Synthesis, Structural Investigations, Molecular Docking, and Anticancer Activity of Some Novel Schiff Bases and Their Uranyl Complexes
Abstract
:1. Introduction
2. Experimental
2.1. Materials and Methods
2.2. Synthesis of Schiff Base Ligands
- i. (4-bromo-2-((pyrazin-2-ylimino)methyl)phenol) (L1)
- i. (4-nitro-2-((pyrazin-2-ylimino)methyl)phenol) (L2)
- (2-((pyraiz-2-ylimino)methyl)phenol) (L3)
2.3. Synthesis of Metal Complexes
2.3.1. Metal Complexes of Schiff Base (1:2)
- i. Synthesis of [UO2(L1)2]·2H2O
2.3.2. Metal Complexes of Schiff Bases (1:1)
- i. Synthesis of [UO2(L2)OAc]·2H2O
- i. Synthesis of [UO2(L3)OAc]·2H2O
2.4. Mass Spectra
2.5. Molar Ratio
2.6. Computational Details
2.7. DNA Binding Studies
2.8. Molecular Docking
2.9. Anticancer and Toxicological Studies
3. Results and Discussion
3.1. IR Studies
3.2. 1H NMR Spectra for Ligands
3.3. Conductance Measurements
3.4. Molar Ratios
3.5. The Electronic Spectra
3.6. Thermal Analysis
3.7. Kinetic Studies
3.8. Suggested Structures
3.9. Computational Calculations
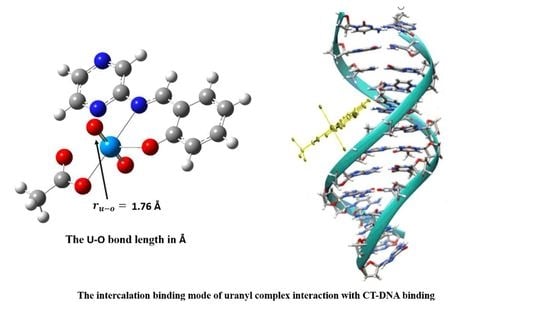
3.10. DNA-Binding Studies
3.11. Molecular Docking
3.12. Anticancer and Toxicological Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fidler, M.M.; Bray, F.; Soerjomataram, I. The global cancer burden and human development: A review. Scand. J. Public Health 2018, 46, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Tüzün, B. Investigation of pyrazoly derivatives schiff base ligands and their metal complexes used as anti-cancer drug. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 227, 117663–117674. [Google Scholar] [CrossRef] [PubMed]
- Staicu, C.E.; Predescu, D.-V.; Rusu, C.M.; Radu, B.M.; Cretoiu, D.; Suciu, N.; Crețoiu, S.M.; Voinea, S.-C. Role of microRNAs as Clinical Cancer Biomarkers for Ovarian Cancer: A Short Overview. Cells 2020, 9, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linck-Paulus, L.; Hellerbrand, C.; Bosserhoff, A.K.; Dietrich, P. Dissimilar Appearances Are Deceptive–Common microRNAs and Therapeutic Strategies in Liver Cancer and Melanoma. Cells 2020, 9, 114. [Google Scholar] [CrossRef] [Green Version]
- Hariprasath, K.; Deepthi, B.; Babu, I.S.; Venkatesh, P.; Sharfudeen, S.; Soumya, V. Metal complexes in drug research-a review. J. Chem. Pharm. Res. 2010, 2, 496–499. [Google Scholar]
- Mumtaz, A.; Khan, F.A.; Ahmad, S.; Najam-Ul-Haq, M.; Atif, M.; Khan, S.A.; Maalik, A.; Azhar, S.; Murtaza, G. Recent pharmacological advancements in schiff bases: A review. Acta Pol. Pharm. Drug Res. 2014, 71, 531–535. [Google Scholar]
- Mishra, V.R.; Ghanavatkar, C.W.; Mali, S.N.; Chaudhari, H.; Sekar, N. Schiff base clubbed benzothiazole: Synthesis, potent antimicrobial and MCF-7 anticancer activity, DNA cleavage and computational study. J. Biomol. Struct. Dyn. 2019, 38, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Asadi, Z.; Asadi, M.; Zeinali, A.; Ranjkeshshorkaei, M.; Fejfarová, K.; Eigner, V.; Dušek, M.; Dehnokhalaji, A. Synthesis, structural investigation and kinetic studies of uranyl(VI) unsymmetrical Schiff base complexes. J. Chem. Sci. 2014, 126, 1673–1683. [Google Scholar] [CrossRef]
- Ebrahimipour, S.Y.; Mohamadi, M.; Mahani, M.T.; Simpson, J.; Mague, J.T.; Sheikhshoaei, I. Synthesis and structure elucidation of novel salophen-based dioxo-uranium(VI) com-plexes: In-vitro and in-silico studies of their DNA/BSA-binding properties and anticancer activity. Eur. J. Med. Chem. 2017, 140, 172–186. [Google Scholar] [CrossRef] [PubMed]
- Deghadi, R.G.; Abbas, A.A.; Mohamed, G.G. Theoretical and experimental investigations of new bis (amino triazole) schiff base ligand: Preparation of its UO 2 (II), Er (III), and La (III) complexes, studying of their antibacterial, anticancer, and molecular docking. Appl. Organomet. Chem. 2021, 35, e6292. [Google Scholar] [CrossRef]
- Asadi, Z.; Asadi, M.; Firuzabadi, F.D.; Yousefi, R.; Jamshidi, M. Synthesis, characterization, anticancer activity, thermal and electrochemical studies of some novel uranyl Schiff base complexes. J. Iran. Chem. Soc. 2014, 11, 423–429. [Google Scholar] [CrossRef]
- Mohamadi, M.; Ebrahimipour, S.Y.; Castro, J.; Torkzadeh-Mahani, M. Synthesis, characterization, crystal structure, DNA and BSA binding, molecular docking and in vitro anticancer activities of a mononuclear diox-ido-uranium (VI) complex derived from a tridentate ONO aroylhydrazone. J. Photochem. Photobiol. B Biol. 2016, 158, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Al-Sha’Alan, N.H. Antimicrobial Activity and Spectral, Magnetic and Thermal Studies of Some Transition Metal Complexes of a Schiff Base Hydrazone Containing a Quinoline Moiety. Molecules 2007, 12, 1080–1091. [Google Scholar] [CrossRef]
- Mohamed, G.; Omar, M.M.; Hindy, A.M. Metal complexes of Schiff bases: Preparation, characterization, and biological activity. Turk. J. Chem. 2006, 30, 361–382. [Google Scholar]
- Beck, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Fox, D.J. Gaussian09 Program; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Serezhkina, L.B.; Vologzhanina, A.V.; Klepov, V.; Serezhkin, V.N. Crystal structure of R[UO2(CH3COO)3] (R = NH4+, K+, or Cs+). Crystallogr. Rep. 2010, 55, 773–779. [Google Scholar] [CrossRef]
- Hay, P.J. Gaussian basis sets for molecular calculations. The representation of 3d orbitals in transition-metal atoms. J. Chem. Phys. 1977, 66, 4377–4384. [Google Scholar] [CrossRef]
- Mashat, K.H.; Babgi, B.A.; Hussien, M.A.; Arshad, M.N.; Abdellattif, M.H. Synthesis, structures, DNA-binding and anticancer activities of some copper(I)-phosphine complexes. Polyhedron 2019, 158, 164–172. [Google Scholar] [CrossRef]
- Adegoke, O.A.; Ghosh, M.; Jana, A.; Mukherjee, A. Photo-physical investigation of the binding interac-tions of alumina nanoparticles with calf thymus DNA. Nucleus 2019, 62, 251–257. [Google Scholar]
- Jayakumar, J.; Anishetty, S. Molecular Dynamics simulations of Inhibitor of Apoptosis Proteins and identification of potential small molecule inhibitors. Bioorg. Med. Chem. Lett. 2014, 24, 2098–2104. [Google Scholar] [CrossRef] [PubMed]
- Sait, K.H.W.; Alam, Q.; Anfinan, N.; Al-Ghamdi, O.; Malik, A.; Noor, R.; Jahan, F.; Tarique, M. Structure-based virtual screening and molecular docking for the identification of potential novel EGFRkinase inhibitors against ovarian cancer. Bioinformation 2019, 15, 287–294. [Google Scholar] [CrossRef]
- Hosny, N.M.; Hussien, M.A.; Radwan, F.M.; Nawar, N. Synthesis, spectral characterization and DNA binding of Schiff-base metal complexes derived from 2-amino-3-hydroxyprobanoic acid and acetylacetone. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 132, 121–129. [Google Scholar] [CrossRef]
- Clougherty, L.; Sousa, J.; Wyman, G. C= N stretching frequency in infrared spectra of aromatic azome-thines. J. Org. Chem. 1957, 22, 462. [Google Scholar] [CrossRef]
- El-Dissouky, A.; El-Sonbati, A.Z. Effect of substituants on the structure of dioxouranium (VI) complex-es of 7-carboxaldehyde-8-hydroxyquinoline and some of its Schiff bases. Transit. Met. Chem. 1986, 11, 112–115. [Google Scholar] [CrossRef]
- Kovacic, J. The C=N stretching frequency in the infrared spectra of Schiff’s base complexes—I. Copper complexes of salicylidene anilines. Spectrochim. Acta Part A Mol. Spectrosc. 1967, 23, 183–187. [Google Scholar] [CrossRef]
- Wehling, R.L. Infrared Spectroscopy in Food Analysis; Springer: Boston, MA, USA, 2010; pp. 407–420. [Google Scholar]
- Baasov, T.; Friedman, N.; Sheves, M. Factors affecting the C:N stretching in protonated retinal Schiff base: A model study for bacteriorhodopsin and visual pigments. Biochemistry 1987, 26, 3210–3217. [Google Scholar] [CrossRef]
- Minacheva, L.K.; Ivanova, I.S.; Pyatova, E.N.; Dorokhov, A.V.; Bicherov, A.V.; Burlov, A.S.; Tsivadze, A.Y. Synthesis, Crystal Structure, and Vibrational Spectra of an Azomethine Derivative of Benzo-15-crown-5, N-(4′-Benzo-15-crown-5)-5-bromo-2-hydroxyphenylaldimine. Dokl. Chem. 2004, 395, 68–73. [Google Scholar] [CrossRef]
- Arjunan, V.; Subramanian, S.; Mohan, S. FTIR and FTR spectral studies of 2-amino-6-bromo-3-formylchromone. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2004, 60, 995–1000. [Google Scholar] [CrossRef]
- Shlyapochnikov, V.A.; Khaikin, L.S.; Grikina, O.E.; Bock, C.W.; Vilkov, L.V. The structure of nitro-benzene and the interpretation of the vibrational frequencies of the C-NO2 moiety on the basis of ab initio calculations. J. Mol. Struct. 1994, 326, 1–16. [Google Scholar] [CrossRef]
- Deacon, G.B.; Phillips, R.J. Relationships between the carbon-oxygen stretching frequencies of carboxylato complexes and the type of carboxylate coordination. Coord. Chem. Rev. 1980, 33, 227–250. [Google Scholar] [CrossRef]
- Kakihana, M.; Nagumo, T.; Okamoto, M.; Kakihana, H. Coordination structures for uranyl carboxylate complexes in aqueous solution studied by IR and carbon-13 NMR spectra. J. Phys. Chem. 1987, 91, 6128–6136. [Google Scholar] [CrossRef]
- El-Sonbati, A.Z.; Belal, A.A.M.; El-Wakeel, S.I.; Hussien, M.A. Stereochemistry of new nitrogen containing heterocyclic compounds.: X. Supramolecular structures and stereochemical versatility of polymeric complexes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2004, 60, 965–972. [Google Scholar] [CrossRef]
- Breitmaier, E.; Sinnema, A. Structure Elucidation by NMR in Organic Chemistry: A Practical Guide; Wiley: Chichester, UK; New York, NY, USA; Brisbane, Australia; Toronto, ON, Canada; Singapore, 1993. [Google Scholar]
- Udhayakumari, D.; Velmathi, S. Colorimetric and fluorescent sensor for selective sensing of Hg2+ ions in semi aqueous medium. J. Lumin 2013, 136, 117–121. [Google Scholar] [CrossRef]
- Hansen, P.; Rozwadowski, Z.; Dziembowska, T. NMR Studies of Hydroxy Schiff Bases. Curr. Org. Chem. 2009, 13, 194–215. [Google Scholar] [CrossRef]
- Tirmizi, S.A.; Wattoo, F.H.; Sarwar, S.; Anwar, W.; Wattoo, F.H.; Memon, A.N.; Iqbal, J. Spectrophotometric study of stability constants of famotidine-Cu (II) complex at different temperatures. Arab. J. Sci. Eng. 2009, 34, 43–51. [Google Scholar]
- Hussien, M.A.; Essa, E.; El-Gizawy, S.A. Investigation of the effect of formulation additives on telmisartan dissolution rate: Development of oral disintegrating tablets. Eur. J. Biomed. Pharm. Sci. 2019, 6, 12–20. [Google Scholar]
- Özdemir, Özlem Synthesis and characterization of a new diimine Schiff base and its Cu2+ and Fe3+ complexes: Investigation of their photoluminescence, conductance, spectrophotometric and sensor behaviors. J. Mol. Struct. 2019, 1179, 376–389. [CrossRef]
- Ali, D.I.; Wani, W.; Saleem, K. Empirical Formulae to Molecular Structures of Metal Complexes by Molar Conductance. Synth. React. Inorg. Met. Chem. 2013, 43, 1162–1170. [Google Scholar] [CrossRef]
- El-Behery, M.; El-Twigry, H. Synthesis, magnetic, spectral, and antimicrobial studies of Cu (II), Ni (II) Co (II), Fe (III), and UO2 (II) complexes of a new Schiff base hydrazone derived from 7-chloro-4-hydrazinoquinoline. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2007, 66, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Hosny, N.M.; Hussien, M.A.; Radwan, F.M.; Nawar, N. Synthesis, spectral, thermal and optical proper-ties of Schiff-base complexes derived from 2 (E)-2-((z)-4-hydroxypent-3-en-2-ylideneamino)-5-guanidinopentanoic acid and acetylacetone. J. Mol. Struct. 2017, 1143, 176–183. [Google Scholar] [CrossRef]
- Çanakçı, D. Synthesis, Spectroscopic, Thermodynamics and Kinetics Analysis Study of Novel Polymers Containing Various Azo Chromophore. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emara, A.A.A. Novel Asymmetric Tetradentate Schiff Base Ligands Derived from 6-Metwia-Formyl4-Hydroxy-2-(1H)-Quinolone and Their Metal Complexes. Synth. React. Inorg. Met. Chem. 1999, 29, 87–103. [Google Scholar] [CrossRef]
- Arnold, P.L.; Purkis, J.M.; Rutkauskaite, R.; Kovacs, D.; Love, J.B.; Austin, J. Controlled Photocatalytic Hydrocarbon Oxidation by Uranyl Complexes. ChemCatChem 2019, 11, 3786–3790. [Google Scholar] [CrossRef]
- Abdel-Rahman, L.H.; Ismail, N.M.; Ismael, M.; Abu-Dief, A.M.; Ahmed, E.A.H. Synthesis, characteri-zation, DFT calculations and biological studies of Mn (II), Fe (II), Co (II) and Cd (II) complexes based on a tetradentate ONNO donor Schiff base ligand. J. Mol. Struct. 2017, 1134, 851–862. [Google Scholar] [CrossRef]
- El-Bindary, A.; Mohamed, G.; El-Sonbati, A.; Diab, M.; Hassan, W.; Morgan, S.; Elkholy, A. Geometrical structure, potentiometric, molecular docking and thermodynamic studies of azo dye ligand and its metal complexes. J. Mol. Liq. 2016, 218, 138–149. [Google Scholar] [CrossRef]
- Ponya Utthra, P.; Kumaravel, G.; Senthilkumar, R.; Raman, N. Heteroleptic Schiff base complexes con-taining terpyridine as chemical nucleases and their biological potential: A study of DNA binding and cleaving, an-timicrobial and cytotoxic tendencies. Appl. Organomet. Chem. 2017, 31, e3629–e3630. [Google Scholar] [CrossRef]
- Maheswari, P.U.; Palaniandavar, M. DNA binding and cleavage properties of certain tetrammine ruthe-nium (II) complexes of modified 1, 10-phenanthrolines–effect of hydrogen-bonding on DNA-binding affinity. J. Inorg. Biochem. 2004, 98, 219–230. [Google Scholar] [CrossRef]
- Vijesh, A.; Isloor, A.M.; Telkar, S.; Arulmoli, T.; Fun, H.-K. Molecular docking studies of some new imidazole derivatives for antimicrobial properties. Arab. J. Chem. 2013, 6, 197–204. [Google Scholar] [CrossRef] [Green Version]
- Sharfalddin, A.A.; Hussien, M.A. Bivalence metal complexes of antithyroid drug carbimazole; synthe-sis, characterization, computational simulation, and biological studies. J. Mol. Struct. 2021, 1228, 129725–129736. [Google Scholar] [CrossRef]
- Hajalsiddig, T.T.H.; Osman, A.B.M.; Saeed, A.E.M. 2D-QSAR Modeling and Molecular Docking Studies on 1H-Pyrazole-1-carbothioamide Derivatives as EGFR Kinase Inhibitors. ACS Omega 2020, 5, 18662–18674. [Google Scholar] [CrossRef]
- Zarko Gagic, Dusan Ruzic, Nemanja Djokovic, Teodora Djikic and Katarina NikolicIn silico Methods for Design of Kinase Inhibitors as Anticancer Drugs. Front. Chem. 2019, 7, 873. [CrossRef] [Green Version]
- El-Bindary, E.A.; Toson, K.R.; Shoueir, H.A.; Aljohani, M.M. Abo-Ser, Metal–organic frameworks as efficient materials for drug delivery: Synthesis, characterization, antioxidant, anticancer, antibacterial and molecular docking investigation. Appl. Organomet. Chem. 2020, 34, e5905. [Google Scholar] [CrossRef]
- El-Gammal, O.A.; Mohamed, F.S.; Rezk, G.N.; El-Bindary, A.A. Synthesis, characterization, catalytic, DNA binding and antibacterial activities of Co(II), Ni(II) and Cu(II) complexes with new Schiff base ligand. J. Mol. Liq. 2021, 326, 115223. [Google Scholar] [CrossRef]
- El-Gammal, O.A.; Mohamed, F.S.; Rezk, G.N.; El-Bindary, A.A. Structural characterization and biological activity of a new metal complexes based of Schiff base. J. Mol. Liq. 2021, 330, 115522. [Google Scholar] [CrossRef]
- Kiwaan, H.A.; El-Mowafy, A.S.; El-Bindary, A.A. Synthesis, spectral characterization, DNA binding, catalytic and in vitro cytotoxicity of some metal complexes. J. Mol. Liq. 2021, 326, 115381. [Google Scholar] [CrossRef]
- Abou-Melha, G.A.; Al-Hazmi, I.; Althagafi, A.; Alharbi, F.; Shaaban, N.M.; El-Metwaly, A.A.; El-Bindary, M.A.; El-Bindary, A. Synthesis, characterization, DFT calculation, DNA binding and antimicrobial activities of metal com-plexes of dimedone arylhydrazone. J. Mol. Liq. 2021, 334, 116498–116506. [Google Scholar] [CrossRef]







| Basis Set | UO Bond Length, Å | % Error | |
|---|---|---|---|
| For U Atom | For Other Atoms | ||
| SDD | 6-31G | 1.451 | 17.73 |
| SDD | 6-31G(d,p) | 1.280 | 27.38 |
| SDD | 6-31++G(d,p) | 1.300 | 26.28 |
| SDD | cc-pvdz | 1.294 | 26.63 |
| SDD | SDD | 1.796 | 1.86 |
| Compounds | (C=N) Azomethine | (C=N) Pyrazine Ring | C-O Phenol | C-X | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exp. | DFT | Exp. | DFT | Exp. | DFT | Exp. | DFT | |||
| L1 | 1602 | 1621.94 | 1578.35 | 1588.34 | 1252.98 | 1289.55 | 630.59 | 634.76 | ||
| L2 | 1608.95 | 1798.16 | 1578.35 | 1668.00 | 1252.48 | 1354.52 | νasym | νsym | νasym | νsym |
| 1559.70 | 1344.77 | 1897.89 | 1564.29 | |||||||
| L3 | 1602.98 | 1676.97 | 1561.94 | 1573.69 | 1252.98 | 128529 | ------ | ------ | ||
| [UO2(L1)2]·2H2O | 1639.91 | 1658.90 | 1612.68 | 1583.05 | 1255.97 | 1339.70 | 624.62 | 638.72 | ||
| [UO2(L2)OAc]·2H2O | 1660.36 | 1660.32 | 1601.49 | 1585.48 | 1244.77 | 1348.61 | νasym | νsym | νasym | νsym |
| 1542.50 | 1319.40 | 1471.66 | 1403.04 | |||||||
| [UO2(L3)OAc]·2H2O | 1637.69 | 1647.27 | 1558.95 | 1586.29 | 1250 | 1341.16 | ------ | ------ | ||
| Compounds | O=U=O | |||||
|---|---|---|---|---|---|---|
| IR | DFT | IR | DFT | |||
| νasym | νsym | νasym | νsym | |||
| [UO2(L1)2]·2H2O | 897.76 | 871.64 | 930.81 | 851.02 | 1.74 Å | 1.79 Å |
| [UO2(L2)OAc]·2H2O | 864.99 | 839.81 | 940.17 | 863.68 | 1.75 Å | 1.79 Å |
| [UO2(L3)OAc]·2H2O | 844.97 | 822.89 | 934.45 | 855.20 | 1.76 Å | 1.79 Å |
| Compounds | M.wt | Color | Yield % | M.p °C | Conductivity | Elemental Analysis | |||
|---|---|---|---|---|---|---|---|---|---|
| Found% (Calc.%) | |||||||||
| C | H | N | O | ||||||
| L1 | 278.11 | Orange | 82.91 | 149.6–151.4 | 14 | 47.49 (47.51) | 2.85 (2.90) | 15.17 (15.11) | 5.70 (5.75) |
| L2 | 244.21 | Orange | 93.12 | 210–212.7 | 19 | 53.89 (54.10) | 3.20 (3.30) | 22.90 (22.94) | 19.69 (19.65) |
| L3 | 199.21 | Orange | 47 | 95–100.8 | 11 | 66.05 (66.32) | 4.43 (4.55) | 21.14 (21.09) | 8.37 (8.03) |
| [UO2(L1)2]·2H2O | 824.23 | brown | 83.29 | 122–320 | 43.74 | 31.77 (32.06) | 1.69 (1.71) | 10.35 (10.20) | 7.80 (7.76) |
| [UO2(L2)OAc]·2H2O | 572.27 | yellow | 88 | 291–321 | 54.81 | 25.59 (25.67) | 2.31 (2.32) | 9.14 (9.21) | 23.88 (23.67) |
| [UO2(L3)OAc]·2H2O | 527.28 | Light orange | 69.6 | 204–390 | 26.56 | 27.65 (27.67) | 2.77 (2.86) | 7.69 (7.45) | 19.53 (19.85) |
| Compounds | TG Range (°C) | Mass Loss% Calc. (Found) | Assignment | Metallic Residue |
|---|---|---|---|---|
| L1 | 54.53–248.32248.32–680.62 | 73.71 (73.09)16.6 (16.92) | Loss of (HBr), (2NO), (NH3), 2(C2H2) and (CO)Loss of (CO) | 5C |
| Compounds | Method | Stages | An (s−1) | ΔG (kJ mol−1) | ΔH (kJ mol−1) | ΔS (Jmol−1) | E (kJ mol−1) | R2 |
|---|---|---|---|---|---|---|---|---|
| L1 | CR | 1st | 5.97 × 103 | 1.77 × 105 | 9.22 × 104 | −1.77 × 102 | 9.62 × 104 | 0.99940 |
| HM | 1.23 × 10−2 | 1.44 × 105 | 6.45 × 103 | −2.86 × 102 | 1.05 × 105 | 0.99952 | ||
| Average | 2.99 × 103 | 1.61 × 105 | 4.93 × 104 | −2.31 × 102 | 5.33 × 104 | 0.99946 | ||
| CR | 2nd | 4.57 × 101 | 2.30 × 105 | 5.95 × 104 | −2.21 × 102 | 6.59 × 104 | 0.98963 | |
| HM | 6.16 × 102 | 2.26 × 105 | 7.27 × 104 | −1.99 × 102 | 7.91 × 104 | 0.99247 | ||
| Average | 3.31 × 102 | 2.28 × 105 | 6.61 × 104 | −2.10 × 102 | 7.25 × 104 | 0.99105 |
| Compounds | Steps | Coast–Redfern Method | Horowitz–Metzger Method |
|---|---|---|---|
| L1 | 1st |  |  |
| 2nd |  |  |
| Quantum Chemical Parameters | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compounds | HOMO | LUMO | ΔE | χ | η | i | σ | S | ω | ΔNmax |
| L1 | −6.25 | −2.11 | 4.14 | 4.18 | 2.07 | −4.18 | 0.48 | 0.24 | 4.22 | 2.02 |
| L2 | −6.59 | −2.25 | 4.34 | 4.42 | 2.17 | −4.42 | 0.46 | 0.23 | 4.50 | 2.04 |
| L3 | −6.24 | −1.97 | 4.27 | 4.11 | 2.14 | −4.11 | 0.47 | 0.23 | 3.95 | 1.92 |
| [UO2(L1)2]·2H2O | −6.10 | −3.18 | 2.92 | 4.64 | 1.46 | −4.64 | 0.68 | 0.34 | 7.37 | 3.18 |
| [UO2(L2)OAc]·2H2O | −7.02 | −3.74 | 3.28 | 5.38 | 1.64 | −5.38 | 0.61 | 0.30 | 8.82 | 3.28 |
| [UO2(L3)OAc]·2H2O | −6.30 | −3.23 | 3.07 | 4.77 | 1.54 | −4.77 | 0.47 | 0.33 | 7.40 | 3.10 |
| Compounds | Kb (M−1) | Λmax Free (nm) | λmax Bound (nm) | Type of Chromism |
|---|---|---|---|---|
| L1 | 1 × 106 | 286 | 279 blue-shift | Hyperchromic |
| L2 | 6.67 × 105 | 358 | 353 blue-shift | Hyperchromic |
| L3 | 8 × 105 | 321 | 315 blue-shift | Hyperchromic |
| [UO2(L1)2]·2H2O | 3.33 × 105 | 317 | 312 blue-shift | Hyperchromic |
| [UO2(L2)OAc]·2H2O | 4 × 105 | 376 | 361 blue-shift | Hyperchromic |
| [UO2(L3)OAc]·2H2O | 5 × 105 | 327 | 322 blue-shift | Hyperchromic |
| Types of Protein | Compounds | S | Rmsd_Refine | E_conf | E_place | E_refine | E_score2 |
|---|---|---|---|---|---|---|---|
| Ovarian cancer 3W2S protein | L1 | −5.66 | 1.22 | −6.44 | −63.70 | −10.96 | −21.16 |
| [UO2(L1)2]·2H2O | −6.42 | 2.68 | −3969.01 | −19.27 | −16.38 | 94.20 | |
| L2 | −5.54 | 1.65 | 5.19 | −66.33 | −11.20 | −26.01 | |
| [UO2(L2)OAc]·2H2O | −6.73 | 5.30 | −2648.45 | −71.11 | −14.08 | 7.20 | |
| L3 | −5.40 | 0.98 | 53.32 | −65.04 | −9.60 | −20.48 | |
| [UO2(L3)OAc]·2H2O | −6.19 | 1.51 | −1446.87 | −41.42 | −13.54 | −19.76 | |
| Melanoma cancer 2OPZ protein | L1 | −6.03 | 1.43 | −15.88 | −48.82 | −9.86 | −21.53 |
| [UO2(L1)2]·2H2O | −6.68 | 2.53 | −4015.41 | −64.82 | −12.28 | −8.31 | |
| L2 | −6.14 | 3.84 | 6.52 | −44.09 | −9.28 | −26.42 | |
| [UO2(L2)OAc]·2H2O | −6.77 | 2.96 | −2676.53 | −26.45 | −8.46 | −4.73 | |
| L3 | −5.90 | 1.72 | 47.12 | −60.04 | −9.39 | −18.68 | |
| [UO2(L3)OAc]·2H2O | −6.41 | 6.21 | −1566.02 | −34.58 | −9.59 | −39.59 |
| Types of Protein | Comp. | Ligand | Receptor | Interaction | Distance | E (kcal/mol) |
|---|---|---|---|---|---|---|
| Melanoma | L1 | C 3 | 5-ring TRP 323 (A) | π-H | 4.24 | −0.8 |
| C 3 | 6-ring TRP 323 (A) | π-H | 4.17 | −0.8 | ||
| 6-ring | CA LEU 307 (A) | π-H | 4.80 | −0.5 | ||
| [UO2(L1)2]·2H2O | O 2 | OG1 THR 308 (A) | H-acceptor | 3.16 | −4.8 | |
| 6-ring | N THR 308 (A) | π-H | 4.20 | −1.7 | ||
| 6-ring | CB THR 308 (A) | π-H | 3.64 | −0.6 | ||
| L2 | O 24 | N THR 308 (A) | H-acceptor | 3.21 | −1.7 | |
| 3-ring | CD LYS 297 (A) | π-H | 3.81 | −0.7 | ||
| 6-ring | NZ LYS 297 (A) | π-cation | 4.37 | −3.0 | ||
| [UO2(L2)OAc]·2H2O | C 9 | OE2 GLU 314 (A) | H-donor | 3.37 | −1.4 | |
| L3 | No measurable interactions | |||||
| [UO2(L3)OAc]·2H2O | O 3 | NZ LYS 322 (A) | H-acceptor | 2.66 | −7.9 | |
| O 7 | NZ LYS 322 (A) | H-acceptor | 2.88 | −3.4 | ||
| U 2 | NZ LYS 322 (A) | Ionic | 2.96 | −4.8 | ||
| O 3 | NZ LYS 322 (A) | Ionic | 2.66 | −7.2 | ||
| Ovarian | L1 | 6-ring | CB ASP 837 (A) | π-H | 3.75 | −0.5 |
| [UO2(L1)2]·2H2O | C 15 | OD1 ASP 837 (A) | H-donor | 3.22 | −1.2 | |
| O 2 | N GLY 724 (A) | H-acceptor | 2.98 | −26.8 | ||
| L2 | C 11 | NZ LYS 745 (A) | Ionic | 3.21 | −3.2 | |
| 6-ring | NZ LYS 745 (A) | π-cation | 4.01 | −0.6 | ||
| [UO2(L2)OAc]·2H2O | O 3 | CA GLY 796 (A) | H-acceptor | 3.27 | −0.6 | |
| O 5 | CA GLY 719 (A) | H-acceptor | 3.71 | −1.3 | ||
| L3 | 6-ring | CB LYS 745 (A) | π-H | 3.70 | −1.0 | |
| [UO2(L3)OAc]·2H2O | C 20 | OD1 ASP 837 (A) | H-donor | 3.28 | −0.7 | |
| N 19 | NE ARG 841 (A) | H-acceptor | 3.58 | −0.7 | ||
| U 2 | NZ LYS 745 (A) | Ionic | 3.06 | −4.1 | ||
| O 3 | NZ LYS 745 (A) | Ionic | 2.57 | −8.1 | ||
| Melanoma 2OPZ Result | Ovarian Cancer 3W2S Result | ||
|---|---|---|---|
| L1 | 2D |  |  |
| 3D |  |  | |
| [UO2(L1)2]·2H2O | 2D |  |  |
| 3D |  |  | |
| L2 | 2D |  |  |
| 3D |  |  | |
| [UO2(L2)OAc]·2H2O | 2D |  |  |
| 3D |  |  | |
| L3 | 2D |  |  |
| 3D |  |  | |
| [UO2(L3)OAc]·2H2O | 2D |  |  |
| 3D |  |  | |
 | |||
| Compounds | Ovar3 (Ovarian) | M14 (Melanoma) | ||
|---|---|---|---|---|
| IC50 μg/ml | SD | IC50 μg/ml | SD | |
| L1 | 7.10 | 0.05 | 6.49 | 0.08 |
| L2 | 7.01 | 0.05 | 6.35 | 0.08 |
| L3 | 7.51 | 0.05 | 6.71 | 0.08 |
| [UO2(L1)2]·2H2O | 6.33 | 0.03 | 5.17 | 0.04 |
| [UO2(L2)OAc]·2H2O | 6.19 | 0.03 | 5.01 | 0.04 |
| [UO2(L3)OAc]·2H2O | 6.16 | 0.02 | 5.33 | 0.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Howsaui, H.B.; Basaleh, A.S.; Abdellattif, M.H.; Hassan, W.M.I.; Hussien, M.A. Synthesis, Structural Investigations, Molecular Docking, and Anticancer Activity of Some Novel Schiff Bases and Their Uranyl Complexes. Biomolecules 2021, 11, 1138. https://doi.org/10.3390/biom11081138
Howsaui HB, Basaleh AS, Abdellattif MH, Hassan WMI, Hussien MA. Synthesis, Structural Investigations, Molecular Docking, and Anticancer Activity of Some Novel Schiff Bases and Their Uranyl Complexes. Biomolecules. 2021; 11(8):1138. https://doi.org/10.3390/biom11081138
Chicago/Turabian StyleHowsaui, Hanan B., Amal S. Basaleh, Magda H. Abdellattif, Walid M. I. Hassan, and Mostafa A. Hussien. 2021. "Synthesis, Structural Investigations, Molecular Docking, and Anticancer Activity of Some Novel Schiff Bases and Their Uranyl Complexes" Biomolecules 11, no. 8: 1138. https://doi.org/10.3390/biom11081138
APA StyleHowsaui, H. B., Basaleh, A. S., Abdellattif, M. H., Hassan, W. M. I., & Hussien, M. A. (2021). Synthesis, Structural Investigations, Molecular Docking, and Anticancer Activity of Some Novel Schiff Bases and Their Uranyl Complexes. Biomolecules, 11(8), 1138. https://doi.org/10.3390/biom11081138