Identification of Biomolecules Involved in the Adaptation to the Environment of Cold-Loving Microorganisms and Metabolic Pathways for Their Production
Abstract
:1. Introduction
2. Biomolecules That Are Protective against the Cold
2.1. Antifreeze Molecules
2.2. Pigments
2.3. Proteins
2.3.1. Enzymes
2.3.2. Other Proteins
2.4. Lipids
3. Metabolic Pathways to Produce Biomolecules
3.1. Production of Antimicrobial Compounds
3.1.1. Genome Mining
3.1.2. CRISPR-Cas
3.2. Production of Pigments
3.2.1. Carotenoids
3.2.2. Melanins
3.3. Production of Hopanoids
4. Cultures in Different Media and Environmental Conditions
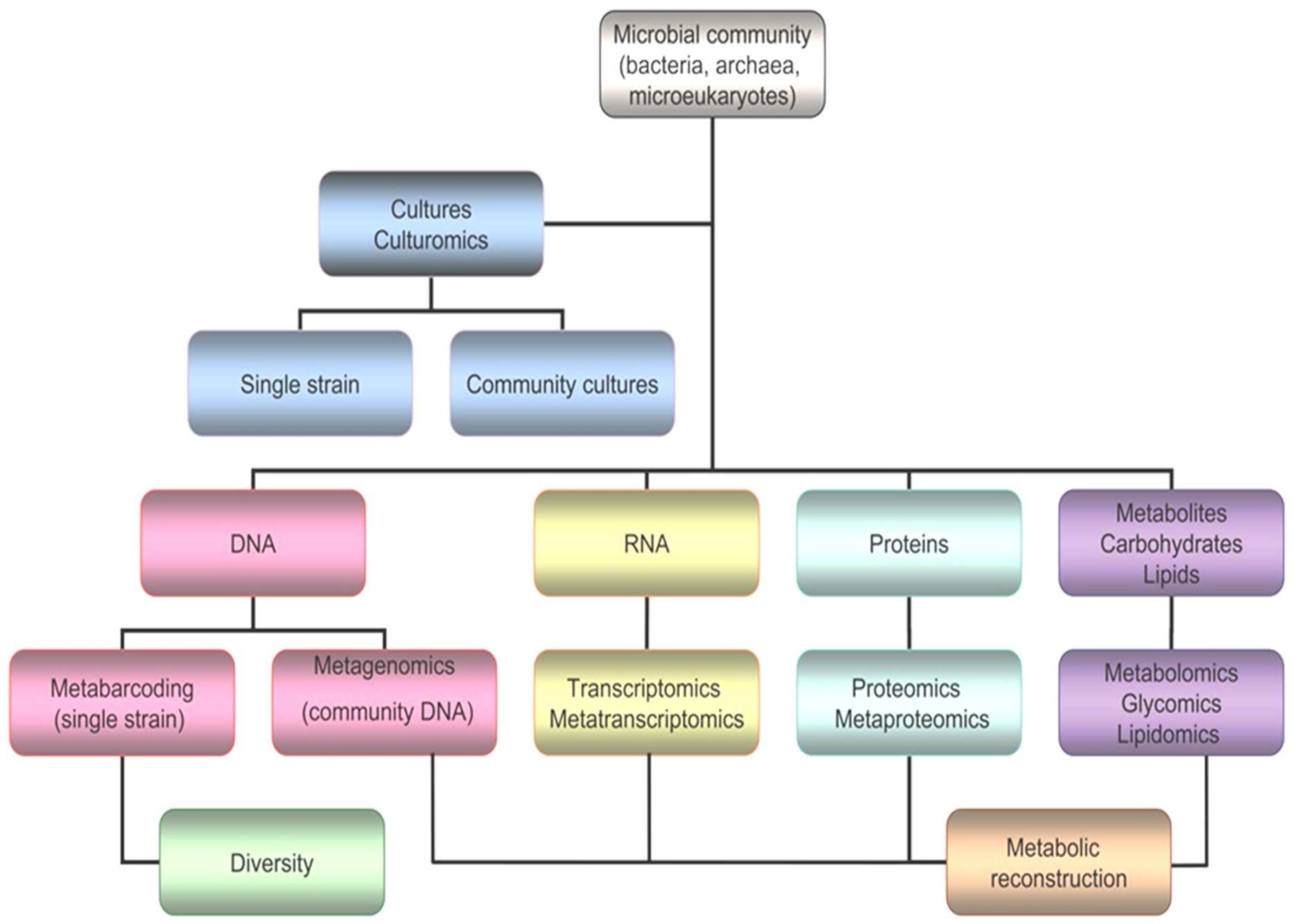
5. Omics in Cold-Loving Microorganisms
5.1. Genomics
5.1.1. Metabarcoding
5.1.2. Metagenomics
5.2. Transcriptomics and Metatranscriptomics
5.3. Proteomics and Metaproteomics
5.4. Glycomics and Lipidomics
5.5. Metabolomics
6. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boetius, A.; Anesio, A.M.; Deming, J.W.; Mikucki, J.A.; Rapp, J. Microbial ecology of the cryosphere: Sea ice and glacial habitats. Nat. Rev. Genet. 2015, 13, 677–690. [Google Scholar] [CrossRef] [PubMed]
- Craig, L.; Moyer, R.; Collins, R.; Morita, R.Y. Psychrophiles and Psychrotrophs. Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar] [CrossRef]
- Priscu, J.C.; Christner, B.C. Earth’s icy biosphere. In Microbial Diversity and Bioprospecting; Bull, A., Ed.; ASM Press: Washington, DC, USA, 2004; pp. 130–145. [Google Scholar]
- Alcazar, A.; Garcia-Descalzo, L.; Cid, C. Microbial evolution and adaptation in icy worlds. Astrobiology: Physical Origin, Biological Evolution and Spatial Distribution; Springer: New York, NY, USA, 2010; pp. 81–95. [Google Scholar]
- Irwin, J.A.; Baird, A.W. Extremophiles and their application to veterinary medicine. Ir. Veter. J. 2004, 57, 348–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margesin, R. (Ed.) Psychrophiles: From Biodiversity to Biotechnology; Springer: Cham, Germany, 2017. [Google Scholar] [CrossRef]
- Singh, O.V. (Ed.) Bio-Pigmentation and Biotechnological Implementations; Wiley-Blackwell: Hoboken, NJ, USA, 2016. [Google Scholar]
- Singh, O.V. Extremophiles: Sustainable Resources and Biotechnological Implications; Wiley-Blackwell: Hoboken, NJ, USA, 2012; pp. 159–174. [Google Scholar]
- Decho, A.W. Microbial exopolymer secretions in ocean environments: Their role(s) in food webs and marine processes. In Oceanography and Marine Biology: An Annual Review; Aberdeen Univ Press: Aberdeen, UK, 1990; pp. 73–153. [Google Scholar]
- Cócera, M.; López, O.; Sabés, M.; Parra, J.L.; Guinea, J.; De La Maza, A. Assembly properties and applications of a new exopolymeric compound excreted by Pseudoalteromonas antarctica NF3. J. Biomater. Sci. Polym. Ed. 2001, 12, 409–427. [Google Scholar] [CrossRef]
- Nichols, C.M.; Garon, S.; Bowman, J.; Raguenes, G.; Guezennec, J. Production of exopolysaccharides by Antarctic marine bacterial isolates. J. Appl. Microbiol. 2004, 96, 1057–1066. [Google Scholar] [CrossRef] [Green Version]
- Qin, G.; Zhu, L.; Chen, X.; Wang, P.G.; Zhang, Y. Structural characterization and ecological roles of a novel exopolysaccharide from the deep-sea psychrotolerant bacterium Pseudoalteromonas sp. SM9913. Microbiology 2007, 153, 1566–1572. [Google Scholar] [CrossRef] [Green Version]
- Li, W.W.; Zhou, W.Z.; Zhang, Y.Z.; Wang, J.; Zhu, X.B. Flocculation behavior and mechanism of an exopolysaccharide from the deep-sea psychrophilic bacterium Pseudoalteromonas sp. SM9913. Bioresour. Technol. 2008, 99, 6893–6899. [Google Scholar] [CrossRef]
- Das, D.; Hervé, M.; Feuerhelm, J.; Farr, C.L.; Chiu, H.-J.; Elsliger, M.-A.; Knuth, M.W.; Klock, H.E.; Miller, M.D.; Godzik, A.; et al. Structure and Function of the First Full-Length Murein Peptide Ligase (Mpl) Cell Wall Recycling Protein. PLoS ONE 2011, 6, e17624. [Google Scholar] [CrossRef] [Green Version]
- García-Descalzo, L.; Alcazar, A.; Baquero, F.; Cid, C. Biotechnological Applications of Cold-Adapted Bacteria. Extremophiles: Sustainable Resources and Biotechnological Implications; Wiley-Blackwell: Hoboken, NJ, USA, 2012; pp. 159–174. [Google Scholar] [CrossRef]
- Casillo, A.; Parrilli, E.; Sannino, F.; Mitchell, D.E.; Gibson, M.I.; Marino, G.; Lanzetta, R.; Parrilli, M.; Cosconati, S.; Novellino, E.; et al. Structure-activity relationship of the exopolysaccharide from a psychrophilic bacterium: A strategy for cryoprotection. Carbohydr. Polym. 2017, 156, 364–371. [Google Scholar] [CrossRef] [Green Version]
- Marx, J.G.; Carpenter, S.D.; Deming, J.W. Production of cryoprotectant extracellular polysaccharide substances (EPS) by the marine psychrophilic bacteriumColwellia psychrerythraeastrain 34H under extreme conditionsThis article is one of a selection of papers in the Special Issue on Polar and Alpine Microbiology. Can. J. Microbiol. 2009, 55, 63–72. [Google Scholar] [CrossRef]
- Jagannadham, M.V.; Chattopadhyay, M.K.; Subbalakshmi, C.; Vairamani, M.; Narayanan, K.; Rao, C.M.; Shivaji, S. Carotenoids of an Antarctic psychrotolerant bacterium, Sphingobacterium antarcticus, and a mesophilic bacterium, Sphingobacterium multivorum. Arch. Microbiol. 2000, 173, 418–424. [Google Scholar] [CrossRef]
- Vollmers, J.; Voget, S.; Dietrich, S.; Gollnow, K.; Smits, M.; Meyer, K.; Brinkhoff, T.; Simon, M.; Daniel, R. Poles Apart: Arctic and Antarctic Octadecabacter strains Share High Genome Plasticity and a New Type of Xanthorhodopsin. PLoS ONE 2013, 8, e63422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Descalzo, L.; Garcia-Lopez, E.; Alcazar, A.; Baquero, F.; Cid, C. Proteomic analysis of the adaptation to warming in the Antarctic bacteria Shewanella frigidimarina. BBA Proteins Proteom. 2014, 1844, 2229–2240. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Sonjak, S.; Zalar, P.; Frisvad, J.; Diderichsen, B.; Plemenitaš, A. Extremophilic fungi in arctic ice: A relationship between adaptation to low temperature and water activity. Phys. Chem. Earth Parts A/B/C 2003, 28, 1273–1278. [Google Scholar] [CrossRef]
- Weber, M.H.W.; Marahiel, M.A. Bacterial Cold Shock Responses. Sci. Prog. 2003, 86, 9–75. [Google Scholar] [CrossRef]
- Muñoz, P.; Márquez, S.L.; González-Nilo, F.D.; Márquez-Miranda, V.; Blamey, J.M. Structure and application of antifreeze proteins from Antarctic bacteria. Microb. Cell Factories 2017, 16, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Gwak, I.G.; Jung, W.S.; Kim, H.J.; Kang, S.-H.; Jin, E. Antifreeze Protein in Antarctic Marine Diatom, Chaetoceros neogracile. Mar. Biotechnol. 2009, 12, 630–639. [Google Scholar] [CrossRef]
- Raymond, J.A.; Fritsen, C.; Shen, K. An ice-binding protein from an Antarctic sea ice bacterium. FEMS Microbiol. Ecol. 2007, 61, 214–221. [Google Scholar] [CrossRef] [Green Version]
- Bayer-Giraldi, M.; Weikusat, I.; Besir, H.; Dieckmann, G. Characterization of an antifreeze protein from the polar diatom Fragilariopsis cylindrus and its relevance in sea ice. Cryobiology 2011, 63, 210–219. [Google Scholar] [CrossRef]
- Raymond, J.A. The ice-binding proteins of a snow alga, Chloromonas brevispina: Probable acquisition by horizontal gene transfer. Extremophiles 2014, 18, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Lonhienne, T.; Zoidakis, J.; Vorgias, C.E.; Feller, G.; Gerday, C.; Bouriotis, V. Modular structure, local flexibility and cold-activity of a novel chitobiase from a psychrophilic antarctic bacterium. J. Mol. Biol. 2001, 310, 291–297. [Google Scholar] [CrossRef]
- Wada, H.; Gombos, Z.; Murata, N. Contribution of membrane lipids to the ability of the photosynthetic machinery to tolerate temperature stress. Proc. Natl. Acad. Sci. USA 1994, 91, 4273–4277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, N.J. Psychrophilic bacteria—Molecular adaptations of membrane lipids. Comp. Biochem. Physiol. Part A Physiol. 1997, 118, 489–493. [Google Scholar] [CrossRef]
- Nichols, C.M.; Guezennec, J.; Bowman, J. Bacterial Exopolysaccharides from Extreme Marine Environments with Special Consideration of the Southern Ocean, Sea Ice, and Deep-Sea Hydrothermal Vents: A Review. Mar. Biotechnol. 2005, 7, 253–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cocera, M.; Lopez, O.; Parra, J.; Mercade, E.; Guinea, J.; de la Maza, A. Protective effect caused by the exopolymer excreted by Pseudoalteromonas antarctica NF3 on liposomes against the action of octyl glucoside. Int. J. Pharm. 2000, 207, 39–47. [Google Scholar] [CrossRef]
- Meiners, K.; Krembs, C.; Gradinger, R. Exopolymer particles: Microbial hotspots of enhanced bacterial activity in Arctic fast ice (Chukchi Sea). Aquat. Microb. Ecol. 2008, 52, 195–207. [Google Scholar] [CrossRef]
- Madigan, M.T.; Martinko, J.M.; Stahl, D.A.; Clark, D.P. Brock Biology of Microorganisms, 13th ed.; Pearson Education, Inc.: San Francisco, CA, USA, 2012. [Google Scholar]
- Siefirmann-Harms, D. The light-harvesting and protective functions of carotenoids in photosynthetic membranes. Physiol. Plant 1987, 69, 501–568. [Google Scholar] [CrossRef]
- Soliev, A.B.; Hosokawa, K.; Enomoto, K. Bioactive Pigments from Marine Bacteria: Applications and Physiological Roles. Evid. Based Complement. Altern. Med. 2011, 2011, 1–17. [Google Scholar] [CrossRef]
- Martin-Cerezo, M.L.; Garcia-Lopez, E.; Cid, C. Isolation and identification of a red pigment from the Antarctic bacterium Shewanella frigidimarina. Protein. Pept. Lett. 2015, 22, 1076–1082. [Google Scholar] [CrossRef]
- Becker-Hapak, M.; Troxtel, E.; Hoerter, J.; Eisenstark, A. RpoS dependent overexpression of carotenoids from Erwinia herbico-la in OXYR-deficient Escherichia coli. Biochem. Biophys. Res. Commun. 1997, 239, 305–309. [Google Scholar] [CrossRef]
- Goodwin, T.W. The Biochemistry of Carotenoids; Chapman and Hall: London, UK, 1980; Volume 1. [Google Scholar]
- García-López, E.; Alcazar, A.; Moreno, A.M.; Cid, C. Color producing extremophiles. Bio-Pigmentation and Biotechnological Implementations; Wiley-Blackwell: Hoboken, NJ, USA, 2016. [Google Scholar]
- Maccario, L.; Sanguino, L.; Vogel, T.M.; Larose, C. Snow and ice ecosystems: Not so extreme. Res. Microbiol. 2015, 166, 782–795. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S.; Cavicchioli, R. Cold-Adapted Enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Amico, S.; Collins, T.; Marx, J.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Hochachka, P.W.; Somero, G.N. Biochemical adaptation. Mechanism and Process in Physical Evolution; Oxford University Press: New York, NY, USA, 2002. [Google Scholar]
- Garcia-Lopez, E.; Rodriguez-Lorente, I.; Alcazar, P.; Cid, C. Microbial Communities in Coastal Glaciers and Tidewater Tongues of Svalbard Archipelago, Norway. Front. Mar. Sci. 2019, 5, 512. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Siddiqui, K.S.; Andrews, D.; Sowers, K.R. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 2002, 13, 253–261. [Google Scholar] [CrossRef]
- Goldstein, J.; Pollitt, N.S.; Inouye, M. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 283–287. [Google Scholar] [CrossRef] [Green Version]
- Leeson, D.T.; Gai, F.; Rodriguez, H.M.; Gregoret, L.M.; Dyer, R.B. Protein folding and unfolding on a complex energy land-scape. Proc. Natl. Acad. Sci. USA 2000, 97, 2527–2532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayer-Giraldi, M.; Uhlig, C.; John, U.; Mock, T.; Valentin, K. Antifreeze proteins in polar sea ice diatoms: Diversity and gene expression in the genus Fragilariopsis. Environ. Microbiol. 2010, 12, 1041–1052. [Google Scholar] [CrossRef]
- Dolhi, J.M.; Maxwell, D.P.; Morgan-Kiss, R.M. Review: The Antarctic Chlamydomonas raudensis: An emerging model for cold adaptation of photosynthesis. Extremophiles 2013, 17, 711–722. [Google Scholar] [CrossRef]
- Feller, G. Life at low temperatures: Is disorder the driving force? Extremophiles 2006, 11, 211–216. [Google Scholar] [CrossRef]
- Rossi, M.; Ciaramella, M.; Cannio, R.; Pisani, F.M.; Moracci, M.; Bartolucci, S. Extremophiles 2002. J. Bacteriol. 2003, 185, 3683–3689. [Google Scholar] [CrossRef] [Green Version]
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic discovery: History, methods and perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382. [Google Scholar] [CrossRef]
- Núñez-Montero, K.; Quezada-Solís, D.; Khalil, Z.G.; Capon, R.J.; Andreote, F.D.; Barrientos, L. Genomic and Metabolomic Analysis of Antarctic Bacteria Revealed Culture and Elicitation Conditions for the Production of Antimicrobial Compounds. Biomolecules 2020, 10, 673. [Google Scholar] [CrossRef] [PubMed]
- Pawluk, A.; Davidson, A.R.; Maxwell, K. Anti-CRISPR: Discovery, mechanism and function. Nat. Rev. Genet. 2017, 16, 12–17. [Google Scholar] [CrossRef]
- Pavan, M.E.; López, N.I.; Pettinari, M.J. Melanin biosynthesis in bacteria, regulation and production perspectives. Appl. Microbiol. Biotechnol. 2019, 104, 1357–1370. [Google Scholar] [CrossRef]
- Toledo, A.V.; Franco, M.E.E.; López, S.M.Y.; Troncozo, M.I.; Saparrat, M.C.N.; Balatti, P.A. Melanins in fungi: Types, locali-zation and putative biological roles. Physiol. Mol. Plant. Path. 2017, 99, 2–6. [Google Scholar] [CrossRef]
- Ghimire, G.P.; Koirala, N.; Sohng, J.K. Activation of Cryptic hop Genes from Streptomyces peucetius ATCC 27952 Involved in Hopanoid Biosynthesis. J. Microbiol. Biotechnol. 2015, 25, 658–661. [Google Scholar] [CrossRef]
- Teske, A. Grand Challenges in Extreme Microbiology. Front. Microbiol. 2010, 1, 111. [Google Scholar] [CrossRef] [Green Version]
- Christner, B.C.; Mosley-Thompson, E.; Thompson, L.G.; Reeve, J.N. Bacterial recovery from ancient glacial ice. Environ. Microbiol. 2003, 5, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Pulschen, A.A.; Bendia, A.; Fricker, A.D.; Pellizari, V.; Galante, D.; Rodrigues, F. Isolation of Uncultured Bacteria from Antarctica Using Long Incubation Periods and Low Nutritional Media. Front. Microbiol. 2017, 8, 1346. [Google Scholar] [CrossRef] [PubMed]
- Mourembou, G.; Yasir, M.; Azhar, E.; Lagier, J.C.; Bibi, F.; Jiman-Fatani, A.A.; Helmy, N.; Robert, C.; Rathored, J.; Fournier, P.-E.; et al. Rise of Microbial Culturomics: Noncontiguous Finished Genome Sequence and Description of Beduini massiliensisgen. nov., sp. nov. OMICS A J. Integr. Biol. 2015, 19, 766–776. [Google Scholar] [CrossRef] [Green Version]
- Lagier, J.-C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Seck, E.H.; et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016, 1, 1–8. [Google Scholar] [CrossRef]
- Martinez-Alonso, E.; Pena-Perez, S.; Serrano, S.; Garcia-Lopez, E.; Alcazar, A.; Cid, C. Taxonomic and functional characterization of a microbial community from a volcanic englacial ecosystem in Deception Island, Antarctica. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Bidle, K.D.; Lee, S.; Marchant, D.R.; Falkowski, P.G. Fossil genes and microbes in the oldest ice on Earth. Proc. Natl. Acad. Sci. USA 2007, 104, 13455–13460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Descalzo, L.; Alcazar, A.; Baquero, F.; Cid, C. Identification of in vivo HSP90-interacting proteins reveals modularity of HSP90 complexes is dependent on the environment in psychrophilic bacteria. Cell Stress Chaperon. 2010, 16, 203–218. [Google Scholar] [CrossRef] [Green Version]
- Maghembe, R.; Damian, D.; Makaranga, A.; Nyandoro, S.S.; Lyantagaye, S.L.; Kusari, S.; Hatti-Kaul, R. Omics for Bioprospecting and Drug Discovery from Bacteria and Microalgae. Antibiotics 2020, 9, 229. [Google Scholar] [CrossRef]
- García-Lopez, E.; Serrano, S.; Calvo, M.; Perez, S.P.; Sanchez-Casanova, S.; García-Descalzo, L.; Cid, C. Microbial Community Structure Driven by a Volcanic Gradient in Glaciers of the Antarctic Archipelago South Shetland. Microorganisms 2021, 9, 392. [Google Scholar] [CrossRef]
- Cao, S.; Zhang, W.; Ding, W.; Wang, M.; Fan, S.; Yang, B.; Mcminn, A.; Wang, M.; Xie, B.-B.; Qin, Q.-L.; et al. Structure and function of the Arctic and Antarctic marine microbiota as revealed by metagenomics. Microbiome 2020, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.-R.; Robeson, M.S.; Hauser, L.J.; Schadt, C.W.; Gorin, A.A. PanFP: Pangenome-based functional profiles for microbial communities. BMC Res. Notes 2015, 8, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwai, S.; Weinmaier, T.; Schmidt, B.L.; Albertson, D.G.; Poloso, N.J.; Dabbagh, K.; DeSantis, T.Z. Piphillin: Improved Prediction of Metagenomic Content by Direct Inference from Human Microbiomes. PLoS ONE 2016, 11, e0166104. [Google Scholar] [CrossRef] [Green Version]
- Wemheuer, F.; Taylor, J.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 2020, 15, 1–12. [Google Scholar] [CrossRef]
- Bowman, J.S.; Ducklow, H.W. Microbial Communities Can Be Described by Metabolic Structure: A General Framework and Application to a Seasonally Variable, Depth-Stratified Microbial Community from the Coastal West Antarctic Peninsula. PLoS ONE 2015, 10, e0135868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raymond-Bouchard, I.; Whyte, L.G. From Transcriptomes to Metatranscriptomes: Cold Adaptation and Active Metabolisms of Psychrophiles from Cold Environments. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Ed.; Springer: Cham, Germany, 2017. [Google Scholar] [CrossRef]
- Heueis, N.; Vockenhuber, M.-P.; Suess, B. Small non-coding RNAs in streptomycetes. RNA Biol. 2014, 11, 464–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adibekian, A.; Stallforth, P.; Hecht, M.-L.; Werz, D.B.; Gagneux, P.; Seeberger, P.H. Comparative bioinformatics analysis of the mammalian and bacterial glycomes. Chem. Sci. 2010, 2, 337–344. [Google Scholar] [CrossRef]
- Garcia-Lopez, E.; Cid, C. Glaciers and Ice Sheets as Analog Environments of Potentially Habitable Icy Worlds. Front. Microbiol. 2017, 8, 1407. [Google Scholar] [CrossRef]
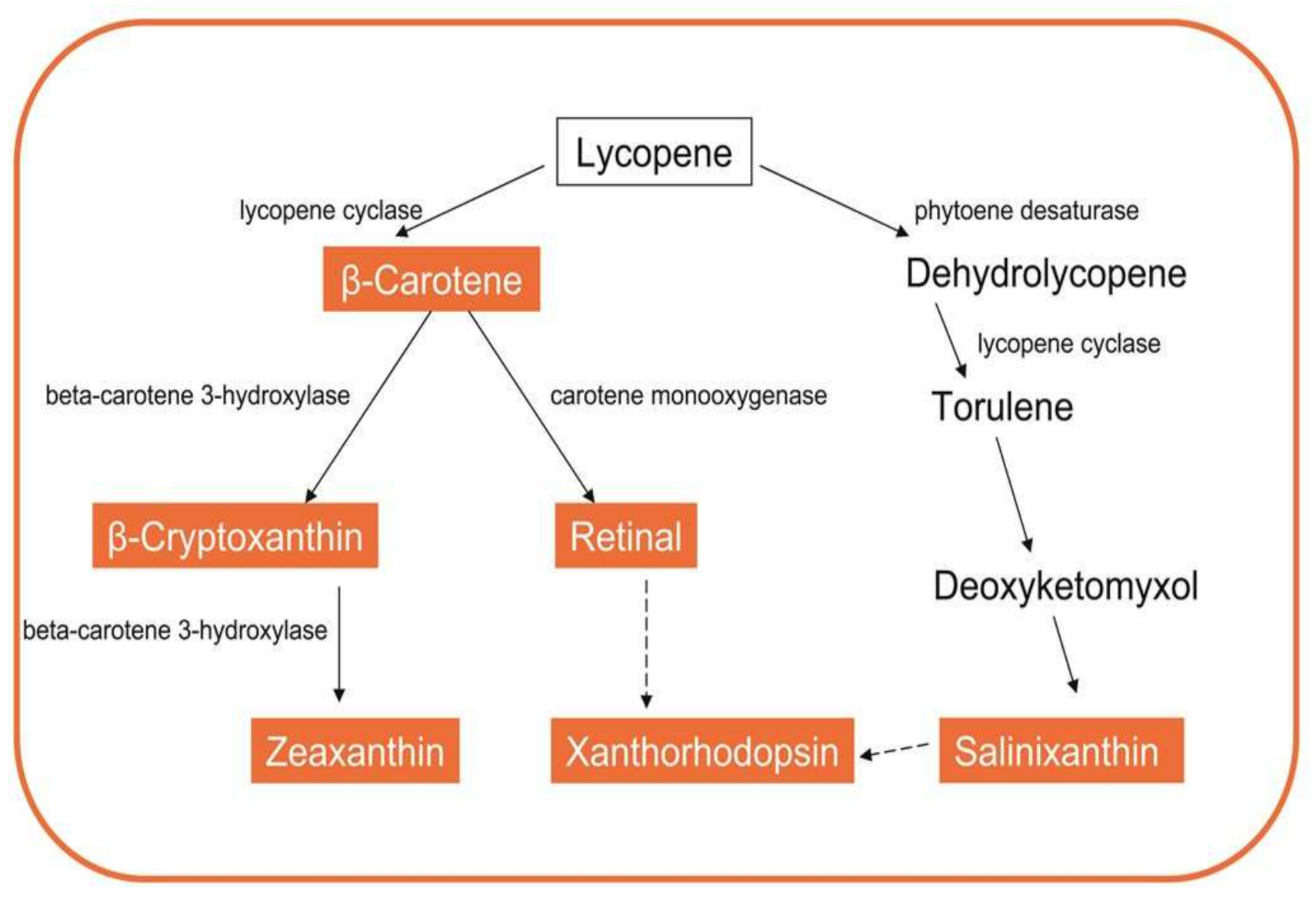
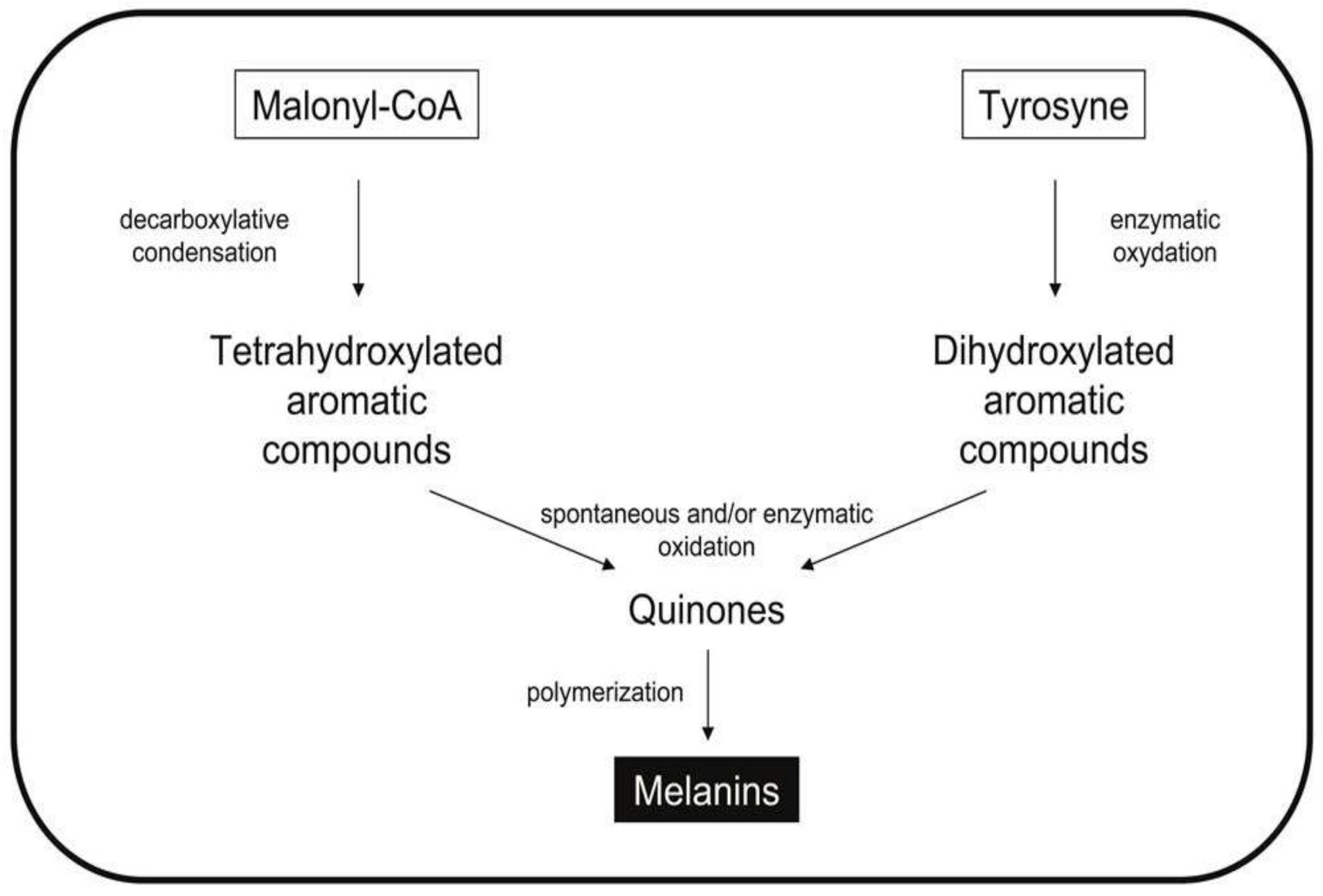
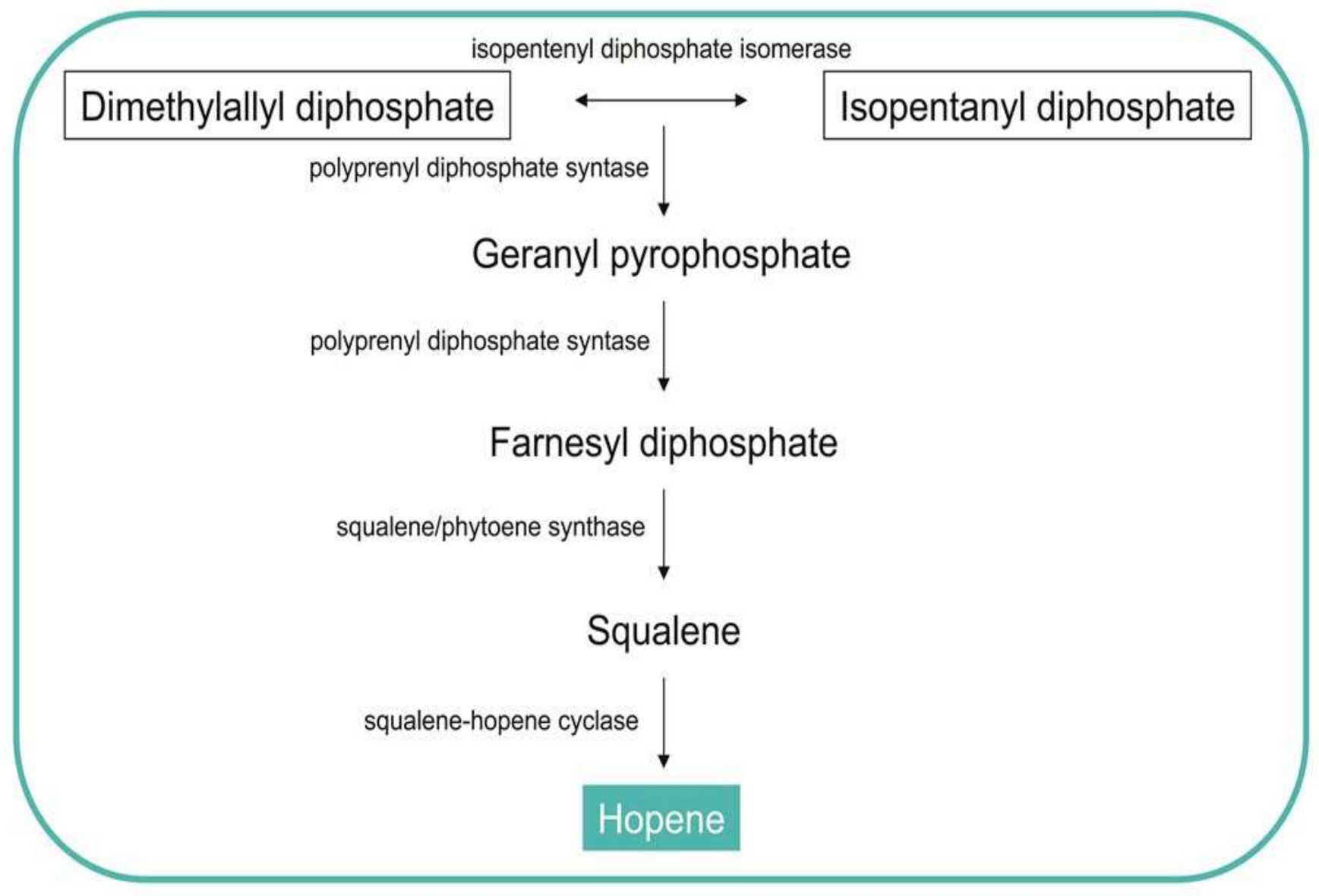
| Microorganism | Biomolecules | References | |
|---|---|---|---|
| Pseudoalteromonas antarctica | EPS | Glycoprotein | [10] |
| Pseudoalteromonas sp. | Sulfated heteropolysaccharide | [11] | |
| Pseudoalteromonas sp. | Linear arrangement of α-(1–6) linkage of glucose with a high degree of acetylation | [12,13] | |
| Psychrobacter arcticus | Peptidoglycan | [14] | |
| Psychrobacter frigidicola | Peptidoglycan | [15] | |
| Colwellia psychrerythraea | N-acetyl quinovosamine and galacturonic acid | [16,17] | |
| Sphingobacterium antarcticus | Pigments | Zeaxanthin, b-cryptoxanthin, b-carotene | [18] |
| Octadecabacter arcticus | Xanthorhodopsin | [19] | |
| Octadecabacter antarcticus | Xanthorhodopsin | [19] | |
| Shewanella frigidimarina | Cytochrome c3 | [20] | |
| Cladosporium (fungi) | Melanin | [21] | |
| Aureobasidium (fungi) | Melanin | [21] | |
| Escherichia coli | Proteins | Csps | [22] |
| Pseudomonas, Arthrobacter | AFP | [23] | |
| Chaetoceros neogracile (diatoms) | AFP | [24] | |
| Chloromonas brevispina (alga) | Ice-binding proteins | [25] | |
| Fragilariopsis cylindrus (diatoms) | Ice-binding proteins | [26] | |
| Colwellia | Ice-binding proteins | [27] | |
| Arthrobacter | Enzymes (chitobiase) | [28] | |
| Synechocystis | Lipids | Diunsaturated fatty acids | [29] |
| Antarctic bacterium, strain JS6 | Polyunsaturated fatty acids | [30] |
| Name | Composition (g/L) | Reference |
|---|---|---|
| M1 | 2 g peptone, 4 g yeast extract, 10 g starch | [54] |
| ISP2 | 4 g yeast extract, 10 g malt extract, 4 g glucose | |
| M2 | 40 g mannitol, 40 g maltose, 10 g yeast extract, 2 g K2HPO4, 0.5 g MgSO4.7H2O, 0.01 g FeSO4.7H2O | |
| SCA | 10 g starch, 0.3 g casein, 2 g KNO3, 2 g NaCl, 2 g K2HPO4, 0.5 g MgSO4.7H2O, 0.02 g CaCO3, 0.01 g FeSO4.7H2O | |
| IMA | 4 g yeast extract, 10 g malt extract, 4 g glucose, mannitol | |
| SDB | p g peptidic digest of animal tissue, 5 g casein pancreatin disgest, 40 g dextrose | |
| CCA | 30 g glicerol, 2 g peptone, 1 g K2HPO4, 1 g NaCl, 0.5 g MgSO4.7H2O, 5 mL trace element solution | |
| YEME | 3 g yeast extract, 5 g peptone, 3 g malt extract, 10 g glucose, 170 g sucrose | |
| GYA | 4 g Yeast extract, 10 g malt extract, 4 g glucose, 2 g CaCO3, 20 g starch | |
| YES | 150 g sucrose, 20 g yeast extract, 0.5 g MgSO4.7H2O, 0.01 g ZnSO4.7H2O, 0.005 g CuSO4.5H2O | |
| ISP4 | 10 g starch, 2 g CaCO3, 2 g (NH4)2SO4, 1 g K2HPO4, 1 g MgSO4.7H2O, 1 g NaCl, 1 mg FeSO4.7H2O, 1 mg MnCl2.7H2O, 1 mg ZnSO4.7H2O | |
| R2YE | 103 g sucrose, 0.25 g K2SO4, 0.12 g MgCl2.6H2O, 10 g Glucose, 0.1 g casaminoacids, 5 mL yeast extract (10%), 1 mL KH2PO4 (0.5%), 8 mL CaCl2.2H2O (3.68%), 1.5 mL L-proline (20%), 10 mL TES buffer (5.73%), 0.2 mL trace element solution, 0.5 mL NaOH (1N) | [58] |
| T2 | 5 g peptone, 0.15 g ferric ammonium citrate, 0.2 g MgSO4.7H2O, 0.05 g CaCl2, 0.05 g MnSO4.H2O, 0.01 g FeCl3.6H2O | [64,65] |
| T3 | 1 g glucose, 1 g peptone, 0.5 g yeast extract, 0.2 g MgSO4.7H2O, 0.05 g MnSO4.4H2O | |
| T4 | 1 g glucose, 0.5 g casamino acids, 0.5 g yeast extract, 1 g KH2PO4, 0.5 g CaCl2.2H2O, 0.5 g MnCl2.4H2O | |
| T6 | R2A | |
| T7 | Marine broth 2216 | [66] |
| T8 | Trypticase soy broth |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Lopez, E.; Alcazar, P.; Cid, C. Identification of Biomolecules Involved in the Adaptation to the Environment of Cold-Loving Microorganisms and Metabolic Pathways for Their Production. Biomolecules 2021, 11, 1155. https://doi.org/10.3390/biom11081155
Garcia-Lopez E, Alcazar P, Cid C. Identification of Biomolecules Involved in the Adaptation to the Environment of Cold-Loving Microorganisms and Metabolic Pathways for Their Production. Biomolecules. 2021; 11(8):1155. https://doi.org/10.3390/biom11081155
Chicago/Turabian StyleGarcia-Lopez, Eva, Paula Alcazar, and Cristina Cid. 2021. "Identification of Biomolecules Involved in the Adaptation to the Environment of Cold-Loving Microorganisms and Metabolic Pathways for Their Production" Biomolecules 11, no. 8: 1155. https://doi.org/10.3390/biom11081155
APA StyleGarcia-Lopez, E., Alcazar, P., & Cid, C. (2021). Identification of Biomolecules Involved in the Adaptation to the Environment of Cold-Loving Microorganisms and Metabolic Pathways for Their Production. Biomolecules, 11(8), 1155. https://doi.org/10.3390/biom11081155






