Microbe 2011, 9, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Castel, B.; Ngou, P.-M.; Cevik, V.; Redkar, A.; Kim, D.-S.; Yang, Y.; Ding, P.; Jones, J.D.G. Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol. 2019, 222, 966–980. [Google Scholar] [CrossRef]
- Wu, Z.; Li, M.; Dong, O.X.; Xia, S.; Liang, W.; Bao, Y.; Wasteneys, G.; Li, X. Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol. 2019, 222, 938–953. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Stuttmann, J.; Rietz, S.; Guerois, R.; Brunstein, E.; Bautor, J.; Niefind, K.; Parker, J.E. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host Microbe 2013, 14, 619–630. [Google Scholar] [CrossRef] [Green Version]
- Lapin, D.; Kovacova, V.; Sun, X.; Dongus, J.A.; Bhandari, D.; von Born, P.; Bautor, J.; Guarneri, N.; Rzemieniewski, J.; Stuttmann, J.; et al. A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 2019, 31, 2430–2455. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; van Wersch, R.; Zhang, Y. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant Microbe Interact. 2017, 31, 403–409. [Google Scholar] [CrossRef] [Green Version]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose Deposition: A Multifaceted Plant Defense Response. Mol. Plant Microbe Interact. 2010, 24, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Torres, M.A. ROS in biotic interactions. Physiol. Plant 2010, 138, 414–429. [Google Scholar] [CrossRef]
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Xu, X.; Liu, X.; Yan, Y.; Wang, W.; Gebretsadik, K.; Qi, X.; Xu, Q.; Chen, X. Comparative proteomic analysis of cucumber powdery mildew resistance between a single-segment substitution line and its recurrent parent. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahir, J.; Rashid, M.; Afzal, A.J. Post-translational modifications in effectors and plant proteins involved in host–pathogen conflicts. Plant Pathol. 2019, 68, 628–644. [Google Scholar] [CrossRef]
- Withers, J.; Dong, X. Post-translational regulation of plant immunity. Curr. Opin. Plant Biol. 2017, 38, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Noor, J.J.; Gohain, B.; Gulabani, H.; Dnyaneshwar, I.K.; Singla, A. Post-translational modifications in regulation of pathogen surveillance and signaling in plants: The inside- (and perturbations from) outside story. IUBMB Life 2015, 67, 524–532. [Google Scholar] [CrossRef] [Green Version]
- Bigeard, J.; Hirt, H. Nuclear signaling of plant MAPKs. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Li, B.; Lu, D.; Chen, S.; Zhu, N.; He, P.; Shan, L. Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates arabidopsis innate immunity. Proc. Natl. Acad. Sci. USA 2014, 111, 3632–3637. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, Z.; Liu, D.; Xu, J.; Wei, X.; Yan, L.; Yang, C.; Lou, Z.; Shui, W. Assessment of BAK1 activity in different plant receptor-like kinase complexes by quantitative profiling of phosphorylation patterns. J. Proteom. 2014, 108, 484–493. [Google Scholar] [CrossRef]
- Laluk, K.; Luo, H.; Chai, M.; Dhawan, R.; Lai, Z.; Mengiste, T. Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in Plant Growth, Ethylene Signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell 2011, 23, 2831–2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orosa, B.; Yates, G.; Verma, V.; Srivastava, A.K.; Srivastava, M.; Campanaro, A.; Vega, D.D.; Fernandes, A.; Zhang, C.; Lee, J.; et al. SUMO conjugation to the pattern recognition receptor FLS2 triggers intracellular signalling in plant innate immunity. Nat. Commun. 2018, 9, 5185. [Google Scholar] [CrossRef]
- Kadota, Y.; Sklenar, J.; Derbyshire, P.; Stransfeld, L.; Asai, S.; Ntoukakis, V.; Jones, J.D.; Shirasu, K.; Menke, F.; Jones, A.; et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 2014, 54, 43–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Li, M.; Yu, L.; Zhou, Z.; Liang, X.; Liu, Z.; Cai, G.; Gao, L.; Zhang, X.; Wang, Y.; et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 2014, 15, 329–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-W.; Wang, Z.-Y. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 2010, 61, 681–704. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Ma, Y.; Liu, D.; Wei, X.; Sun, Y.; Chen, X.; Zhao, H.; Zhou, J.; Wang, Z.; Shui, W.; et al. Structural basis for the impact of phosphorylation on the activation of plant receptor-like kinase BAK1. Cell Res. 2012, 22, 1304–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Kota, U.; He, K.; Blackburn, K.; Li, J.; Goshe, M.B.; Huber, S.C.; Clouse, S.D. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 2008, 15, 220–235. [Google Scholar] [CrossRef] [Green Version]
- Schwessinger, B.; Roux, M.; Kadota, Y.; Ntoukakis, V.; Sklenar, J.; Jones, A.; Zipfel, C. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011, 7, e1002046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nürnberger, T.; Jones, J.D.G.; Felix, G.; Boller, T. A Flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef]
- Perraki, A.; DeFalco, T.A.; Derbyshire, P.; Avila, J.; Séré, D.; Sklenar, J.; Qi, X.; Stransfeld, L.; Schwessinger, B.; Kadota, Y.; et al. Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 2018, 561, 248–252. [Google Scholar] [CrossRef]
- Jagodzik, P.; Tajdel-Zielinska, M.; Ciesla, A.; Marczak, M.; Ludwikow, A. Mitogen-activated protein kinase cascades in plant hormone signaling. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Bi, G.; Zhou, Z.; Wang, W.; Li, L.; Rao, S.; Wu, Y.; Zhang, X.; Menke, F.L.H.; Chen, S.; Zhou, J.-M. Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 2018, 30, 1543–1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berriri, S.; Garcia, A.V.; dit Frey, N.F.; Rozhon, W.; Pateyron, S.; Leonhardt, N.; Montillet, J.-L.; Leung, J.; Hirt, H.; Colcombet, J. Constitutively Active Mitogen-Activated Protein Kinase Versions Reveal Functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 2012, 24, 4281–4293. [Google Scholar] [CrossRef] [Green Version]
- Genot, B.; Lang, J.; Berriri, S.; Garmier, M.; Gilard, F.; Pateyron, S.; Haustraete, K.; Van Der Straeten, D.; Hirt, H.; Colcombet, J. Constitutively Active Arabidopsis MAP Kinase 3 Triggers Defense Responses Involving Salicylic Acid and SUMM2 resistance protein1. Plant Physiol. 2017, 174, 1238–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, Q.; Qu, N.; Gao, M.; Zhang, Z.; Ding, X.; Yang, F.; Li, Y.; Dong, O.X.; Chen, S.; Li, X.; et al. The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 2012, 24, 2225–2236. [Google Scholar] [CrossRef] [Green Version]
- Petersen, M.; Brodersen, P.; Naested, H.; Andreasson, E.; Lindhart, U.; Johansen, B.; Nielsen, H.B.; Lacy, M.; Austin, M.J.; Parker, J.E.; et al. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 2000, 103, 1111–1120. [Google Scholar] [CrossRef] [Green Version]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, N.; Tootle, T.L.; Glazebrook, J. Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 1999, 11, 2419–2428. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Yang, L.; Zhu, Q.; Wu, H.; He, Y.; Liu, Y.; Xu, J.; Jiang, D.; Zhang, S. Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector-triggered immunity. PLoS Biol. 2018, 16, e2004122. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, Y.; Gao, M.; Zhang, J.; Kong, Q.; Liu, Y.; Ba, H.; Zhou, J.; Zhang, Y. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 2012, 11, 253–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Liu, Y.; Huang, H.; Gao, M.; Wu, D.; Kong, Q.; Zhang, Y. The NLR protein SUMM2 senses the disruption of an immune signaling MAP kinase cascade via CRCK3. EMBO Rep. 2017, 18, 292–302. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Feng, B.; Zhou, J.-M.; Tang, D. Plant immune signaling: Advancing on two frontiers. J. Integr. Plant Biol. 2020, 62, 2–24. [Google Scholar] [CrossRef] [Green Version]
- Segonzac, C.; Macho, A.P.; Sanmartín, M.; Ntoukakis, V.; Sánchez-Serrano, J.J.; Zipfel, C. Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J. 2014, 33, 2069–2079. [Google Scholar] [CrossRef] [Green Version]
- Couto, D.; Niebergall, R.; Liang, X.; Bücherl, C.A.; Sklenar, J.; Macho, A.P.; Ntoukakis, V.; Derbyshire, P.; Altenbach, D.; Maclean, D.; et al. The Arabidopsis protein phosphatase PP2C38 negatively regulates the central immune kinase BIK1. PLOS Pathog. 2016, 12, e1005811. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Liu, B.; Chen, S.; Gong, B.-Q.; Chen, L.; Zhou, Q.; Xiong, F.; Wang, M.; Feng, D.; Li, J.-F.; et al. A Tyrosine phosphorylation cycle regulates fungal activation of a plant receptor Ser/Thr kinase. Cell Host Microbe 2018, 23, 241–253.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schweighofer, A.; Kazanaviciute, V.; Scheikl, E.; Teige, M.; Doczi, R.; Hirt, H.; Schwanninger, M.; Kant, M.; Schuurink, R.; Mauch, F.; et al. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 2007, 19, 2213–2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shubchynskyy, V.; Boniecka, J.; Schweighofer, A.; Simulis, J.; Kvederaviciute, K.; Stumpe, M.; Mauch, F.; Balazadeh, S.; Mueller-Roeber, B.; Boutrot, F.; et al. Protein phosphatase AP2C1 negatively regulates basal resistance and defense responses to Pseudomonas syringae. J. Exp. Bot. 2017, 68, 1169–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, J.C.; Bartels, S.; Besteiro, M.A.G.; Shahollari, B.; Ulm, R.; Peck, S.C. Arabidopsis MAP kinase phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J. 2011, 67, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Bartels, S.; Anderson, J.C.; González Besteiro, M.A.; Carreri, A.; Hirt, H.; Buchala, A.; Métraux, J.-P.; Peck, S.C.; Ulm, R. MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 2009, 21, 2884–2897. [Google Scholar] [CrossRef] [Green Version]
- Park, H.C.; Song, E.H.; Nguyen, X.C.; Lee, K.; Kim, K.E.; Kim, H.S.; Lee, S.M.; Kim, S.H.; Bae, D.W.; Yun, D.-J.; et al. Arabidopsis MAP kinase phosphatase 1 is phosphorylated and activated by its substrate AtMPK6. Plant Cell Rep. 2011, 30, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Callis, J. The ubiquitination machinery of the ubiquitin system. Arabidopsis Book 2014, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [Green Version]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell. Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Zhou, B.; Zeng, L. Conventional and unconventional ubiquitination in plant immunity. Mol. Plant Pathol. 2017, 18, 1313–1330. [Google Scholar] [CrossRef] [PubMed]
- Kachewar, N.R.; Gupta, V.; Ranjan, A.; Patel, H.K.; Sonti, R.V. Overexpression of OsPUB41, a rice E3 ubiquitin ligase induced by cell wall degrading enzymes, enhances immune responses in rice and Arabidopsis. BMC Plant Biol. 2019, 19, 530. [Google Scholar] [CrossRef]
- Becker, F.; Buschfeld, E.; Schell, J.; Bachmair, A. Altered response to viral infection by tobacco plants perturbed in ubiquitin system. Plant J. 1993, 3, 875–881. [Google Scholar] [CrossRef]
- Goritschnig, S.; Zhang, Y.; Li, X. The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 2007, 49, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Saeki, Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017, 161, 113–124. [Google Scholar] [CrossRef]
- Duplan, V.; Rivas, S. E3 Ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Lu, D.; Xu, G.; Finlayson, S.A.; He, P.; Shan, L. The dominant negative ARM domain uncovers multiple functions of PUB13 in Arabidopsis immunity, flowering, and senescence. J. Exp. Bot. 2015, 66, 3353–3366. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Cao, Y.; Sun, X.; Espinoza, C.; Nguyen, C.T.; Liang, Y.; Stacey, G. Arabidopsis E3 ubiquitin ligase plant U-BOX13 (PUB13) regulates chitin receptor lysin motif receptor kinase5 (LYK5) protein abundance. New Phytol. 2017, 214, 1646–1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Grubb, L.E.; Wang, J.; Liang, X.; Li, L.; Gao, C.; Ma, M.; Feng, F.; Li, M.; Li, L.; et al. A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 2018, 69, 493–504.e6. [Google Scholar] [CrossRef] [Green Version]
- Desaki, Y.; Takahashi, S.; Sato, K.; Maeda, K.; Matsui, S.; Yoshimi, I.; Miura, T.; Jumonji, J.-I.; Takeda, J.; Yashima, K.; et al. PUB4, a CERK1-interacting ubiquitin ligase, positively regulates MAMP-triggered immunity in Arabidopsis. Plant Cell Physiol. 2019, 60, 2573–2583. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF COI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Xie, D.-X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091–1094. [Google Scholar] [CrossRef]
- Xu, L.; Liu, F.; Lechner, E.; Genschik, P.; Crosby, W.L.; Ma, H.; Peng, W.; Huang, D.; Xie, D. The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 2002, 14, 1919–1935. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Chen, X.; Zhong, X.; Li, M.; Ao, K.; Huang, J.; Li, X. Plant TRAF proteins regulate NLR immune receptor turnover. Cell Host Microbe 2016, 19, 204–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzucotelli, E.; Belloni, S.; Marone, D.; De Leonardis, A.; Guerra, D.; Di Fonzo, N.; Cattivelli, L.; Mastrangelo, A. The E3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genom. 2006, 7, 509–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vierstra, R.D. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 2012, 160, 2–14. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Tong, M.; Tian, L.; Zhu, C.; Liu, X.; Zhang, Y.; Li, X. Plant E3 ligases SNIPER1 and SNIPER2 broadly regulate the homeostasis of sensor NLR immune receptors. EMBO J. 2020, e104915. [Google Scholar] [CrossRef]
- Nijman, S.M.B.; Luna-Vargas, M.P.A.; Velds, A.; Brummelkamp, T.R.; Dirac, A.M.G.; Sixma, T.K.; Bernards, R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005, 123, 773–786. [Google Scholar] [CrossRef] [Green Version]
- Ewan, R.; Pangestuti, R.; Thornber, S.; Craig, A.; Carr, C.; O’Donnell, L.; Zhang, C.; Sadanandom, A. Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol. 2011, 191, 92–106. [Google Scholar] [CrossRef]
- Jeong, J.S.; Jung, C.; Seo, J.S.; Kim, J.-K.; Chua, N.-H. The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell 2017, 29, 1406–1424. [Google Scholar] [CrossRef] [Green Version]
- Bailey, M.; Srivastava, A.; Conti, L.; Nelis, S.; Zhang, C.; Florance, H.; Love, A.; Milner, J.; Napier, R.; Grant, M.; et al. Stability of small ubiquitin-like modifier (SUMO) proteases overly tolerant to salt1 and -2 modulates salicylic acid signalling and SUMO1/2 conjugation in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 353–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colignon, B.; Delaive, E.; Dieu, M.; Demazy, C.; Muhovski, Y.; Wallon, C.; Raes, M.; Mauro, S. Proteomics analysis of the endogenous, constitutive, leaf SUMOylome. J. Proteom. 2017, 150, 268–280. [Google Scholar] [CrossRef]
- Ingole, K.D.; Dahale, S.K.; Bhattacharjee, S. Proteomic analysis of SUMO1-SUMOylome changes during defense elicitation in Arabidopsis. J. Proteom. 2021, 232, 104054. [Google Scholar] [CrossRef]
- Miller, M.J.; Barrett-Wilt, G.A.; Hua, Z.; Vierstra, R.D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 16512–16517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Gao, F.; Bhattacharjee, S.; Adiasor, J.A.; Nam, J.C.; Gassmann, W. The Arabidopsis Resistance-Like Gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 2010, 6, e1001172. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Halane, M.K.; Kim, S.H.; Gassmann, W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 2011, 334, 1405–1408. [Google Scholar] [CrossRef] [PubMed]
- Hammoudi, V.; Vlachakis, G.; Schranz, M.E.; van den Burg, H.A. Whole-genome duplications followed by tandem duplications drive diversification of the protein modifier SUMO in angiosperms. New Phytol. 2016, 211, 172–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurepa, J.; Walker, J.M.; Smalle, J.; Gosink, M.M.; Davis, S.J.; Durham, T.L.; Sung, D.-Y.; Vierstra, R.D. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis accumulation of sumo1 and -2 conjugates is increased by stress. J. Biol. Chem. 2003, 278, 6862–6872. [Google Scholar] [CrossRef] [Green Version]
- Van denBurg, H.A.; Kini, R.K.; Schuurink, R.C.; Takken, F.L.W. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 2010, 22, 1998–2016. [Google Scholar] [CrossRef] [Green Version]
- Merrill, J.C.; Melhuish, T.A.; Kagey, M.H.; Yang, S.-H.; Sharrocks, A.D.; Wotton, D. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS ONE 2010, 5, e8794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villajuana-Bonequi, M.; Elrouby, N.; Nordström, K.; Griebel, T.; Bachmair, A.; Coupland, G. Elevated salicylic acid levels conferred by increased expression of isochorismate synthase 1 Contribute to Hyperaccumulation of SUMO1 conjugates in the Arabidopsis mutant early in short days 4. Plant J. 2014, 79, 206–219. [Google Scholar] [CrossRef]
- Lee, J.; Nam, J.; Park, H.C.; Na, G.; Miura, K.; Jin, J.B.; Yoo, C.Y.; Baek, D.; Kim, D.H.; Jeong, J.C.; et al. Salicylic acid-mediated innate immunity in arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007, 49, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Gou, M.; Huang, Q.; Qian, W.; Zhang, Z.; Jia, Z.; Hua, J. Sumoylation E3 Ligase SIZ1 modulates plant immunity partly through the immune receptor gene SNC1 in Arabidopsis. Mol. Plant Microbe Interact. 2017, 30, 334–342. [Google Scholar] [CrossRef] [Green Version]
- Hammoudi, V.; Fokkens, L.; Beerens, B.; Vlachakis, G.; Chatterjee, S.; Arroyo-Mateos, M.; Wackers, P.F.K.; Jonker, M.J.; van den Burg, H.A. The Arabidopsis SUMO E3 Ligase SIZ1 mediates the temperature dependent trade-off between plant immunity and growth. PLoS Genet. 2018, 14, e1007157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, D.; Lin, X.-L.; Kong, X.; Qu, G.-P.; Cai, B.; Lee, J.; Jin, J.B. SIZ1-Mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis. Mol. Plant 2019, 12, 215–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gou, M.; Shi, Z.; Zhu, Y.; Bao, Z.; Wang, G.; Hua, J. The F-Box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J. 2012, 69, 411–420. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, L. Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity. Plant Commun. 2020, 100041. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Orosa, B.; Singh, P.; Cummins, I.; Walsh, C.; Zhang, C.; Grant, M.; Roberts, M.R.; Anand, G.S.; Fitches, E.; et al. SUMO Suppresses the activity of the jasmonic acid receptor coronatine insensitive1. Plant Cell 2018, 30, 2099–2115. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Aceti, D.J.; Sabat, G.; Song, J.; Makino, S.-I.; Fox, B.G.; Bent, A.F. Mutations in FLS2 Ser-938 dissect signaling activation in FLS2-mediated Arabidopsis immunity. PLoS Pathog. 2013, 9, e1003313. [Google Scholar] [CrossRef] [Green Version]
- Chinchilla, D.; Bauer, Z.; Regenass, M.; Boller, T.; Felix, G. The Arabidopsis receptor kinase FLS2 binds Flg22 and determines the specificity of flagellin perception. Plant Cell 2006, 18, 465–476. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Li, L.; Macho, A.P.; Han, Z.; Hu, Z.; Zipfel, C.; Zhou, J.-M.; Chai, J. Structural basis for Flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 2013, 342, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Claus, L.A.N.; Leslie, M.E.; Tao, K.; Wu, Z.; Liu, J.; Yu, X.; Li, B.; Zhou, J.; Savatin, D.V.; et al. Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 2020, 581, 199–203. [Google Scholar] [CrossRef]
- Monaghan, J.; Matschi, S.; Romeis, T.; Zipfel, C. The calcium-dependent protein kinase CPK28 negatively regulates the BIK1-mediated PAMP-induced calcium burst. Plant Signal. Behav. 2015, 10. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.; Yi, H.; Chen, X.; Wang, J. Post-translational modifications of proteins have versatile roles in regulating plant immune responses. Int. J. Mol. Sci. 2019, 20, 2807. [Google Scholar] [CrossRef] [Green Version]
- Withers, J.; Dong, X. Posttranslational Modifications of NPR1: A single protein playing multiple roles in plant immunity and physiology. PLoS Pathog. 2016, 12, e1005707. [Google Scholar] [CrossRef]
- Després, C.; DeLong, C.; Glaze, S.; Liu, E.; Fobert, P.R. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of BZIP transcription factors. Plant Cell 2000, 12, 279–290. [Google Scholar] [CrossRef] [Green Version]
- Saleh, A.; Withers, J.; Mohan, R.; Marqués, J.; Gu, Y.; Yan, S.; Zavaliev, R.; Nomoto, M.; Tada, Y.; Dong, X. Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 2015, 18, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Spoel, S.H.; Mou, Z.; Tada, Y.; Spivey, N.W.; Genschik, P.; Dong, X. Proteasome-mediated turnover of the transcription co-activator NPR1 plays dual roles in regulating plant immunity. Cell 2009, 137, 860–872. [Google Scholar] [CrossRef] [Green Version]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Huang, T.; Ao, K.; Yan, X.; Huang, Y. Sumoylation, phosphorylation, and acetylation fine-tune the turnover of plant immunity components mediated by ubiquitination. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backer, R.; Naidoo, S.; van den Berg, N. The nonexpressor of pathogenesis-related genes 1 (NPR1) and related family: Mechanistic insights in plant disease resistance. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, G.-H.; Hoey, T.; Zhu, S.; Clavel, M.; Yu, K.; Navarre, D.; Kachroo, A.; Deragon, J.-M.; Kachroo, P. COP1, a Negative regulator of photomorphogenesis, positively regulates plant disease resistance via double-stranded RNA binding proteins. PLoS Pathog. 2018, 14, e1006894. [Google Scholar] [CrossRef]
- Lin, X.-L.; Niu, D.; Hu, Z.-L.; Kim, D.H.; Jin, Y.H.; Cai, B.; Liu, P.; Miura, K.; Yun, D.-J.; Kim, W.-Y.; et al. An Arabidopsis SUMO E3 ligase, SIZ1, negatively regulates photomorphogenesis by promoting COP1 activity. PLoS Genet. 2016, 12, e1006016. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Jang, I.-C.; Seo, H.S. COP1 controls abiotic stress responses by modulating AtSIZ1 function through its E3 ubiquitin ligase activity. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Van den Burg, H.A.; Takken, F.L. SUMO-, MAPK- and resistance protein-signaling converge at transcription complexes that regulate plant innate immunity. Plant Signal. Behav. 2010, 5, 1597–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, V.; Srivastava, A.K.; Gough, C.; Campanaro, A.; Srivastava, M.; Morrell, R.; Joyce, J.; Bailey, M.; Krysan, P.J.; Sadanandom, A. SUMO enables substrate selectivity by mitogen-activated protein kinases to regulate immunity in plants. Proc. Nat. Acad. Sci. USA 2021, 118, e2021351118. [Google Scholar] [CrossRef]
- Willems, P.; Horne, A.; Parys, T.V.; Goormachtig, S.; Smet, I.D.; Botzki, A.; Breusegem, F.V.; Gevaert, K. The plant PTM viewer, a central resource for exploring plant protein modifications. Plant J. 2019, 99, 752–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hotson, A.; Chosed, R.; Shu, H.; Orth, K.; Mudgett, M.B. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 2003, 50, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; Shirsekar, G.; Bellizzi, M.; Chen, S.; Songkumarn, P.; Xie, X.; Shi, X.; Ning, Y.; Zhou, B.; Suttiviriya, P.; et al. The E3 ligase APIP10 connects the effector AvrPiz-t to the NLR receptor Piz-t in rice. PLoS Pathog. 2016, 12, e1005529. [Google Scholar] [CrossRef] [Green Version]
- Yin, Z.; Chen, J.; Zeng, L.; Goh, M.; Leung, H.; Khush, G.S.; Wang, G.L. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 2000, 13, 869–876. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.-R.; Qu, S.; Bordeos, A.; Yang, C.; Baraoidan, M.; Yan, H.; Xie, Q.; Nahm, B.H.; Leung, H.; Wang, G.-L. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-Box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 2004, 16, 2795–2808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, B.; Zeng, L. The tomato U-Box type E3 ligase PUB13 acts with group III ubiquitin E2 enzymes to modulate FLS2-mediated immune signaling. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.P.; Dalal, V.; Singh, N.K.; Sharma, T.R. Comparative Analysis of the 100 Kb region containing the Pi-Kh locus between Indica and Japonica rice lines. Genom. Proteom. Bioinform. 2007, 5, 35–44. [Google Scholar] [CrossRef]
- Vasudevan, K.; Gruissem, W.; Bhullar, N.K. Identification of novel alleles of the rice blast resistance gene Pi54. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Chern, M.; Canlas, P.E.; Jiang, C.; Ruan, D.; Cao, P.; Ronald, P.C. A conserved threonine residue in the juxtamembrane domain of the XA21 pattern recognition receptor is critical for kinase autophosphorylation and XA21-mediated immunity. J. Biol. Chem. 2010, 285, 10454–10463. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-S.; Pi, L.-Y.; Chen, X.; Chakrabarty, P.K.; Jiang, J.; Leon, A.L.D.; Liu, G.-Z.; Li, L.; Benny, U.; Oard, J.; et al. Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 2006, 18, 3635–3646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luu, D.D.; Joe, A.; Chen, Y.; Parys, K.; Bahar, O.; Pruitt, R.; Chan, L.J.G.; Petzold, C.J.; Long, K.; Adamchak, C.; et al. Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor. Proc. Natl. Acad. Sci. USA 2019, 116, 8525–8534. [Google Scholar] [CrossRef] [Green Version]
- Park, C.-J.; Han, S.-W.; Chen, X.; Ronald, P.C. Elucidation of XA21-mediated innate immunity. Cell Microbiol. 2010, 12, 1017–1025. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Liu, X.; Chen, X.; Snyder, A.; Song, W.-Y. Members of the XB3 family from diverse plant species induce programmed cell death in Nicotiana benthamiana. PLoS ONE 2013, 8, e63868. [Google Scholar] [CrossRef]
- Gimenez-Ibanez, S.; Hann, D.R.; Ntoukakis, V.; Petutschnig, E.; Lipka, V.; Rathjen, J.P. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 2009, 19, 423–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Göhre, V.; Spallek, T.; Häweker, H.; Mersmann, S.; Mentzel, T.; Boller, T.; de Torres, M.; Mansfield, J.W.; Robatzek, S. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr. Biol. 2008, 18, 1824–1832. [Google Scholar] [CrossRef] [Green Version]
- Rosebrock, T.R.; Zeng, L.; Brady, J.J.; Abramovitch, R.B.; Xiao, F.; Martin, G.B. A Bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 2007, 448, 370–374. [Google Scholar] [CrossRef]
- Mathieu, J.; Schwizer, S.; Martin, G.B. Pto Kinase Binds Two Domains of AvrPtoB and Its Proximity to the Effector E3 ligase determines if it evades degradation and activates plant immunity. PLoS Pathog. 2014, 10, e1004227. [Google Scholar] [CrossRef]
- Luo, Y.; Caldwell, K.S.; Wroblewski, T.; Wright, M.E.; Michelmore, R.W. Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell 2009, 21, 2458–2472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, T.; Zong, N.; Zou, Y.; Wu, Y.; Zhang, J.; Xing, W.; Li, Y.; Tang, X.; Zhu, L.; Chai, J.; et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 2008, 18, 74–80. [Google Scholar] [CrossRef] [Green Version]
- Mackey, D.; Holt, B.F.; Wiig, A.; Dangl, J.L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 2002, 108, 743–754. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wu, S.; Chen, X.; Liu, C.; Sheen, J.; Shan, L.; He, P. Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J. 2014, 77, 235–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilton, M.; Subramaniam, R.; Elmore, J.; Felsensteiner, C.; Coaker, G.; Desveaux, D. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc. Natl. Acad. Sci. USA 2010, 107, 2349–2354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, D.K.; Calviño, M.; Brauer, E.K.; Fernandez-Pozo, N.; Strickler, S.; Yalamanchili, R.; Suzuki, H.; Aoki, K.; Shibata, D.; Stratmann, J.W.; et al. The Tomato Kinome and the Tomato Kinase Library ORFeome: Novel resources for the study of kinases and signal transduction in tomato and solanaceae species. Mol. Plant Microbe Interact. 2013, 27, 7–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, J.; Hou, S.; Wang, X.; Li, Y.; Ren, D.; Chen, S.; Tang, X.; Zhou, J.-M. A Pseudomonas syringae ADP-ribosyltransferase inhibits arabidopsis mitogen-activated protein kinase kinases. Plant Cell 2010, 22, 2033–2044. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Shao, F.; Li, Y.; Cui, H.; Chen, L.; Li, H.; Zou, Y.; Long, C.; Lan, L.; Chai, J.; et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 2007, 1, 175–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teper, D.; Girija, A.M.; Bosis, E.; Popov, G.; Savidor, A.; Sessa, G. The Xanthomonas euvesicatoria type III effector XopAU is an active protein kinase that manipulates plant MAP kinase signaling. PLoS Pathog. 2018, 14, e1006880. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Zhou, X.; Sun, L.; Wang, K.; Yang, F.; Liao, H.; Rong, W.; Yin, J.; Chen, H.; Chen, X.; et al. The Xanthomonas effector XopK harbours E3 ubiquitin-ligase activity that is required for virulence. New Phytol. 2018, 220, 219–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-G.; Stork, W.; Mudgett, M.B. Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 2013, 13, 143–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruneda, J.N.; Durkin, C.H.; Geurink, P.P.; Ovaa, H.; Santhanam, B.; Holden, D.W.; Komander, D. The molecular basis for ubiquitin and ubiquitin-like specificities in bacterial effector proteases. Mol. Cell 2016, 63, 261–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.; Rong, W.; Luo, H.; Chen, Y.; He, C. The Xanthomonas campestris effector protein XopDXcc8004 triggers plant disease tolerance by targeting DELLA proteins. New Phytol. 2014, 204, 595–608. [Google Scholar] [CrossRef]
- Üstün, S.; Börnke, F. Interactions of Xanthomonas type-III effector proteins with the plant ubiquitin and ubiquitin-like pathways. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Cheong, M.S.; Kirik, A.; Kim, J.-G.; Frame, K.; Kirik, V.; Mudgett, M.B. AvrBsT acetylates Arabidopsis ACIP1, a protein that associates with microtubules and is required for immunity. PLoS Pathog. 2014, 10, e1003952. [Google Scholar] [CrossRef] [Green Version]
- Szczesny, R.; Büttner, D.; Escolar, L.; Schulze, S.; Seiferth, A.; Bonas, U. Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol. 2010, 187, 1058–1074. [Google Scholar] [CrossRef]
- Roden, J.; Eardley, L.; Hotson, A.; Cao, Y.; Mudgett, M.B. Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant Microbe Interact. 2004, 17, 633–643. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Chen, J.; Li, M.; Chang, M.; Xu, K.; Shang, Z.; Zhao, Y.; Palmer, I.; Zhang, Y.; McGill, J.; et al. A bacterial type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe 2017, 22, 777–788.e7. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Kim, P.; Yu, L.; Cai, G.; Chen, S.; Alfano, J.R.; Zhou, J.-M. Activation-dependent destruction of a co-receptor by a Pseudomonas syringae effector dampens plant immunity. Cell Host Microbe 2016, 20, 504–514. [Google Scholar] [CrossRef] [Green Version]
- Macho, A.P.; Zipfel, C. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr. Opin. Microbiol. 2015, 23, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Macho, A.P.; Schwessinger, B.; Ntoukakis, V.; Brutus, A.; Segonzac, C.; Roy, S.; Kadota, Y.; Oh, M.-H.; Sklenar, J.; Derbyshire, P.; et al. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 2014, 343, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.-H.; El-Kasmi, F.; He, Y.; Loehr, A.; Dangl, J.L. A plant phosphoswitch platform repeatedly targeted by type III effector proteins regulates the output of both tiers of plant immune receptors. Cell Host Microbe 2014, 16, 484–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, S.K.; Macoy, D.M.; Kim, W.-Y.; Lee, S.Y.; Kim, M.G. Role of RIN4 in Regulating PAMP-triggered immunity and effector-triggered immunity: Current status and future perspectives. Mol. Cells 2019, 42, 503–511. [Google Scholar] [CrossRef]
- Lee, D.; Bourdais, G.; Yu, G.; Robatzek, S.; Coaker, G. Phosphorylation of the plant immune regulator RPM1-interacting protein4 enhances plant plasma membrane H+-ATPase activity and inhibits flagellin-triggered immune responses in Arabidopsis. Plant Cell 2015, 27, 2042–2056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Elmore, J.M.; Lin, Z.-J.D.; Coaker, G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 2011, 9, 137–146. [Google Scholar] [CrossRef] [Green Version]
- Coaker, G.; Falick, A.; Staskawicz, B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 2005, 308, 548–550. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Ma, X.; Chiang, Y.-H.; Yadeta, K.A.; Ding, P.; Dong, L.; Zhao, Y.; Li, X.; Yu, Y.; Zhang, L.; et al. Proline isomerization of the immune receptor-interacting protein RIN4 by a cyclophilin inhibits effector-triggered immunity in Arabidopsis. Cell Host Microbe 2014, 16, 473–483. [Google Scholar] [CrossRef] [Green Version]
- Mackey, D.; Belkhadir, Y.; Alonso, J.M.; Ecker, J.R.; Dangl, J.L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Chung, E.-H.; da Cunha, L.; Wu, A.-J.; Gao, Z.; Cherkis, K.; Afzal, A.J.; Mackey, D.; Dangl, J.L. Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 2011, 9, 125–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toruño, T.Y.; Shen, M.; Coaker, G.; Mackey, D. Regulated disorder: Posttranslational modifications control the RIN4 plant immune signaling hub. Mol. Plant Microbe Interact. 2018, 32, 56–64. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Greenwood, D.R.; Templeton, M.D.; Libich, D.S.; McGhie, T.K.; Xue, B.; Yoon, M.; Cui, W.; Kirk, C.A.; Jones, W.T.; et al. The Intrinsically Disordered Structural Platform of the Plant Defence Hub Protein RPM1-interacting protein 4 provides insights into its mode of action in the host-pathogen interface and evolution of the nitrate-induced domain protein family. FEBS J. 2014, 281, 3955–3979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popov, G.; Majhi, B.B.; Sessa, G. Effector gene XopAE of Xanthomonas Euvesicatoria 85-10 is part of an operon and encodes an E3 ubiquitin ligase. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.M.; Li, M.-Y.; Yang, P.-Y.; Chang, S.H.; Ho, Y.-P.; Lin, H.; Deng, W.-L.; Yang, J.-Y. Arabidopsis HFR1 is a potential nuclear substrate regulated by the Xanthomonas type III effector XopDXcc8004. PLoS ONE 2015, 10, e0117067. [Google Scholar] [CrossRef]
- Navarro, L.; Bari, R.; Achard, P.; Lisón, P.; Nemri, A.; Harberd, N.P.; Jones, J.D.G. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 2008, 18, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.I.B.; Armstrong, M.R.; Gilroy, E.M.; Boevink, P.C.; Hein, I.; Taylor, R.M.; Zhendong, T.; Engelhardt, S.; Vetukuri, R.R.; Harrower, B.; et al. Phytophthora Infestans Effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA 2010, 107, 9909–9914. [Google Scholar] [CrossRef] [Green Version]
- Park, C.-H.; Chen, S.; Shirsekar, G.; Zhou, B.; Khang, C.H.; Songkumarn, P.; Afzal, A.J.; Ning, Y.; Wang, R.; Bellizzi, M.; et al. The Magnaporthe Oryzae effector AvrPiz-t Targets the RING E3 Ubiquitin Ligase APIP6 to suppress pathogen-associated molecular pattern–triggered immunity in rice. Plant Cell 2012, 24, 4748–4762. [Google Scholar] [CrossRef] [Green Version]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [Green Version]
- Tian, D.; Traw, M.B.; Chen, J.Q.; Kreitman, M.; Bergelson, J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 2003, 423, 74–77. [Google Scholar] [CrossRef]
- Herms, D.A.; Mattson, W.J. The dilemma of plants: To grow or defend. Q. Rev. Biol. 1992, 67, 283–335. [Google Scholar] [CrossRef] [Green Version]
- Campos, M.L.; Yoshida, Y.; Major, I.T.; de Oliveira Ferreira, D.; Weraduwage, S.M.; Froehlich, J.E.; Johnson, B.F.; Kramer, D.M.; Jander, G.; Sharkey, T.D.; et al. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichmann, R.; Schäfer, P. Growth versus Immunity—A Redirection of the Cell Cycle? Curr. Opin. Plant Biol. 2015, 26, 106–112. [Google Scholar] [CrossRef] [Green Version]
- Kliebenstein, D.J. False idolatry of the mythical growth versus immunity tradeoff in molecular systems plant pathology. Physiol. Mol. Plant Pathol. 2016, 95, 55–59. [Google Scholar] [CrossRef] [Green Version]
- Margalha, L.; Confraria, A.; Baena-González, E. SnRK1 and TOR: Modulating growth–defense trade-offs in plant stress responses. J. Exp. Bot. 2019, 70, 2261–2274. [Google Scholar] [CrossRef]
- Dobrenel, T.; Caldana, C.; Hanson, J.; Robaglia, C.; Vincentz, M.; Veit, B.; Meyer, C. TOR Signaling and nutrient sensing. Annu. Rev. Plant Biol. 2016, 67, 261–285. [Google Scholar] [CrossRef] [PubMed]
- Robaglia, C.; Thomas, M.; Meyer, C. Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 2012, 15, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Lastdrager, J.; Hanson, J.; Smeekens, S. Sugar Signals and the Control of Plant Growth and Development. J. Exp. Bot. 2014, 65, 799–807. [Google Scholar] [CrossRef]
- Nukarinen, E.; Nägele, T.; Pedrotti, L.; Wurzinger, B.; Mair, A.; Landgraf, R.; Börnke, F.; Hanson, J.; Teige, M.; Baena-Gonzalez, E.; et al. Quantitative Phosphoproteomics Reveals the Role of the AMPK Plant Ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 2016, 6, 31697. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Sheen, J. Novel links in the plant TOR kinase signaling network. Curr. Opin. Plant Biol. 2015, 28, 83–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halford, N.G.; Hardie, D.G. SNF1-related protein kinases: Global regulators of carbon metabolism in plants? Plant Mol. Biol. 1998, 37, 735–748. [Google Scholar] [CrossRef]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 protein kinases—Key regulators of plant response to abiotic stresses. OMICS 2011, 15, 859–872. [Google Scholar] [CrossRef]
- Baena-González, E.; Sheen, J. Convergent energy and stress signaling. Trends Plant Sci. 2008, 13, 474–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Wang, H.; Sunter, G.; Bisaro, D.M. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 2003, 15, 1034–1048. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-R.; Lee, K.-W.; Chen, C.-Y.; Hong, Y.-F.; Chen, J.-L.; Lu, C.-A.; Chen, K.-T.; Ho, T.-H.D.; Yu, S.-M. SnRK1A-Interacting Negative Regulators Modulate the Nutrient Starvation Signaling Sensor SnRK1 in source-sink communication in cereal seedlings under abiotic stress. Plant Cell 2014, 26, 808–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwachtje, J.; Minchin, P.E.H.; Jahnke, S.; van Dongen, J.T.; Schittko, U.; Baldwin, I.T. SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc. Natl. Acad. Sci. USA 2006, 103, 12935–12940. [Google Scholar] [CrossRef] [Green Version]
- Sheen, J. Master regulators in plant glucose signaling networks. J. Plant Biol. 2014, 57, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Polge, C.; Jossier, M.; Crozet, P.; Gissot, L.; Thomas, M. β-subunits of the SnRK1 Complexes Share a Common Ancestral Function Together with Expression and Function Specificities; Physical interaction with nitrate reductase specifically occurs via AKINβ1-subunit. Plant Physiol. 2008, 148, 1570–1582. [Google Scholar] [CrossRef] [Green Version]
- Sugden, C.; Donaghy, P.G.; Halford, N.G.; Hardie, D.G. Two SNF1-Related Protein Kinases from Spinach Leaf Phosphorylate and Inactivate 3-Hydroxy-3-methylglutaryl-coenzyme a reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 1999, 120, 257–274. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; McCormack, M.; Li, L.; Hall, Q.; Xiang, C.; Sheen, J. Glc-TOR signalling leads transcriptome reprogramming and meristem activation. Nature 2013, 496, 181–186. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.-H.; Liu, M.-J.; Xiong, Y.; Sheen, J.; Wu, S.-H. TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 2018, 115, 12823–12828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Y.; Sheen, J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 2012, 287, 2836–2842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.-C.; Liu, X.; Fu, L.; Hou, Y.-J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 2018, 69, 100–112.e6. [Google Scholar] [CrossRef] [Green Version]
- Jamsheer, M.K.; Jindal, S.; Laxmi, A. Evolution of TOR–SnRK dynamics in green plants and its integration with phytohormone signaling networks. J. Exp. Bot. 2019, 70, 2239–2259. [Google Scholar] [CrossRef]
- Weiste, C.; Pedrotti, L.; Selvanayagam, J.; Muralidhara, P.; Fröschel, C.; Novák, O.; Ljung, K.; Hanson, J.; Dröge-Laser, W. The Arabidopsis BZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PLoS Genet. 2017, 13, e1006607. [Google Scholar] [CrossRef]
- Schepetilnikov, M.; Makarian, J.; Srour, O.; Geldreich, A.; Yang, Z.; Chicher, J.; Hammann, P.; Ryabova, L.A. GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J. 2017, 36, 886–903. [Google Scholar] [CrossRef] [PubMed]
- Hey, S.; Mayerhofer, H.; Halford, N.G.; Dickinson, J.R. DNA Sequences from Arabidopsis, which encode protein kinases and function as upstream regulators of Snf1 in yeast. J. Biol. Chem. 2007, 282, 10472–10479. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, A.; Adamo, M.; Crozet, P.; Margalha, L.; Confraria, A.; Martinho, C.; Elias, A.; Rabissi, A.; Lumbreras, V.; González-Guzmán, M.; et al. ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell 2013, 25, 3871–3884. [Google Scholar] [CrossRef] [Green Version]
- Aznar, N.R.; Consolo, V.F.; Salerno, G.L.; Martínez-Noël, G.M.A. TOR signaling downregulation increases resistance to the cereal killer fusarium graminearum. Plant Signal. Behav. 2018, 13, e1414120. [Google Scholar] [CrossRef] [Green Version]
- Vleesschauwer, D.D.; Filipe, O.; Hoffman, G.; Seifi, H.S.; Haeck, A.; Canlas, P.; Bockhaven, J.V.; Waele, E.D.; Demeestere, K.; Ronald, P.; et al. Target of rapamycin signaling orchestrates growth–defense trade-offs in plants. New Phytol. 2018, 217, 305–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filipe, O.; De Vleesschauwer, D.; Haeck, A.; Demeestere, K.; Höfte, M. The energy sensor OsSnRK1a confers broad-spectrum disease resistance in rice. Sci. Rep. 2018, 8, 3864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, W.; Bobay, B.G.; Greeley, L.A.; Reyes, M.I.; Rajabu, C.A.; Blackburn, R.K.; Dallas, M.B.; Goshe, M.B.; Ascencio-Ibáñez, J.T.; Hanley-Bowdoin, L. Sucrose Nonfermenting 1-Related Protein Kinase 1 Phosphorylates a geminivirus rep protein to impair viral replication and infection. Plant Physiol. 2018, 178, 372–389. [Google Scholar] [CrossRef] [Green Version]
- Janse van Rensburg, H.C.; Van den Ende, W.; Signorelli, S. Autophagy in plants: Both a puppet and a puppet master of sugars. Front. Plant Sci. 2019, 10, 14. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Primavesi, L.F.; Jhurreea, D.; Andralojc, P.J.; Mitchell, R.A.C.; Powers, S.J.; Schluepmann, H.; Delatte, T.; Wingler, A.; Paul, M.J. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 2009, 149, 1860–1871. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Zhang, L.; Zhao, L.; Xue, P.; Qi, T.; Zhang, C.; Yuan, H.; Zhou, L.; Wang, D.; Qiu, J.; et al. SnRK1 Phosphorylates and destabilizes WRKY3 to enhance barley immunity to powdery mildew. Plant Commun. 2020, 1. [Google Scholar] [CrossRef]
- Lee, H.-J.; Park, Y.-J.; Seo, P.J.; Kim, J.-H.; Sim, H.-J.; Kim, S.-G.; Park, C.-M. Systemic immunity requires SnRK2.8-mediated nuclear import of NPR1 in Arabidopsis. Plant Cell 2015, 27, 3425–3438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef] [Green Version]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Hei, R.; Yang, Y.; Zhang, S.; Wang, Q.; Wang, W.; Zhang, Q.; Yan, M.; Zhu, G.; Huang, P.; et al. An orphan protein of Fusarium graminearum modulates host immunity by mediating proteasomal degradation of TaSnRK1α. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Schepetilnikov, M.; Kobayashi, K.; Geldreich, A.; Caranta, C.; Robaglia, C.; Keller, M.; Ryabova, L.A. Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO J. 2011, 30, 1343–1356. [Google Scholar] [CrossRef] [Green Version]
- Popa, C.; Li, L.; Gil, S.; Tatjer, L.; Hashii, K.; Tabuchi, M.; Coll, N.S.; Ariño, J.; Valls, M. The effector AWR5 from the plant pathogen ralstonia solanacearum is an inhibitor of the TOR signalling pathway. Sci. Rep. 2016, 6, 27058. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.-Y.; Neufeld, T.P. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell 2009, 20, 2004–2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soto-Burgos, J.; Bassham, D.C. SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS ONE 2017, 12, e0182591. [Google Scholar] [CrossRef] [Green Version]
- Lai, Z.; Wang, F.; Zheng, Z.; Fan, B.; Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011, 66, 953–968. [Google Scholar] [CrossRef] [PubMed]
- Leary, A.Y.; Sanguankiattichai, N.; Duggan, C.; Tumtas, Y.; Pandey, P.; Segretin, M.E.; Salguero Linares, J.; Savage, Z.D.; Yow, R.J.; Bozkurt, T.O. Modulation of plant autophagy during pathogen attack. J. Exp. Bot. 2018, 69, 1325–1333. [Google Scholar] [CrossRef] [Green Version]
- Crozet, P.; Margalha, L.; Butowt, R.; Fernandes, N.; Elias, C.A.; Orosa, B.; Tomanov, K.; Teige, M.; Bachmair, A.; Sadanandom, A.; et al. SUMOylation represses SnRK1 signaling in Arabidopsis. Plant J. 2016, 85, 120–133. [Google Scholar] [CrossRef] [Green Version]
- Ulrich, H.D. The fast-growing business of SUMO chains. Mol. Cell 2008, 32, 301–305. [Google Scholar] [CrossRef]
- Nietzsche, M.; Landgraf, R.; Tohge, T.; Börnke, F. A protein–protein interaction network linking the energy-sensor kinase SnRK1 to multiple signaling pathways in Arabidopsis thaliana. Curr. Plant Biol. 2016, 5, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Zentella, R.; Zhang, Z.-L.; Park, M.; Thomas, S.G.; Endo, A.; Murase, K.; Fleet, C.M.; Jikumaru, Y.; Nambara, E.; Kamiya, Y.; et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007, 19, 3037–3057. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, Y.; Hu, Y.; Liu, H.; He, M.; Yang, Z.; Kong, F.; Liu, X.; Hou, X. DELLA and EDS1 form a feedback regulatory module to fine-tune plant growth–defense tradeoff in Arabidopsis. Mol. Plant 2019, 12, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Cheng, H.; Grauwe, L.D.; Decat, J.; Schoutteten, H.; Moritz, T.; Straeten, D.V.D.; Peng, J.; Harberd, N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Touriñán, N.; Serrano-Mislata, A.; Alabadí, D. Regulation of DELLA proteins by post-translational modifications. Plant Cell Physiol. 2020, 61, 1891–1901. [Google Scholar] [CrossRef]
- Claeys, H.; De Bodt, S.; Inzé, D. Gibberellins and DELLAs: Central nodes in growth regulatory networks. Trends Plant Sci. 2014, 19, 231–239. [Google Scholar] [CrossRef]
- Dill, A.; Thomas, S.G.; Hu, J.; Steber, C.M.; Sun, T.-P. The Arabidopsis F-Box Protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 2004, 16, 1392–1405. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Richards, D.E.; Fleck, B.; Xie, D.; Burton, N.; Harberd, N.P. The Arabidopsis Mutant Sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 2004, 16, 1406–1418. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, J.; Murase, K.; Rieu, I.; Zentella, R.; Zhang, Z.-L.; Powers, S.J.; Gong, F.; Phillips, A.L.; Hedden, P.; Sun, T.; et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 2006, 18, 3399–3414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murase, K.; Hirano, Y.; Sun, T.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.-P.; Gubler, F. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef] [Green Version]
- Ueguchi-Tanaka, M.; Ashikari, M.; Nakajima, M.; Itoh, H.; Katoh, E.; Kobayashi, M.; Chow, T.; Hsing, Y.C.; Kitano, H.; Yamaguchi, I.; et al. Gibberellin insensitive dwarf1 encodes a soluble receptor for gibberellin. Nature 2005, 437, 693–698. [Google Scholar] [CrossRef]
- Willige, B.C.; Ghosh, S.; Nill, C.; Zourelidou, M.; Dohmann, E.M.N.; Maier, A.; Schwechheimer, C. The DELLA domain of GA insensitive mediates the interaction with the GA insensitive dwarf1A gibberellin receptor of Arabidopsis. Plant Cell 2007, 19, 1209–1220. [Google Scholar] [CrossRef] [Green Version]
- Curaba, J.; Moritz, T.; Blervaque, R.; Parcy, F.; Raz, V.; Herzog, M.; Vachon, G. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by leafy cotyledon2 and fusca3 in Arabidopsis. Plant Physiol. 2004, 136, 3660–3669. [Google Scholar] [CrossRef] [Green Version]
- Gazzarrini, S.; Tsuchiya, Y.; Lumba, S.; Okamoto, M.; McCourt, P. The transcription factor fusca3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 2004, 7, 373–385. [Google Scholar] [CrossRef] [Green Version]
- Tsai, A.Y.-L.; Gazzarrini, S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012, 69, 809–821. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; McFarlane, H.E.; Obata, T.; Richter, A.S.; Lohse, M.; Grimm, B.; Persson, S.; Fernie, A.R.; Giavalisco, P. Inhibition of TOR represses nutrient consumption, which improves greening after extended periods of etiolation1. Plant Physiol. 2018, 178, 101–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, X.; Hu, W.-W.; Shen, L.; Lee, L.Y.C.; Tao, Z.; Han, J.-H.; Yu, H. Global identification of DELLA target genes during Arabidopsis flower development. Plant Physiol. 2008, 147, 1126–1142. [Google Scholar] [CrossRef] [Green Version]
- Vleesschauwer, D.D.; Seifi, H.S.; Filipe, O.; Haeck, A.; Huu, S.N.; Demeestere, K.; Höfte, M. The DELLA protein SLR1 integrates and amplifies salicylic acid- and jasmonic acid-dependent innate immunity in rice. Plant Physiol. 2016, 170, 1831–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Liu, W.; Li, B.; Liu, G.; Wei, Y.; He, C.; Shi, H. Identification and functional analysis of cassava DELLA proteins in plant disease resistance against cassava bacterial blight. Plant Physiol. Biochem. 2018, 124, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Conti, L.; Nelis, S.; Zhang, C.; Woodcock, A.; Swarup, R.; Galbiati, M.; Tonelli, C.; Napier, R.; Hedden, P.; Bennett, M.; et al. Small ubiquitin-like modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev. Cell 2014, 28, 102–110. [Google Scholar] [CrossRef] [Green Version]
- Nelis, S.; Conti, L.; Zhang, C.; Sadanandom, A. A functional small ubiquitin-like modifier (SUMO) interacting motif (SIM) in the Gibberellin Hormone Receptor GID1 is conserved in cereal crops and disrupting this motif does not abolish hormone dependency of the DELLA-GID1 interaction. Plant Signal. Behav. 2015, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campanaro, A.; Battaglia, R.; Galbiati, M.; Sadanandom, A.; Tonelli, C.; Conti, L. SUMO proteases OTS1 and 2 control filament elongation through a DELLA-dependent mechanism. Plant Reprod. 2016, 29, 287–290. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, N.M.; Fernandes, T.; Nunes, C.; Rosa, M.T.G.; Matiolli, C.C.; Rodrigues, M.A.A.; Barros, P.M.; Oliveira, M.M.; Abreu, I.A. SUMOylation of rice DELLA SLR1 modulates transcriptional responses and improves yield under salt stress. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dai, C.; Xue, H.-W. Rice Early Flowering1, a CKI, Phosphorylates DELLA Protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010, 29, 1916–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Q.; Wang, W.; Guo, X.; Yue, J.; Huang, Y.; Xu, X.; Li, J.; Hou, S. Arabidopsis DELLA protein degradation is controlled by a type-one protein phosphatase, TOPP4. PLoS Genet. 2014, 10, e1004464. [Google Scholar] [CrossRef] [Green Version]
- Wild, M.; Davière, J.-M.; Cheminant, S.; Regnault, T.; Baumberger, N.; Heintz, D.; Baltz, R.; Genschik, P.; Achard, P. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 2012, 24, 3307–3319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, X.; Lee, L.Y.C.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.-L.; Yao, J.; Mei, C.-S.; Tong, X.-H.; Zeng, L.-J.; Li, Q.; Xiao, L.-T.; Sun, T.; Li, J.; Deng, X.-W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.-J.; An, X.-H.; Liu, X.; Hu, D.-G.; Wang, X.-F.; You, C.-X.; Hao, Y.-J. MdSnRK1.1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. J. Exp. Bot. 2017, 68, 2977–2990. [Google Scholar] [CrossRef] [Green Version]
- Coello, P.; Hey, S.J.; Halford, N.G. The sucrose non-fermenting-1-related (SnRK) family of protein kinases: Potential for manipulation to improve stress tolerance and increase yield. J. Exp. Bot. 2011, 62, 883–893. [Google Scholar] [CrossRef] [Green Version]
- Karasov, T.L.; Chae, E.; Herman, J.J.; Bergelson, J. Mechanisms to mitigate the trade-off between growth and defense. Plant Cell 2017, 29, 666–680. [Google Scholar] [CrossRef] [Green Version]
- Walters, D.; Heil, M. Costs and trade-offs associated with induced resistance. Physiol. Mol. Plant Pathol. 2007, 71, 3–17. [Google Scholar] [CrossRef]
- de Vega, D.; Newton, A.C.; Sadanandom, A. Post-translational modifications in priming the plant immune system: Ripe for exploitation? FEBS Lett. 2018, 592, 1929–1936. [Google Scholar] [CrossRef] [Green Version]
- Luna, E.; van Hulten, M.; Zhang, Y.; Berkowitz, O.; López, A.; Pétriacq, P.; Sellwood, M.A.; Chen, B.; Burrell, M.; van de Meene, A.; et al. Plant perception of β-aminobutyric acid is mediated by an aspartyl-TRNA synthetase. Nat. Chem. Biol. 2014, 10, 450–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gent, L.; Forde, B.G. How do plants sense their nitrogen status? J. Exp. Bot. 2017, 68, 2531–2539. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.-L.; Yang, Y.; He, Z. Roles of plant hormones and their interplay in rice immunity. Mol. Plant 2013, 6, 675–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckers, G.J.M.; Spoel, S.H. Fine-tuning plant defence signalling: Salicylate versus jasmonate. Plant Biol. 2006, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, D.-L.; Sun, L.; Li, Q.; Mao, B.; He, Z. The systemic acquired resistance regulator OsNPR1 attenuates growth by repressing auxin signaling through promoting IAA-amido synthase expression1. Plant Physiol. 2016, 172, 546–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Pajerowska-Mukhtar, K.; Culler, A.H.; Dong, X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 2007, 17, 1784–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Turner, J.G. Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 2008, 3, e3699. [Google Scholar] [CrossRef]
- Garavaglia, B.S.; Thomas, L.; Gottig, N.; Zimaro, T.; Garofalo, C.G.; Gehring, C.; Ottado, J. Shedding light on the role of photosynthesis in pathogen colonization and host defense. Commun. Integr. Biol. 2010, 3, 382–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrecht, C.; Boutrot, F.; Segonzac, C.; Schwessinger, B.; Gimenez-Ibanez, S.; Chinchilla, D.; Rathjen, J.P.; de Vries, S.C.; Zipfel, C. Brassinosteroids inhibit pathogen-associated molecular pattern–triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 2012, 109, 303–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakashita, H.; Yasuda, M.; Nitta, T.; Asami, T.; Fujioka, S.; Arai, Y.; Sekimata, K.; Takatsuto, S.; Yamaguchi, I.; Yoshida, S. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003, 33, 887–898. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.-W.; Deng, X.-G.; Fu, F.-Q.; Lin, H.-H. Induction of plant virus defense response by brassinosteroids and brassinosteroid signaling in Arabidopsis thaliana. Planta 2015, 241, 875–885. [Google Scholar] [CrossRef]
- Miyaji, T.; Yamagami, A.; Kume, N.; Sakuta, M.; Osada, H.; Asami, T.; Arimoto, Y.; Nakano, T. Brassinosteroid-related transcription factor BIL1/BZR1 increases plant resistance to insect feeding. Biosci. Biotechnol. Biochem. 2014, 78, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Lu, D.; Gao, X.; Jiang, S.; Ma, X.; Wang, Z.; Mengiste, T.; He, P.; Shan, L. Inverse Modulation of Plant Immune and Brassinosteroid Signaling Pathways by the Receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 2013, 110, 12114–12119. [Google Scholar] [CrossRef] [Green Version]
- Jaillais, Y.; Belkhadir, Y.; Balsemão-Pires, E.; Dangl, J.L.; Chory, J. Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc. Natl. Acad. Sci. USA 2011, 108, 8503–8507. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-R.; Ramos, J.M.; Hizon, R.J.M.; Ashikari, M.; Virk, P.S.; Torres, E.A.; Nissila, E.; Jena, K.K. Introgression of a functional epigenetic OsSPL 1 4 WFP allele into elite indica rice genomes greatly improved panicle traits and grain yield. Sci. Rep. 2018, 8, 3833. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.; Zhu, X.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Yu, H.; Xiong, G.; Wang, J.; Jiao, Y.; Liu, G.; Jing, Y.; Meng, X.; Hu, X.; Qian, Q.; et al. Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 2013, 25, 3743–3759. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yu, H.; Xiong, G.; Lu, Z.; Jiao, Y.; Meng, X.; Liu, G.; Chen, X.; Wang, Y.; Li, J. Tissue-specific ubiquitination by IPA1 interacting protein1 modulates IPA1 protein levels to regulate plant architecture in rice. Plant Cell 2017, 29, 697–707. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, H.; Ma, B.; Liu, G.; Wang, J.; Wang, J.; Gao, R.; Li, J.; Liu, J.; Xu, J.; et al. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 2017, 8, 14789. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Shi, Z.; Zhang, X.; Wang, M.; Zhang, L.; Zheng, K.; Liu, J.; Hu, X.; Di, C.; Qian, Q.; et al. Inducible overexpression of ideal plant architecture1 improves both yield and disease resistance in rice. Nat. Plants 2019, 5, 389–400. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Liu, K.; Song, W. Pleiotropic function of SQUAMOSA PROMOTER-BINDING PROTEIN-BOX gene TaSPL14 on plant architecture of wheat. Planta 2020, 253, 44. [Google Scholar] [CrossRef]
- Wei, H.; Zhao, Y.; Xie, Y.; Wang, H. Exploiting SPL genes to improve maize plant architecture tailored for high-density planting. J. Exp. Bot. 2018, 69, 4675–4688. [Google Scholar] [CrossRef]
- Xie, Y.; Liu, Y.; Ma, M.; Zhou, Q.; Zhao, Y.; Zhao, B.; Wang, B.; Wei, H.; Wang, H. Arabidopsis FHY3 and FAR1 integrate light and strigolactone signaling to regulate branching. Nat. Commun. 2020, 11, 1955. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Hong, G.; Li, L.; Zhang, X.; Kong, Y.; Sun, Z.; Li, J.; Chen, J.; He, Y. MiR156/SPL9 regulates reactive oxygen species accumulation and immune response in Arabidopsis thaliana. Phytopathology 2018, 109, 632–642. [Google Scholar] [CrossRef] [Green Version]
- Olsen, K.M.; Wendel, J.F. A Bountiful harvest: Genomic insights into crop domestication phenotypes. Annu. Rev. Plant Biol. 2013, 64, 47–70. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.K.M. Yield penalties of disease resistance in crops. Curr. Opin. Plant Biol. 2002, 5, 339–344. [Google Scholar] [CrossRef]
- Ning, Y.; Liu, W.; Wang, G.-L. Balancing immunity and yield in crop plants. Trends Plant Sci. 2017, 22, 1069–1079. [Google Scholar] [CrossRef]
- Da Silva, A.C.; de Lima, M.F.; Eloy, N.B.; Thiebaut, F.; Montessoro, P.; Hemerly, A.S.; Ferreira, P.C.G. The yin and yang in plant breeding: The trade-off between plant growth yield and tolerance to stresses. Biotechnol. Res. Innov. 2019, 3, 73–79. [Google Scholar] [CrossRef]
- Schenke, D.; Cai, D. Applications of CRISPR/Cas to Improve Crop Disease Resistance: Beyond inactivation of susceptibility factors. iScience 2020, 23. [Google Scholar] [CrossRef]
- Chujo, T.; Miyamoto, K.; Ogawa, S.; Masuda, Y.; Shimizu, T.; Kishi-Kaboshi, M.; Takahashi, A.; Nishizawa, Y.; Minami, E.; Nojiri, H.; et al. Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS ONE 2014, 9, e98737. [Google Scholar] [CrossRef] [Green Version]
- Pearlman, S.M.; Serber, Z.; Ferrell, J.E. A mechanism for the evolution of phosphorylation sites. Cell 2011, 147, 934–946. [Google Scholar] [CrossRef] [Green Version]
- Mansueto, L.; Fuentes, R.R.; Borja, F.N.; Detras, J.; Abriol-Santos, J.M.; Chebotarov, D.; Sanciangco, M.; Palis, K.; Copetti, D.; Poliakov, A.; et al. Rice SNP-seek database update: New SNPs, indels, and queries. Nucleic Acids Res. 2017, 45, D1075–D1081. [Google Scholar] [CrossRef]
- Tareke Woldegiorgis, S.; Wang, S.; He, Y.; Xu, Z.; Chen, L.; Tao, H.; Zhang, Y.; Zou, Y.; Harrison, A.; Zhang, L.; et al. Rice stress-resistant SNP database. Rice 2019, 12. [Google Scholar] [CrossRef] [Green Version]
- Ferro, A.M.; Ramos, P.; Guerreiro, O.; Jerónimo, E.; Pires, I.; Capel, C.; Capel, J.; Lozano, R.; Duarte, M.F.; Oliveira, M.M.; et al. Impact of novel SNPs identified in cynara cardunculus genes on functionality of proteins regulating phenylpropanoid pathway and their association with biological activities. BMC Genom. 2017, 18, 183. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Kang, C.; Min, B.; Yi, G.-S. Detection and analysis of disease-associated single nucleotide polymorphism influencing post-translational modification. BMC Med. Genom. 2015, 8 (Suppl 2), S7. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Peng, X.; Ying, P.; Tian, J.; Li, J.; Ke, J.; Zhu, Y.; Gong, Y.; Zou, D.; Yang, N.; et al. AWESOME: A database of SNPs that affect protein post-translational modifications. Nucleic Acids Res. 2019, 47, D874–D880. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering and Medicine. Genetically Engineered Crops: Experiences and Prospects; The National Academies Press: Washington, DC, USA, 2016. [CrossRef]
- Simó, C.; Ibáñez, C.; Valdés, A.; Cifuentes, A.; García-Cañas, V. Metabolomics of genetically modified crops. Int. J. Mol. Sci. 2014, 15, 18941–18966. [Google Scholar] [CrossRef] [Green Version]
- Spoel, S.H. Orchestrating the proteome with post-translational modifications. J. Exp. Bot. 2018, 69, 4499–4503. [Google Scholar] [CrossRef] [Green Version]
- Yang, B. Grand challenges in genome editing in plants. Front. Genome Ed. 2020, 2. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [Green Version]
- Mishra, R.; Joshi, R.K.; Zhao, K. Base editing in crops: Current advances, limitations and future implications. Plant Biotechnol. J. 2020, 18, 20–31. [Google Scholar] [CrossRef]
- Ren, B.; Liu, L.; Li, S.; Kuang, Y.; Wang, J.; Zhang, D.; Zhou, X.; Lin, H.; Zhou, H. Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice. Mol. Plant 2019, 12, 1015–1026. [Google Scholar] [CrossRef]
- Chen, X.; Zuo, S.; Schwessinger, B.; Chern, M.; Canlas, P.E.; Ruan, D.; Zhou, X.; Wang, J.; Daudi, A.; Petzold, C.J.; et al. An XA21-associated kinase (OsSERK2) regulates immunity mediated by the XA21 and XA3 immune receptors. Mol. Plant 2014, 7, 874–892. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Zu, S.-H.; Jiang, Y.-T.; Hu, L.-Q.; Zhang, Y.-J.; Chang, J.-H.; Xue, H.-W.; Lin, W.-H. Effective modulating brassinosteroids signal to study their specific regulation of reproductive development and enhance yield. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Jia, C.; Zhang, M.; Chen, D.; Chen, S.; Guo, R.; Guo, D.; Wang, Q. Ectopic expression of a BZR1-1D transcription factor in brassinosteroid signalling enhances carotenoid accumulation and fruit quality attributes in tomato. Plant Biotechnol J. 2014, 12, 105–115. [Google Scholar] [CrossRef]
- Wu, C.; Trieu, A.; Radhakrishnan, P.; Kwok, S.F.; Harris, S.; Zhang, K.; Wang, J.; Wan, J.; Zhai, H.; Takatsuto, S.; et al. Brassinosteroids regulate grain filling in rice. Plant Cell 2008, 20, 2130–2145. [Google Scholar] [CrossRef] [Green Version]
- Zuo, S.; Zhou, X.; Chen, M.; Zhang, S.; Schwessinger, B.; Ruan, D.; Yuan, C.; Wang, J.; Chen, X.; Ronald, P.C. OsSERK1 regulates rice development but not immunity to Xanthomonas oryzae Pv. oryzae or Magnaporthe oryzae. J. Integr. Plant Biol. 2014, 56, 1179–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Lu, P.; Guo, L.; Wang, Z.; Li, B.; Li, J.; Li, Y.; Qiu, D.; Shi, W.; Yang, L.; Wang, N.; et al. A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 2020, 11, 680. [Google Scholar] [CrossRef] [Green Version]
- Vijayan, J.; Devanna, B.N.; Singh, N.K.; Sharma, T.R. Cloning and functional validation of early inducible magnaporthe oryzae responsive CYP76M7 promoter from rice. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Pajerowska-Mukhtar, K.M.; Wang, W.; Tada, Y.; Oka, N.; Tucker, C.L.; Fonseca, J.P.; Dong, X. The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 2012, 22, 103–112. [Google Scholar] [CrossRef] [Green Version]
- Blanvillain-Baufumé, S.; Reschke, M.; Solé, M.; Auguy, F.; Doucoure, H.; Szurek, B.; Meynard, D.; Portefaix, M.; Cunnac, S.; Guiderdoni, E.; et al. Targeted promoter editing for rice resistance to Xanthomonas oryzae Pv. oryzae reveals differential activities for SWEET14-inducing TAL effectors. Plant Biotechnol. J. 2017, 15, 306–317. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Wang, B.; Li, N.; Tang, Y.; Yang, S.; Yang, T.; Xu, J.; Guo, C.; Yan, P.; Wang, Q.; et al. CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Gentzel, I.N.; Park, C.H.; Bellizzi, M.; Xiao, G.; Gadhave, K.R.; Murphree, C.; Yang, Q.; LaMantia, J.; Redinbaugh, M.G.; Balint-Kurti, P.; et al. A CRISPR/DCas9 toolkit for functional analysis of maize genes. Plant Methods 2020, 16, 133. [Google Scholar] [CrossRef] [PubMed]
- Castro, P.H.; Santos, M.Â.; Freitas, S.; Cana-Quijada, P.; Lourenço, T.; Rodrigues, M.A.A.; Fonseca, F.; Ruiz-Albert, J.; Azevedo, J.E.; Tavares, R.M.; et al. Arabidopsis thaliana SPF1 and SPF2 are nuclear-located ULP2-like SUMO proteases that act downstream of SIZ1 in plant development. J. Exp. Bot. 2018, 69, 4633–4649. [Google Scholar] [CrossRef] [PubMed]
- Garrido, E.; Srivastava, A.K.; Sadanandom, A. Exploiting protein modification systems to boost crop productivity: SUMO proteases in focus. J. Exp. Bot. 2018, 69, 4625–4632. [Google Scholar] [CrossRef] [Green Version]
- Yates, G.; Srivastava, A.K.; Sadanandom, A. SUMO Proteases: Uncovering the roles of DeSUMOylation in plants. J. Exp. Bot. 2016, 67, 2541–2548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Y.; Botella, J.R.; Liu, Y.; Zhu, J.-K. Gene editing in plants: Progress and challenges. Natl. Sci. Rev. 2019, 6, 421–437. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Duan, L.; Zhou, B.; Yu, H.; Zhu, H.; Cao, Y.; Zhang, Z. Interplay of pathogen-induced defense responses and symbiotic establishment in medicago truncatula. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Ferreira, M.A.; Huang, M.; Camargos, L.F.; Yu, X.; Teixeira, R.M.; Carpinetti, P.A.; Mendes, G.C.; Gouveia-Mageste, B.C.; Liu, C.; et al. The receptor-like kinase NIK1 targets FLS2/BAK1 immune complex and inversely modulates antiviral and antibacterial immunity. Nat. Commun. 2019, 10, 4996. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.; Wiesner-Hanks, T.; Wisser, R.; Balint-Kurti, P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2018, 19, 21–33. [Google Scholar] [CrossRef]
- McGrann, G.R.D.; Stavrinides, A.; Russell, J.; Corbitt, M.M.; Booth, A.; Chartrain, L.; Thomas, W.T.B.; Brown, J.K.M. A Trade off between Mlo Resistance to powdery mildew and increased susceptibility of barley to a newly important disease, ramularia leaf spot. J. Exp. Bot. 2014, 65, 1025–1037. [Google Scholar] [CrossRef] [Green Version]
- Mitra, S.K.; Chen, R.; Dhandaydham, M.; Wang, X.; Blackburn, R.K.; Kota, U.; Goshe, M.B.; Schwartz, D.; Huber, S.C.; Clouse, S.D. An autophosphorylation site database for leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. Plant J. 2015, 82, 1042–1060. [Google Scholar] [CrossRef]
- Chang, C.-C.; Tung, C.-H.; Chen, C.-W.; Tu, C.-H.; Chu, Y.-W. SUMOgo: Prediction of sumoylation sites on lysines by motif screening models and the effects of various post-translational modifications. Sci. Rep. 2018, 8, 15512. [Google Scholar] [CrossRef] [PubMed]
- Mosharaf, P.; Hassan, M.; Ahmed, F.F.; Khatun, S.; Moni, M.A.; Mollah, N.H. Computational prediction of protein ubiquitination sites mapping on Arabidopsis thaliana. Comput. Biol. Chem. 2020, 85, 107238. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Luo, Z.-Q. Post-translational regulation of ubiquitin signaling. J. Cell. Biol. 2019, 218, 1776–1786. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhou, J.; Lin, S.; Deng, W.; Zhang, Y.; Xue, Y. PLMD: An updated data resource of protein lysine modifications. J. Genet. Genom. 2017, 44, 243–250. [Google Scholar] [CrossRef]
- Song, G.; Walley, J.W. Dynamic protein acetylation in plant–pathogen interactions. Front. Plant Sci. 2016, 7, 421. [Google Scholar] [CrossRef] [Green Version]
- Walley, J.W.; Shen, Z.; McReynolds, M.R.; Schmelz, E.A.; Briggs, S.P. Fungal-induced protein hyperacetylation in maize identified by acetylome profiling. Proc. Natl. Acad. Sci. USA 2018, 115, 210–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swinnen, G.; Goossens, A.; Colinas, M. Metabolic editing: Small measures, great impact. Curr. Opin. Biotechnol. 2019, 59, 16–23. [Google Scholar] [CrossRef] [PubMed]

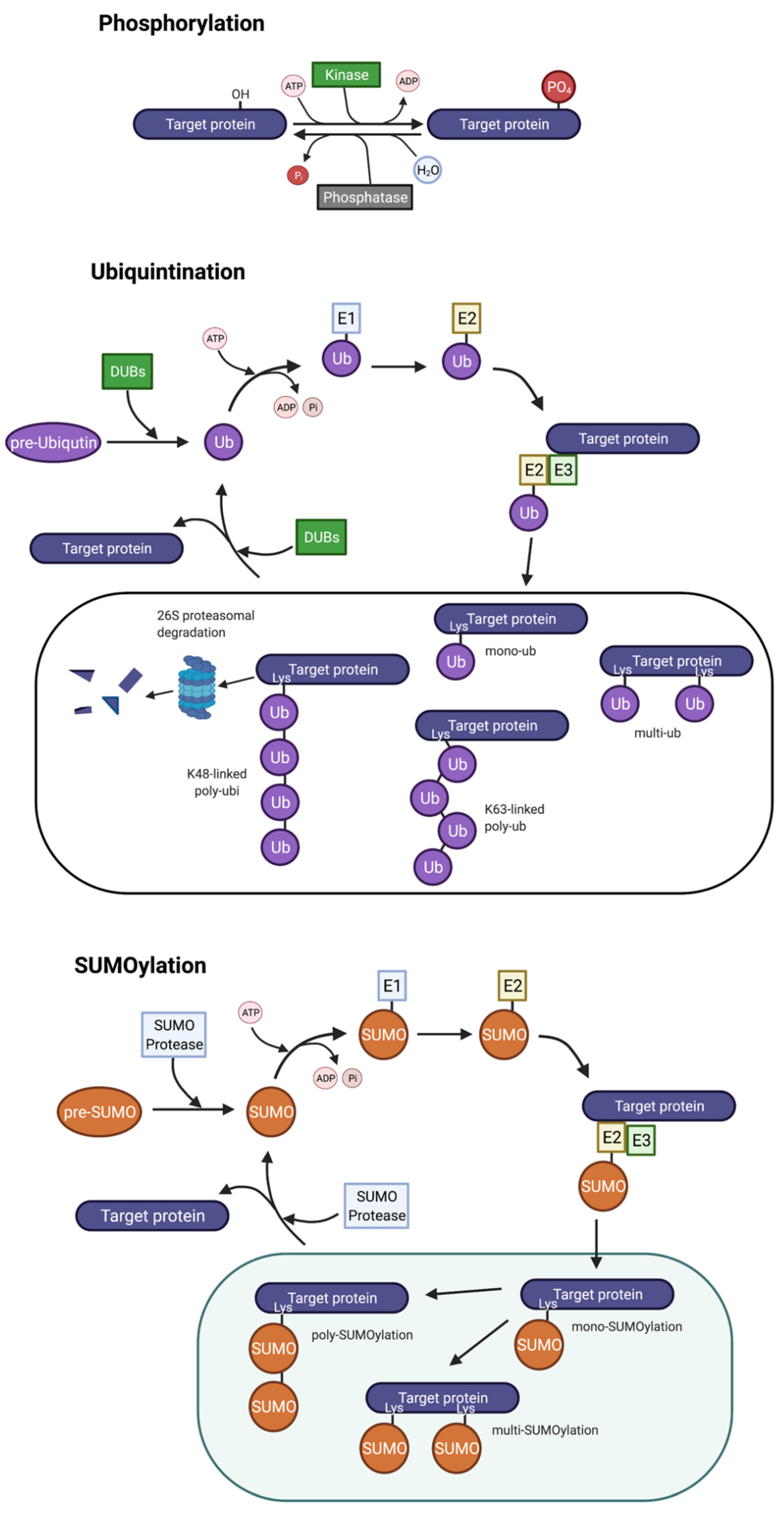
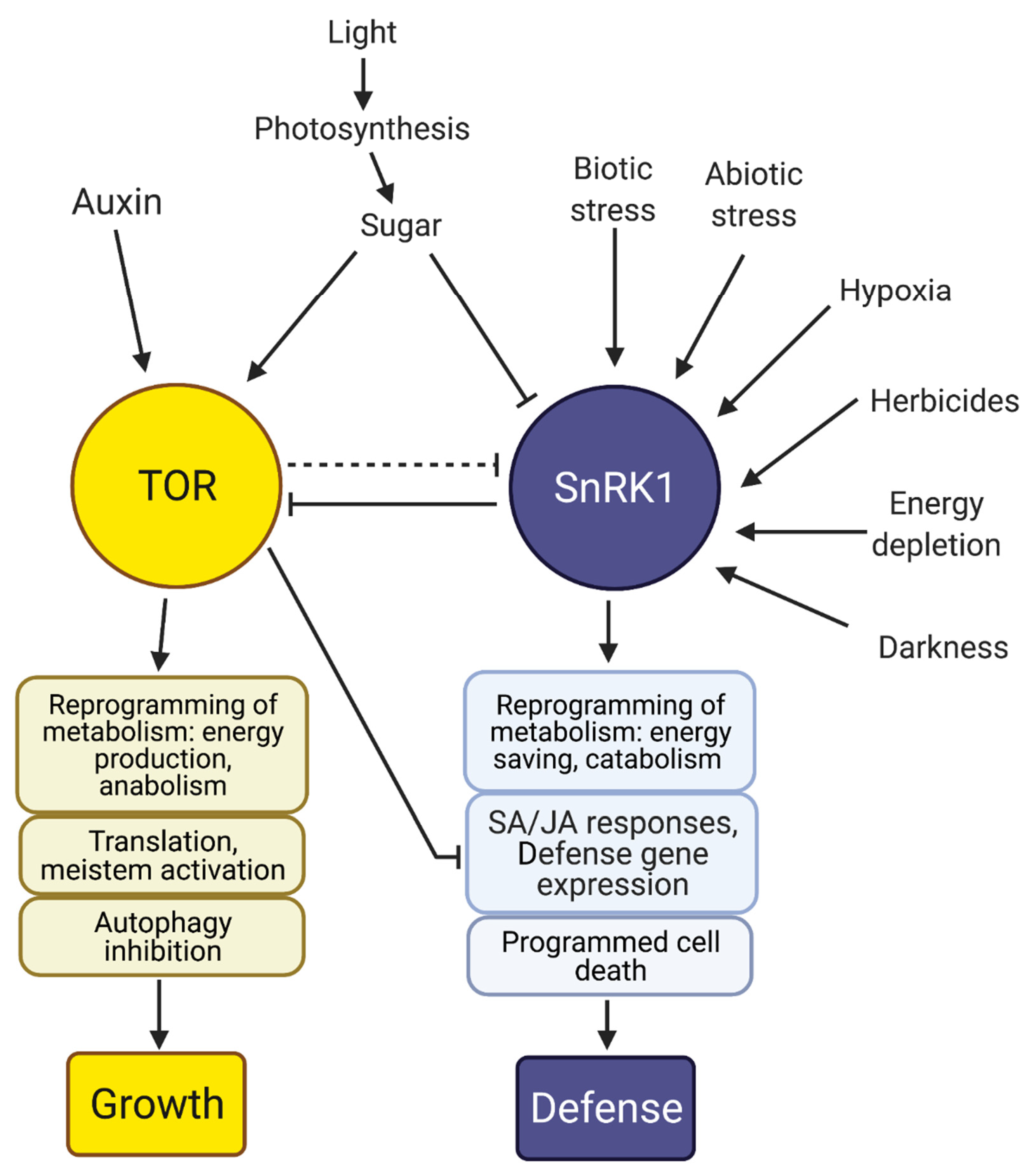
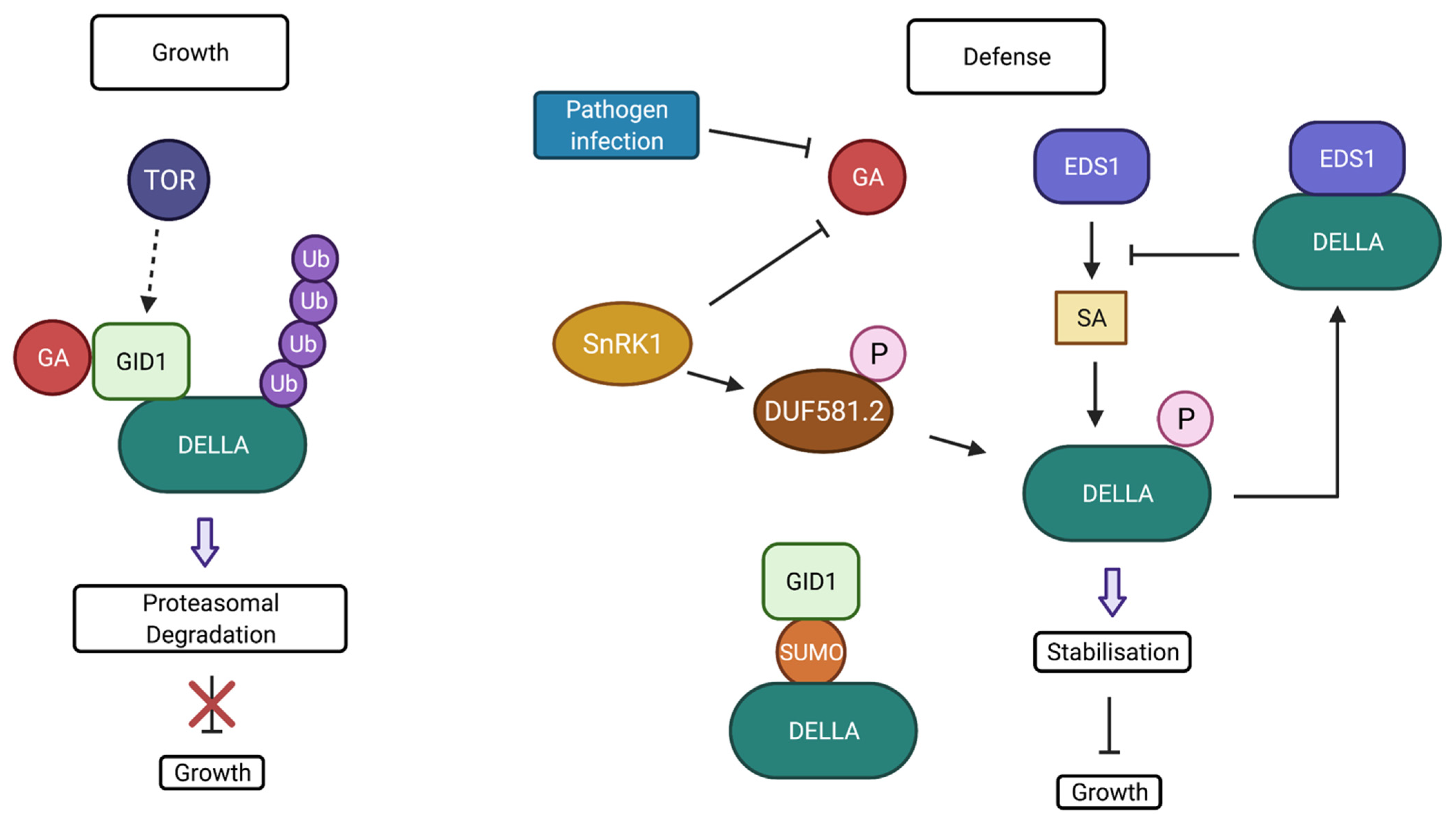

| Effector | Pathogen | Target/Host | Function | References |
|---|---|---|---|---|
| AvrPtoB | P. syringae | FLS2, BAK1, CERK1 (Arabidopsis), FEN (Tomato) | E3 ubiquitin ligase | [152,153,154,155] |
| AvrPto | P. syringae | FLS2, EFR (Arabidopsis) LeFLS2, RIN4 (tomato) | Kinase inhibitor | [156,157] |
| AvrRpm1 | P. syringae | RIN4 (Arabidopsis) | Induces phosphorylation | [158] |
| AvrB | P. syringae | RIN4 (Arabidopsis) | Induces phosphorylation | [158] |
| HopF2 | P. syringae | RIN4, BAK1, MKK5 (Arabidopsis), MPK6 (Tomato) | ADP-ribosylation | [159,160,161,162] |
| HopAI1 | P. syringae | MPK4, MPK6 (Arabidopsis and tomato) | Phosphotheonine lyase | [161,163] |
| XopAU | Xanthomonas sp. | MKK2 (tomato) | Protein kinase | [164] |
| XopK | Xanthomonas sp. | OsSERK2 (rice) | E3 ubiquitin ligase | [165] |
| XopDXcv85-10 | Xanthomonas sp. | ERF4 (tomato) | SUMO protease, ubiquitin protease | [140,166,167] |
| XopDXcc8004 | Xanthomonas sp. | RGA (Arabidopsis) | SUMO protease? | [168] |
| AvrBsT | Xanthomonas sp. | SnRK1, ACETYLATED INTERACTING PROTEIN1 (ACIP1), proteasomal subunit RPN8 (pepper) | Acetyltransferase | [169,170,171] |
| AvrXv4 | Xanthomonas sp. | Unknown targets | SUMO protease? | [169,172] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gough, C.; Sadanandom, A. Understanding and Exploiting Post-Translational Modifications for Plant Disease Resistance. Biomolecules 2021, 11, 1122. https://doi.org/10.3390/biom11081122
Gough C, Sadanandom A. Understanding and Exploiting Post-Translational Modifications for Plant Disease Resistance. Biomolecules. 2021; 11(8):1122. https://doi.org/10.3390/biom11081122
Chicago/Turabian StyleGough, Catherine, and Ari Sadanandom. 2021. "Understanding and Exploiting Post-Translational Modifications for Plant Disease Resistance" Biomolecules 11, no. 8: 1122. https://doi.org/10.3390/biom11081122
APA StyleGough, C., & Sadanandom, A. (2021). Understanding and Exploiting Post-Translational Modifications for Plant Disease Resistance. Biomolecules, 11(8), 1122. https://doi.org/10.3390/biom11081122






