Comparative Genomic Analysis of the DUF34 Protein Family Suggests Role as a Metal Ion Chaperone or Insertase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Capture of Literature, Structural, and Essentiality Data
2.2. Domain Analysis
2.3. Absence-Presence, Phyletic Patterns & Homolog/Paralog Co-Occurrence
2.4. Physical Clustering Analysis
2.5. Coexpression, Covariation Data Acquisition & Enrichment Analysis
2.6. Fusion Analysis
2.7. Strain Construction & List
2.8. dT Sensitivity Assay
2.9. dT Essentiality Complementation Assay
3. Results and Discussion
3.1. Extensive Literature Capture and Analysis Confirms Pleiotropic Role of DUF34 Family Members
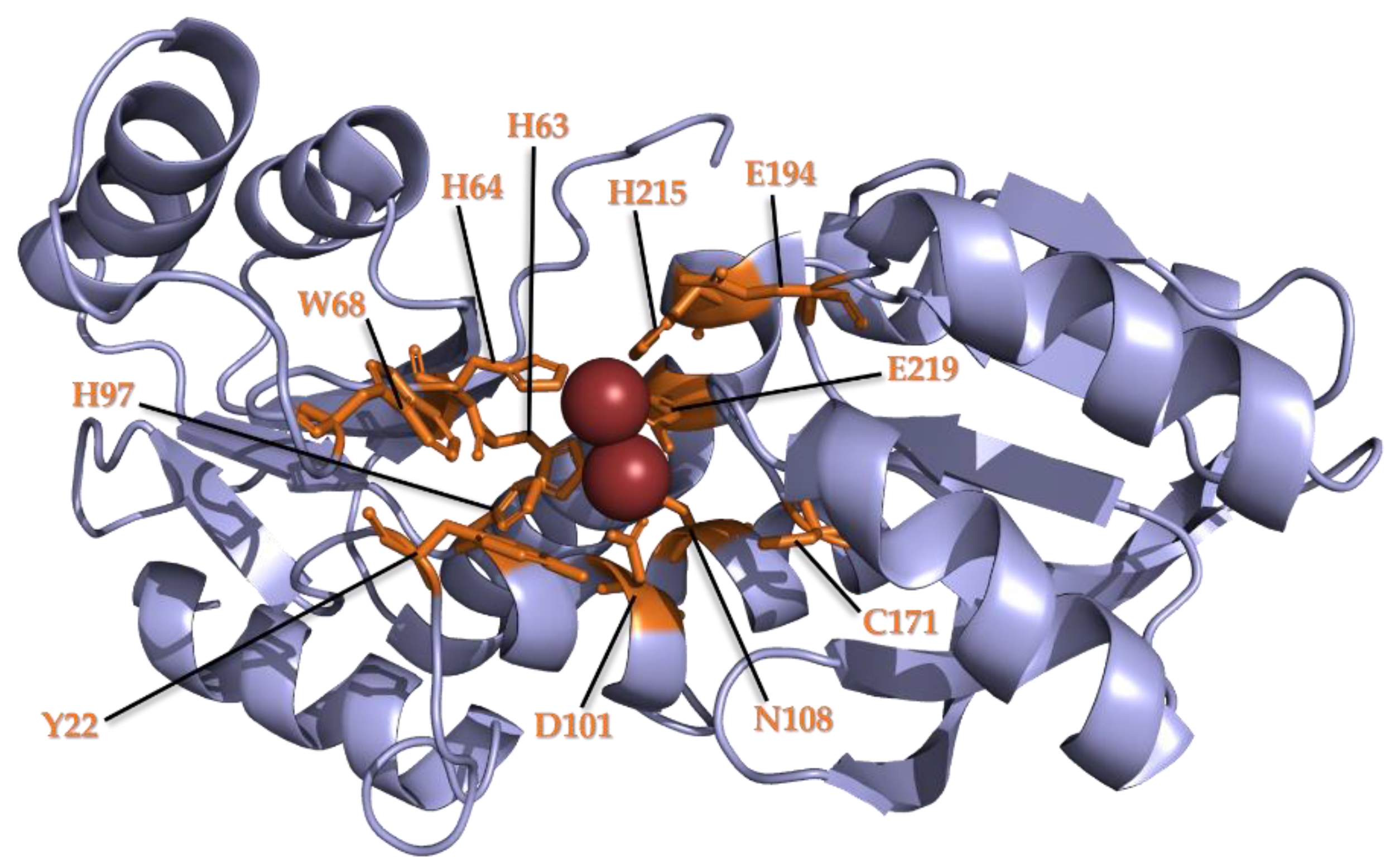
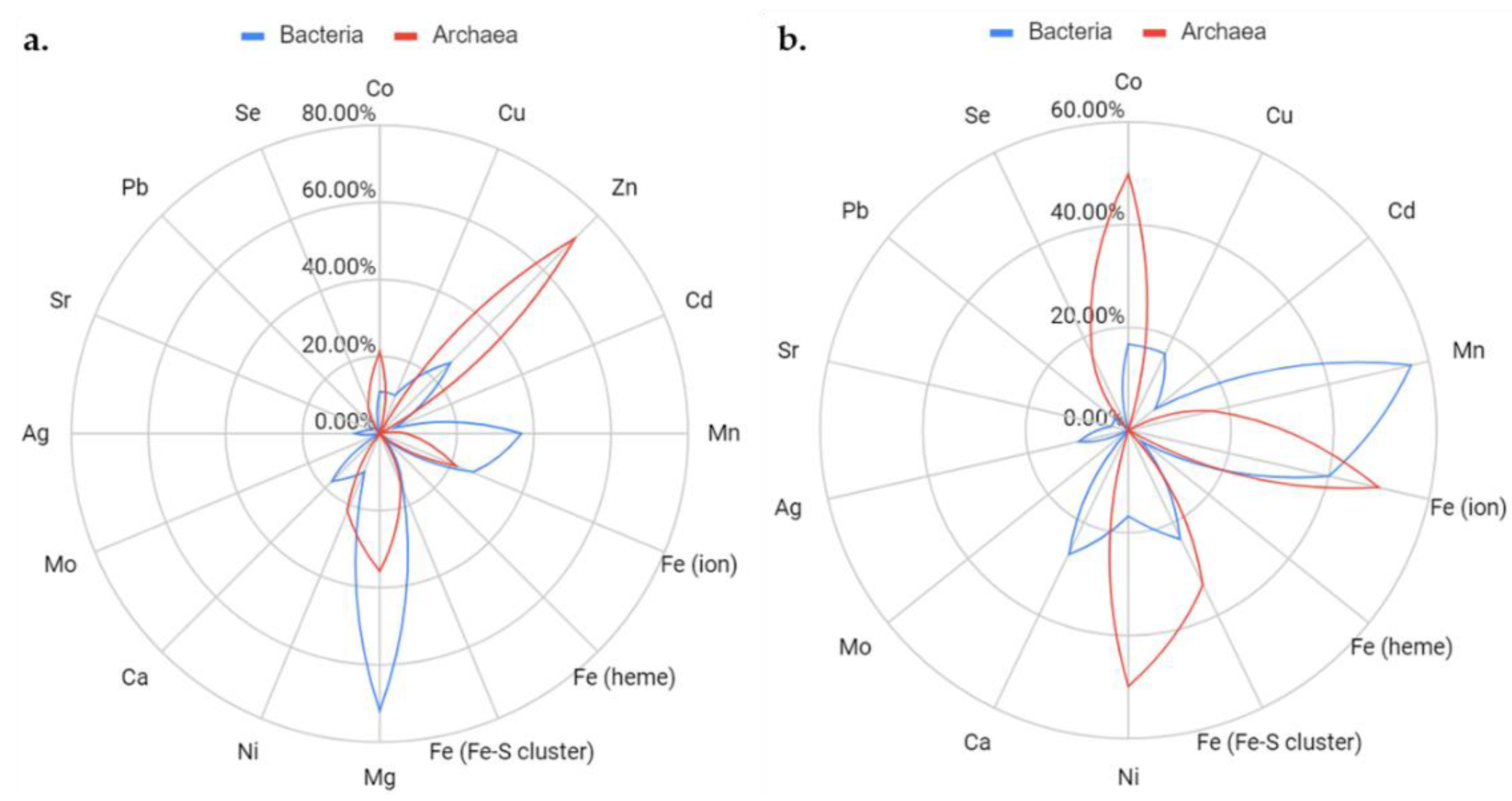
3.2. Conservation of Metal Binding Site but Variability of Metal Identity across DUF34 Structures
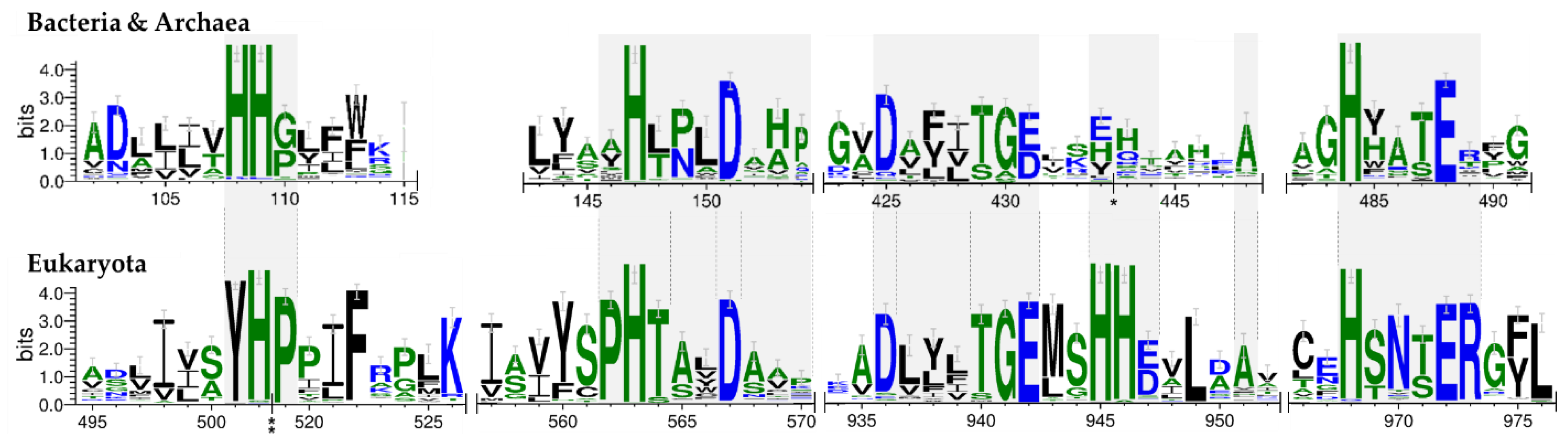
3.3. Family Wide and Superkingdom-Specific Signature Motifs
3.4. A Variable Central Insertion Occurs in Some DUF34 Family Members
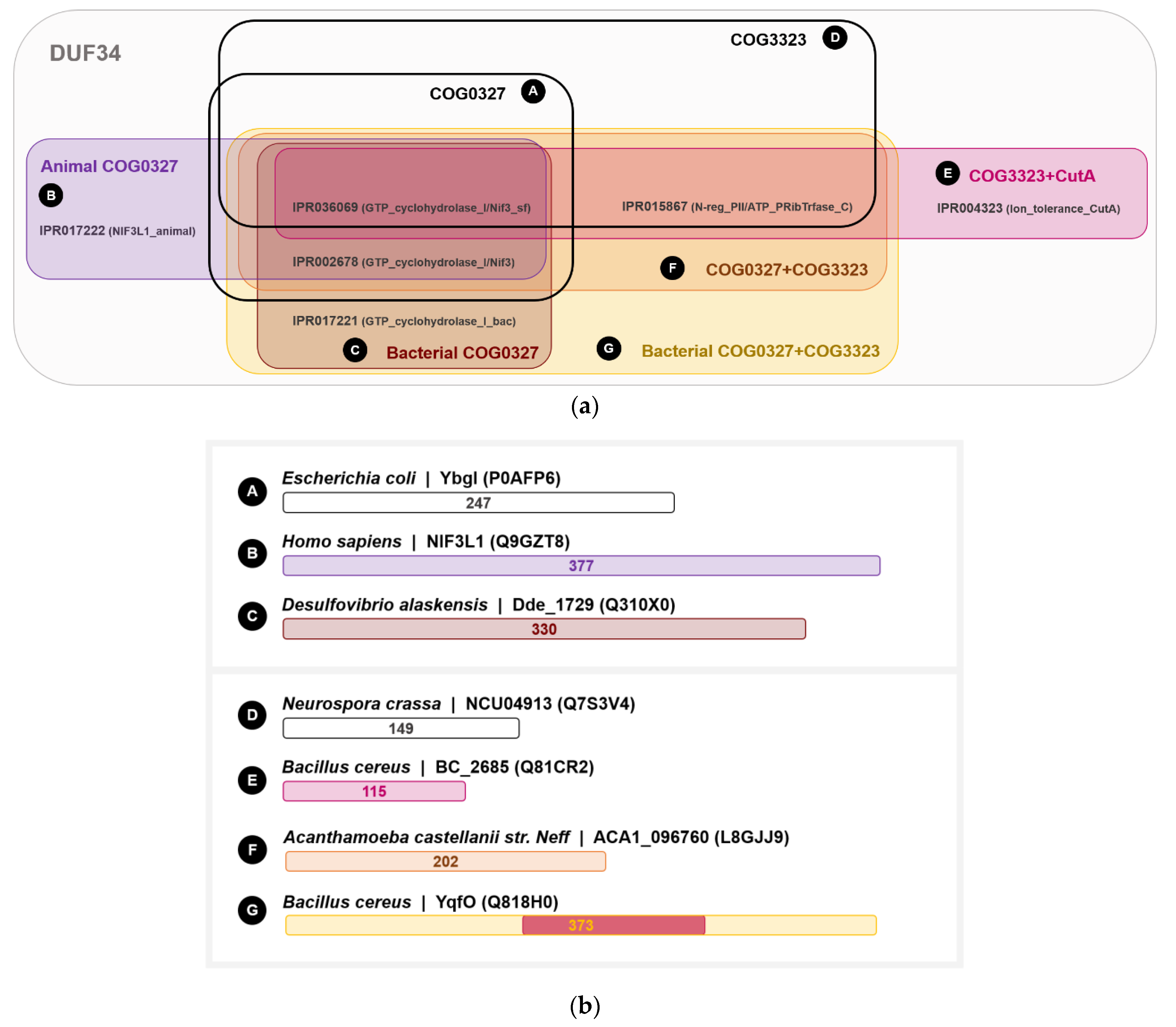
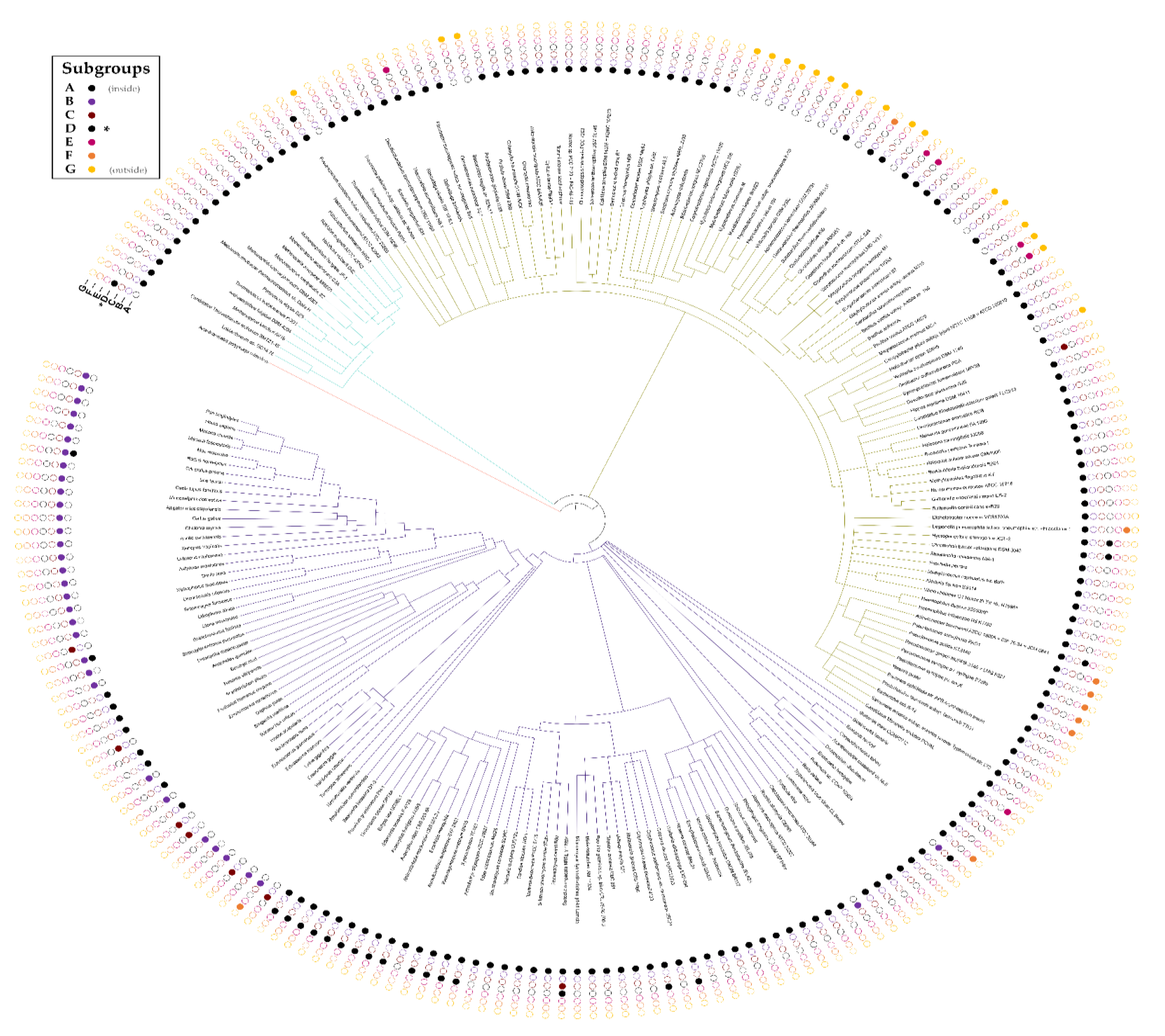
3.5. The DUF34 Family Can Be Split into Eight Interconnected Subgroups
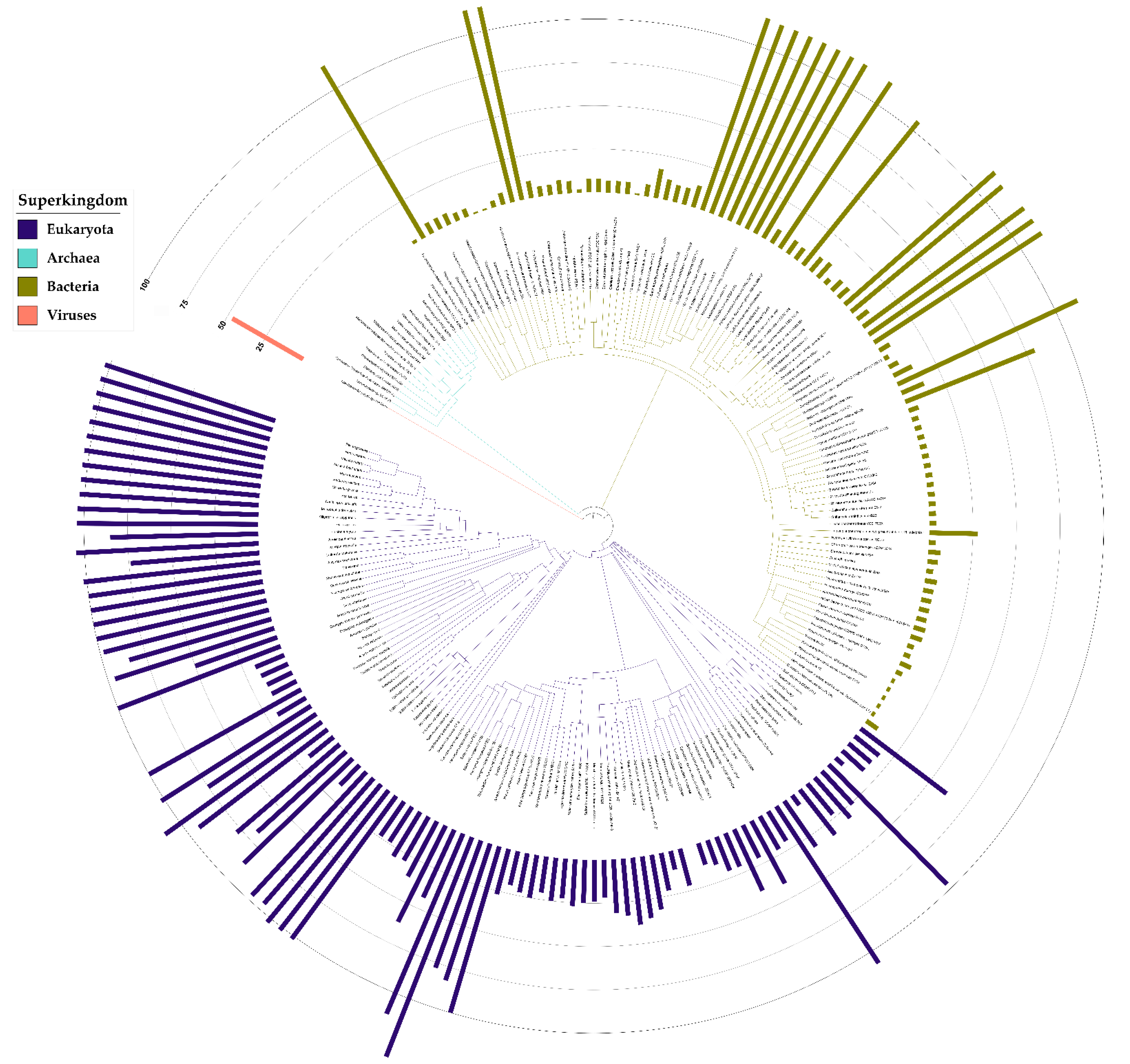
3.6. Taxonomic Distribution Suggests That the NIF3 (COG0327) and YqfO-like (COG3323) Domains Have Different Functions
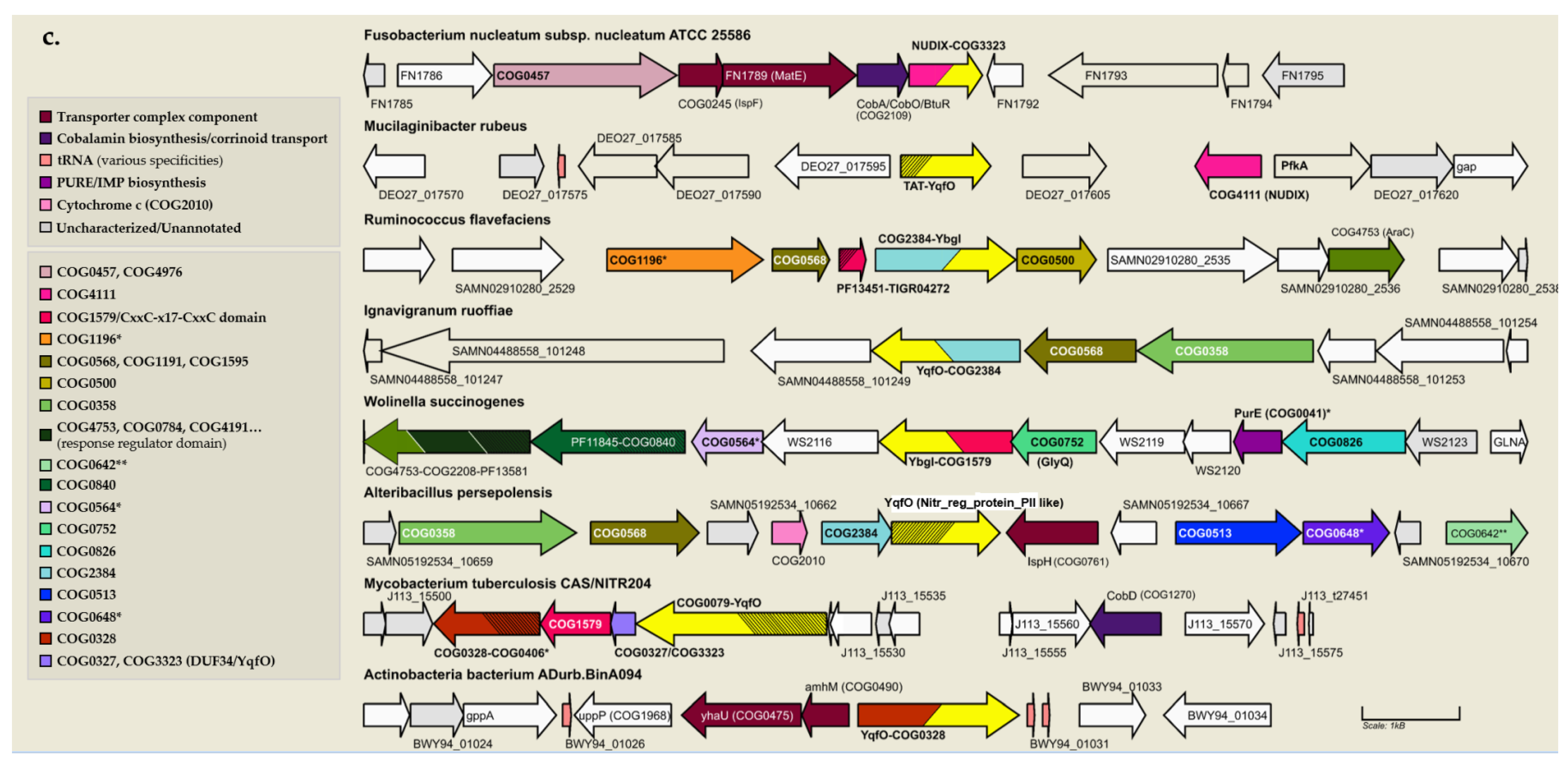
3.7. Physical Clustering and Co-Expression Further Link the DUF34 Family to Metal Ion Homeostasis and Iron Sulfur-Cluster Metabolism
3.8. DUF34 Fusions Fortify Links to Metals and Metallocofactors, Most Notably Fe-S Clusters
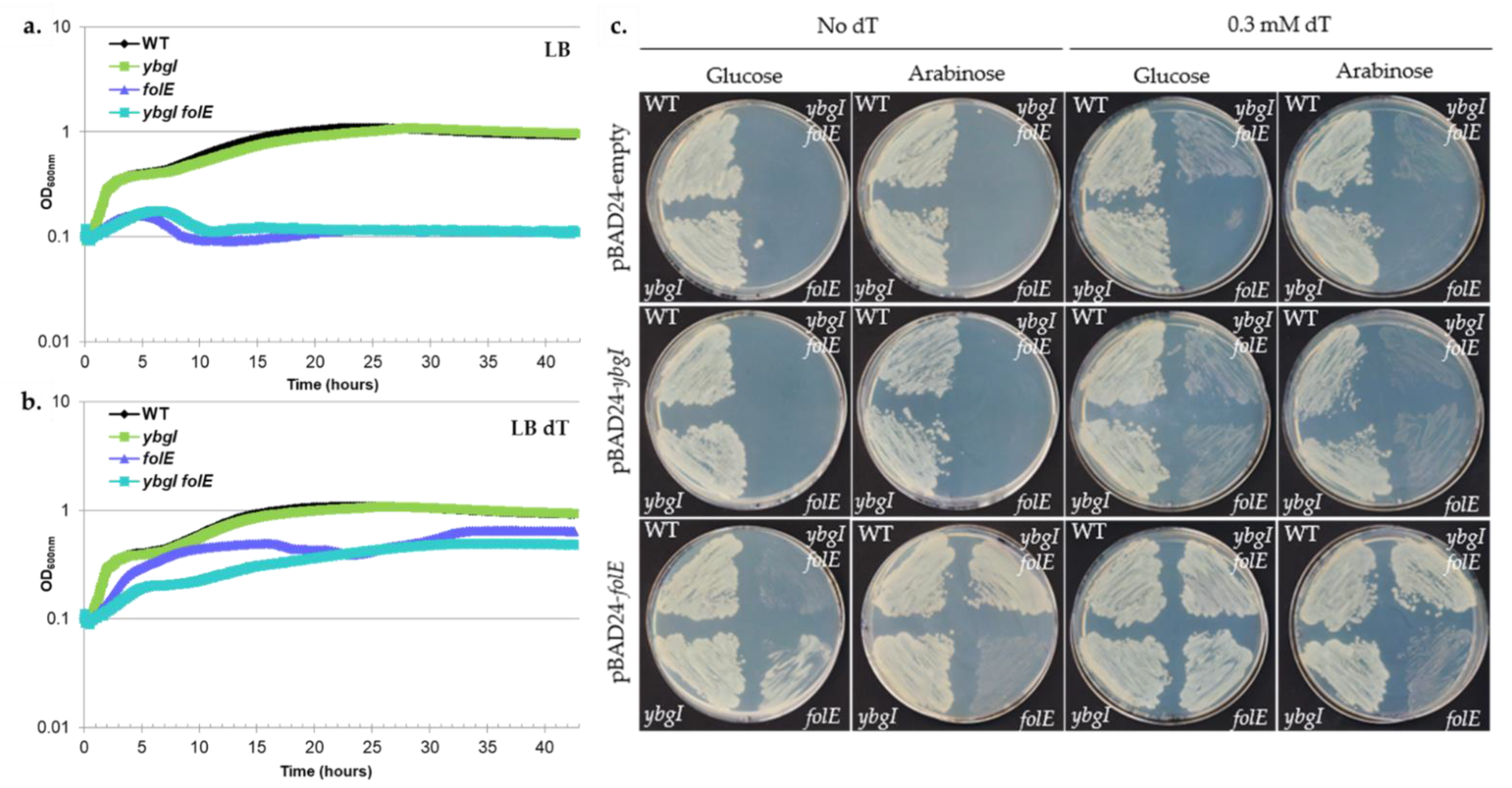
3.9. A Role of the DUF34 Family Protein in Folate Synthesis Is Precluded by Bioinformatic and Experimental Evidence
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Danchin, A.; Fang, G. Unknown unknowns: Essential genes in quest for function. Microb. Biotechnol. 2016, 9, 530–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niehaus, T.D.; Thamm, A.M.; de Crécy-Lagard, V.; Hanson, A.D. Proteins of unknown biochemical function—A persistent problem and a roadmap to help overcome it. Plant Physiol. 2015, 169, 1436–1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Crécy-Lagard, V.; Haas, D.; Hanson, A.D. Newly-discovered enzymes that function in metabolite damage-control. Curr. Opin. Chem. Biol. 2018, 47, 101–108. [Google Scholar] [CrossRef] [PubMed]
- De Crécy-Lagard, V.; Phillips, G.; Grochowski, L.L.; Yacoubi, B.E.; Jenney, F.; Adams, M.W.W.; Murzin, A.G.; White, R.H. Comparative genomics guided discovery of two missing archaeal enzyme families involved in the biosynthesis of the pterin moiety of tetrahydromethanopterin and tetrahydrofolate. ACS Chem. Biol. 2012, 7, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Wetmore, K.M.; Waters, R.J.; Callaghan, M.; Ray, J.; Liu, H.; Kuehl, J.V.; Melnyk, R.A.; Lamson, J.S.; Suh, Y.; et al. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 2018, 557, 503–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolker, E. Identification and functional analysis of “hypothetical” genes expressed in Haemophilus influenzae. Nucleic Acids Res. 2004, 32, 2353–2361. [Google Scholar] [CrossRef] [Green Version]
- Ghodge, S.V. Mechanistic Characterization and Function Discovery of Phosphohydrolase Enzymes from the Amidohydrolase Superfamily; Texas A&M University: College Station, TX, USA, 2015. [Google Scholar]
- Tan, C.L. The absence of universally-conserved protein-coding genes. bioRxiv 2019, 842633. [Google Scholar] [CrossRef]
- Rödelsperger, C.; Prabh, N.; Sommer, R.J. New Gene Origin and Deep Taxon Phylogenomics: Opportunities and Challenges. Trends Genet. 2019, 35, 914–922. [Google Scholar] [CrossRef]
- Alam, M.T.; Takano, E.; Breitling, R. Prioritizing orphan proteins for further study using phylogenomics and gene expression profiles in Streptomyces coelicolor. BMC Res. Notes 2011, 4, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, V.; Lock, A.; Harris, M.A.; Rutherford, K.; Bähler, J.; Oliver, S.G. Hidden in plain sight: What remains to be discovered in the eukaryotic proteome? Open Biol. 2019, 9, 180241. [Google Scholar] [CrossRef] [Green Version]
- Nagy, L.G.; Merényi, Z.; Hegedüs, B.; Bálint, B. Novel phylogenetic methods are needed for understanding gene function in the era of mega-scale genome sequencing. Nucleic Acids Res. 2020, 48, 2209–2219. [Google Scholar] [CrossRef] [Green Version]
- Thiaville, P.C.; Iwata-Reuyl, D.; DeCrécy-Lagard, V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t6A), a universal modification of tRNA. RNA Biol. 2014, 11, 1529–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Yacoubi, B.; Hatin, I.; Deutsch, C.; Kahveci, T.; Rousset, J.-P.; Iwata-Reuyl, D.; G Murzin, A.; de Crécy-Lagard, V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011, 30, 882–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Yacoubi, B.; Lyons, B.; Cruz, Y.; Reddy, R.; Nordin, B.; Agnelli, F.; Williamson, J.R.; Schimmel, P.; Swairjo, M.A.; De Crécy-Lagard, V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009, 37, 2894–2909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutherland, D.R.; Abdullah, K.M.; Cyopick, P.; Mellors, A. Cleavage of the cell-surface O-sialoglycoproteins CD34, CD43, CD44, and CD45 by a novel glycoprotease from Pasteurella haemolytica. J. Immunol. 1992, 148, 1458–1464. [Google Scholar]
- Nichols, C.E.; Lamb, H.K.; Thompson, P.; El Omari, K.; Lockyer, M.; Charles, I.; Hawkins, A.R.; Stammers, D.K. Crystal structure of the dimer of two essential Salmonella typhimurium proteins, YgjD & YeaZ and calorimetric evidence for the formation of a ternary YgjD-YeaZ-YjeE complex. Protein Sci. 2013, 22, 628–640. [Google Scholar] [CrossRef] [Green Version]
- Edvardson, S.; Prunetti, L.; Arraf, A.; Haas, D.; Bacusmo, J.M.; Hu, J.F.; Ta-Shma, A.; Dedon, P.C.; de Crécy-Lagard, V.; Elpeleg, O. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur. J. Hum. Genet. 2017, 25, 545–551. [Google Scholar] [CrossRef] [Green Version]
- Niehaus, T.D.; Gerdes, S.; Hodge-Hanson, K.; Zhukov, A.; Cooper, A.J.L.; ElBadawi-Sidhu, M.; Fiehn, O.; Downs, D.M.; Hanson, A.D. Genomic and experimental evidence for multiple metabolic functions in the RidA/YjgF/YER057c/UK114 (Rid) protein family. BMC Genom. 2015, 16, 382. [Google Scholar] [CrossRef] [Green Version]
- Downs, D.M.; Ernst, D.C. From microbiology to cancer biology: The Rid protein family prevents cellular damage caused by endogenously generated reactive nitrogen species. Mol. Microbiol. 2015, 96, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Irons, J.L.; Hodge-Hanson, K.; Downs, D.M. RidA Proteins Protect against Metabolic Damage by Reactive Intermediates. Microbiol. Mol. Biol. Rev. 2020, 84, 1–28. [Google Scholar] [CrossRef]
- Lambrecht, J.A.; Schmitz, G.E.; Downs, D.M. RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. mBio 2013, 4, e00033-13. [Google Scholar] [CrossRef] [Green Version]
- Borchert, A.J.; Ernst, D.C.; Downs, D.M. Reactive enamines and imines in vivo: Lessons from the RidA paradigm. Trends Biochem. Sci. 2019, 44, 849–860. [Google Scholar] [CrossRef]
- Tascou, S.; Uedelhoven, J.; Dixkens, C.; Nayernia, K.; Engel, W.; Burfeind, P. Isolation and characterization of a novel human gene, NIF3L1, and its mouse ortholog, Nif3l1, highly conserved from bacteria to mammals. Cytogenet. Genome Res. 2000, 90, 330–336. [Google Scholar] [CrossRef]
- Tascou, S.; Kang, T.W.; Trappe, R.; Engel, W.; Burfeind, P. Identification and characterization of NIF3L1 BP1, a novel cytoplasmic interaction partner of the NIF3L1 protein. Biochem. Biophys. Res. Commun. 2003, 309, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Ladner, J.E.; Obmolova, G.; Teplyakov, A.; Howard, A.J.; Khil, P.P.; Camerini-Otero, R.D.; Gilliland, G.L. Crystal structure of Escherichia coli protein YbgI, a toroidal structure with a dinuclear metal site. BMC Struct. Biol. 2003, 3, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baysal, Ö.; Lai, D.; Xu, H.-H.; Siragusa, M.; Çalışkan, M.; Carimi, F.; da Silva, J.A.T.; Tör, M. A Proteomic Approach Provides New Insights into the Control of Soil-Borne Plant Pathogens by Bacillus Species. PLoS ONE 2013, 8, e53182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Misra, S.; Roote, J.; Lewis, S.E.; Blazej, R.; Davis, T.; Doyle, C.; Galle, R.; George, R.; Harris, N.; et al. An exploration of the sequence of a 2.9-Mb region of the genome of Drosophila melanogaster: The Adh region. Genetics 1999, 153, 179–219. [Google Scholar] [CrossRef]
- Geisler, R.; Bergmann, A.; Hiromi, Y.; Nüsslein-Volhard, C. cactus, a gene involved in dorsoventral pattern formation of Drosophila, is related to the IκB gene family of vertebrates. Cell 1992, 71, 613–621. [Google Scholar] [CrossRef]
- Hadano, S.; Yanagisawa, Y.; Skaug, J.; Fichter, K.; Nasir, J.; Martindale, D.; Koop, B.F.; Scherer, S.W.; Nicholson, D.W.; Rouleau, G.A.; et al. Cloning and characterization of three novel genes, ALS2CR1, ALS2CR2, and ALS2CR3, in the juvenile amyotrophic lateral sclerosis (ALS2) critical region at chromosome 2q33-q34: Candidate genes for ALS2. Genomics 2001, 71, 200–213. [Google Scholar] [CrossRef]
- Merla, G.; Howald, C.; Antonarakis, S.E.; Reymond, A. The subcellular localization of the ChoRE-binding protein, encoded by the Williams–Beuren syndrome critical region gene 14, is regulated by 14-3-3. Hum. Mol. Genet. 2004, 13, 1505–1514. [Google Scholar] [CrossRef]
- Sergeeva, O.V.; Bredikhin, D.O.; Nesterchuk, M.V.; Serebryakova, M.V.; Sergiev, P.V.; Dontsova, O.A. Possible Role of Escherichia coli Protein YbgI. Biochemistry 2018, 83, 270–280. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, baw100. [Google Scholar] [CrossRef]
- Choi, H.-P.; Juarez, S.; Ciordia, S.; Fernandez, M.; Bargiela, R.; Albar, J.P.; Mazumdar, V.; Anton, B.P.; Kasif, S.; Ferrer, M.; et al. Biochemical Characterization of Hypothetical Proteins from Helicobacter pylori. PLoS ONE 2013, 8, e66605. [Google Scholar] [CrossRef] [Green Version]
- Adams, N.E.; Thiaville, J.J.; Proestos, J.; Juárez-Vázquez, A.L.; McCoy, A.J.; Barona-Gómez, F.; Iwata-Reuyl, D.; de Crécy-Lagard, V.; Maurelli, A.T. Promiscuous and adaptable enzymes fill “holes” in the tetrahydrofolate pathway in Chlamydia species. mBio 2014, 5, e01378-14. [Google Scholar] [CrossRef] [Green Version]
- De Crécy-Lagard, V. Variations in metabolic pathways create challenges for automated metabolic reconstructions: Examples from the tetrahydrofolate synthesis pathway. Comput. Struct. Biotechnol. J. 2014, 10, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchison, C.A.; Peterson, S.N.; Gill, S.R.; Cline, R.T.; White, O.; Fraser, C.M.; Smith, H.O.; Venter, J.C. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 1999, 286, 2165–2169. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019, 47, D464–D474. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, F.C.; Koetzle, T.F.; Williams, G.J.B.; Meyer, E.F.; Brice, M.D.; Rodgers, J.R.; Kennard, O.; Shimanouchi, T.; Tasumi, M. The Protein Data Bank. A Computer-Based Archival File for Macromolecular Structures. Eur. J. Biochem. 1977, 80, 319–324. [Google Scholar] [CrossRef]
- Andreini, C.; Cavallaro, G.; Lorenzini, S.; Rosato, A. MetalPDB: A database of metal sites in biological macromolecular structures. Nucleic Acids Res. 2013, 41, 312–319. [Google Scholar] [CrossRef] [Green Version]
- Putignano, V.; Rosato, A.; Banci, L.; Andreini, C. MetalPDB in 2018: A database of metal sites in biological macromolecular structures. Nucleic Acids Res. 2018, 46, D459–D464. [Google Scholar] [CrossRef]
- Luo, H.; Lin, Y.; Gao, F.; Zhang, C.-T.; Zhang, R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements: Table 1. Nucleic Acids Res. 2014, 42, D574–D580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.-H.; Lu, G.; Chen, X.; Zhao, X.-M.; Bork, P. OGEE v2: An update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 2017, 45, D940–D944. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhang, R.R. Putative essential and core-essential genes in Mycoplasma genomes. Sci. Rep. 2011, 1, 53. [Google Scholar] [CrossRef] [Green Version]
- Nevers, Y.; Kress, A.; Defosset, A.; Ripp, R.; Linard, B.; Thompson, J.D.; Poch, O.; Lecompte, O. OrthoInspector 3.0: Open portal for comparative genomics. Nucleic Acids Res. 2019, 47, D411–D418. [Google Scholar] [CrossRef]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Landan, G.; Graur, D. Local reliability measures from sets of co-optimal multiple sequence alignments. Pacific Symp. Biocomput. 2008, 24, 15–24. [Google Scholar] [CrossRef]
- Penn, O.; Privman, E.; Ashkenazy, H.; Landan, G.; Graur, D.; Pupko, T. GUIDANCE: A web server for assessing alignment confidence scores. Nucleic Acids Res. 2010, 38, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Sela, I.; Ashkenazy, H.; Katoh, K.; Pupko, T. GUIDANCE2: Accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015, 43, W7–W14. [Google Scholar] [CrossRef] [Green Version]
- Crooks, G.; Hon, G.; Chandonia, J.; Brenner, S. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
- Minatani, K. Proposal for SVG2DOT: An Interoperable Tactile Graphics Creation System Using SVG outputs from Inkscape. Stud. Health Technol. Inform. 2015, 217, 506–511. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bethesda (MD): National Library of Medicine (US), N.C. for B.I. National Center for Biotechnology Information (NCBI) [Internet]. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 26 August 2021).
- Dehal, P.S.; Joachimiak, M.P.; Price, M.N.; Bates, J.T.; Baumohl, J.K.; Chivian, D.; Friedland, G.D.; Huang, K.H.; Keller, K.; Novichkov, P.S.; et al. MicrobesOnline: An integrated portal for comparative and functional genomics. Nucleic Acids Res. 2009, 38, 396–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Guerrero, C.E.; Ciria, R.; Abreu-Goodger, C.; Moreno-Hagelsieb, G.; Merino, E. GeConT 2: Gene context analysis for orthologous proteins, conserved domains and metabolic pathways. Nucleic Acids Res. 2008, 36, 176–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obayashi, T.; Kagaya, Y.; Aoki, Y.; Tadaka, S.; Kinoshita, K. COXPRESdb v7: A gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 2019, 47, D55–D62. [Google Scholar] [CrossRef]
- Kustatscher, G.; Grabowski, P.; Schrader, T.A.; Passmore, J.B.; Schrader, M.; Rappsilber, J. Co-regulation map of the human proteome enables identification of protein functions. Nat. Biotechnol. 2019, 37, 1361–1371. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Sherman, B.T.; Huang, D.W.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruford, E.A.; Braschi, B.; Denny, P.; Jones, T.E.M.; Seal, R.L.; Tweedie, S. Guidelines for human gene nomenclature. Nat. Genet. 2020, 52, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutinet, G.; Kot, W.; Cui, L.; Hillebrand, R.; Balamkundu, S.; Gnanakalai, S.; Neelakandan, R.; Carstens, A.B.; Fa Lui, C.; Tremblay, D.; et al. 7-Deazaguanine modifications protect phage DNA from host restriction systems. Nat. Commun. 2019, 10, 5442. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [Green Version]
- Martens, J.A.; Genereaux, J.; Saleh, A.; Brandl, C.J. Transcriptional Activation by Yeast PDR1p Is Inhibited by Its Association with NGG1p/ADA3p. J. Biol. Chem. 1996, 271, 15884–15890. [Google Scholar] [CrossRef] [Green Version]
- Gou, Y.; Graff, F.; Kilian, O.; Kafkas, S.; Katuri, J.; Kim, J.H.; Marinos, N.; McEntyre, J.; Morrison, A.; Pi, X.; et al. Europe PMC: A full-text literature database for the life sciences and platform for innovation. Nucleic Acids Res. 2015, 43, D1042–D1048. [Google Scholar] [CrossRef] [Green Version]
- Karniely, S.; Rayzner, A.; Sass, E.; Pines, O. α-Complementation as a probe for dual localization of mitochondrial proteins. Exp. Cell Res. 2006, 312, 3835–3846. [Google Scholar] [CrossRef]
- Chen, J.; Gai, Q.; Lv, Z.; Chen, J.; Nie, Z.; Wu, X.; Zhang, Y. All-trans retinoic acid affects subcellular localization of a novel BmNIF3l protein: Functional deduce and tissue distribution of NIF3l gene from silkworm (Bombyx mori). Arch. Insect Biochem. Physiol. 2010, 74, 217–231. [Google Scholar] [CrossRef]
- Manan, A.; Bazai, Z.; Fan, J.; Yu, H.; Li, L. The Nif3-family protein YqfO03 from Pseudomonas syringae MB03 has multiple nematicidal activities against Caenorhabditis elegans and Meloidogyne incognita. Int. J. Mol. Sci. 2018, 19, 3915. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xie, B.; Jiang, Z.; Yuan, B. Relationship between osteoporosis and osteoarthritis based on DNA methylation. Int. J. Clin. Exp. Pathol. 2019, 12, 3399–3407. [Google Scholar] [PubMed]
- Yu, N.; Shin, S.; Lee, K.-A. First Korean Case of SATB2 -Associated 2q32-q33 Microdeletion Syndrome. Ann. Lab. Med. 2015, 35, 275. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Li, Y.; Chen, Y.; Podsypanina, K.; Chamorro, M.; Olshen, A.B.; Desai, K.V.; Tann, A.; Petersen, D.; Green, J.E.; et al. Changes in gene expression during the development of mammary tumors in MMTV-Wnt-1 transgenic mice. Genome Biol. 2005, 6, R84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jostes, S.V. The bromodomain Inhibitor JQ1 as Novel Therapeutic Option for Type II Testicular Germ Cell Tumours: The Role of SOX2 and SOX17 in Regulating Germ Cell Tumour Pluripotency; Rheinischen Friedrich-Wilhelms-Universität: Bonn, Germany, 2019. [Google Scholar]
- Lin, C.-Y.; Ström, A.; Vega, V.B.; Kong, S.L.; Yeo, A.L.; Thomsen, J.S.; Chan, W.C.; Doray, B.; Bangarusamy, D.K.; Ramasamy, A.; et al. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 2004, 5, R66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, Y.; Riker, A.; Shevde-Samant, L.; Samant, R.; Morris, C.; Gavin, E.; Fodstad, O.; Ju, J. Global comparative gene expression analysis of melanoma patient samples, derived cell lines and corresponding tumor xenografts. Cancer Genom. Proteom. 2011, 5, 1–35. [Google Scholar] [CrossRef]
- Schrader, A.; Meyer, K.; Walther, N.; Stolz, A.; Feist, M.; Hand, E.; von Bonin, F.; Evers, M.; Kohler, C.; Shirneshan, K.; et al. Identification of a new gene regulatory circuit involving B cell receptor activated signaling using a combined analysis of experimental, clinical and global gene expression data. Oncotarget 2016, 7, 47061–47081. [Google Scholar] [CrossRef] [Green Version]
- Uxa, S.; Bernhart, S.H.; Mages, C.F.S.; Fischer, M.; Kohler, R.; Hoffmann, S.; Stadler, P.F.; Engeland, K.; Müller, G.A. DREAM and RB cooperate to induce gene repression and cell-cycle arrest in response to p53 activation. Nucleic Acids Res. 2019, 47, 9087–9103. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Zhang, C.-Q.; Huang, K. Predicting glioblastoma prognosis networks using weighted gene co-expression network analysis on TCGA data. BMC Bioinformatics 2012, 13, S12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cury, S.S.; Lapa, R.M.L.; de Mello, J.B.H.; Marchi, F.A.; Domingues, M.A.C.; Pinto, C.A.L.; Carvalho, R.F.; de Carvalho, G.B.; Kowalski, L.P.; Rogatto, S.R. Increased DSG2 plasmatic levels identified by transcriptomic-based secretome analysis is a potential prognostic biomarker in laryngeal carcinoma. Oral Oncol. 2020, 103, 104592. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Shi, Q.; Xu, J.; Yi, W.; Fan, H. Weighted Gene Coexpression Network Analysis Reveals the Dynamic Transcriptome Regulation and Prognostic Biomarkers of Hepatocellular Carcinoma. Evol. Bioinform. 2020, 16, 117693432092056. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, S.; Xiang, Y.; Qu, X.; Xie, Y.; Zhang, X. Bioinformatic Analysis of Circular RNA-Associated ceRNA Network Associated with Hepatocellular Carcinoma. BioMed Res. Int. 2019, 2019, 8308694. [Google Scholar] [CrossRef] [PubMed]
- Quigley, D.A.; Fiorito, E.; Nord, S.; Van Loo, P.; Alnaes, G.G.; Fleischer, T.; Tost, J.; Moen Vollan, H.K.; Tramm, T.; Overgaard, J.; et al. The 5p12 breast cancer susceptibility locus affects MRPS30 expression in estrogen-receptor positive tumors. Mol. Oncol. 2014, 8, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Kusonmano, K.; Halle, M.K.; Wik, E.; Hoivik, E.A.; Krakstad, C.; Mauland, K.K.; Tangen, I.L.; Berg, A.; Werner, H.M.J.; Trovik, J.; et al. Identification of highly connected and differentially expressed gene subnetworks in metastasizing endometrial cancer. PLoS ONE 2018, 13, e0206665. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, L.; Liu, J.; Wang, J. A gene interaction network-based method to measure the common and heterogeneous mechanisms of gynecological cancer. Mol. Med. Rep. 2018, 18, 230–242. [Google Scholar] [CrossRef] [Green Version]
- Antoniali, G.; Serra, F.; Lirussi, L.; Tanaka, M.; D’Ambrosio, C.; Zhang, S.; Radovic, S.; Dalla, E.; Ciani, Y.; Scaloni, A.; et al. Mammalian APE1 controls miRNA processing and its interactome is linked to cancer RNA metabolism. Nat. Commun. 2017, 8, 797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneeweiss, A.; Hartkopf, A.D.; Müller, V.; Wöckel, A.; Lux, M.P.; Janni, W.; Ettl, J.; Belleville, E.; Huober, J.; Thill, M.; et al. Update Breast Cancer 2020 Part 1 – Early Breast Cancer: Consolidation of Knowledge About Known Therapies. Geburtshilfe Frauenheilkd. 2020, 80, 277–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codrich, M.; Comelli, M.; Malfatti, M.C.; Mio, C.; Ayyildiz, D.; Zhang, C.; Kelley, M.R.; Terrosu, G.; Pucillo, C.E.M.; Tell, G. Inhibition of APE1-endonuclease activity affects cell metabolism in colon cancer cells via a p53-dependent pathway. DNA Repair 2019, 82, 102675. [Google Scholar] [CrossRef]
- Wang, L.-J.; Hsu, C.-W.; Chen, C.-C.; Liang, Y.; Chen, L.-C.; Ojcius, D.M.; Tsang, N.-M.; Hsueh, C.; Wu, C.-C.; Chang, Y.-S. Interactome-wide Analysis Identifies End-binding Protein 1 as a Crucial Component for the Speck-like Particle Formation of Activated Absence in Melanoma 2 (AIM2) Inflammasomes. Mol. Cell. Proteom. 2012, 11, 1230–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, L.; Jenkins, G.D.; Bhise, N.; Feldberg, T.; Mitra-Ghosh, T.; Fridley, B.L.; Lamba, J.K. Genome-wide association analysis identified splicing single nucleotide polymorphism in CFLAR predictive of triptolide chemo-sensitivity. BMC Genom. 2015, 16, 483. [Google Scholar] [CrossRef] [Green Version]
- Kalari, K.R.; Necela, B.M.; Tang, X.; Thompson, K.J.; Lau, M.; Eckel-Passow, J.E.; Kachergus, J.M.; Anderson, S.K.; Sun, Z.; Baheti, S.; et al. An Integrated Model of the Transcriptome of HER2-Positive Breast Cancer. PLoS ONE 2013, 8, e79298. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.S.S.J.; Ahameethunisa, A.R.; Santosh, W.; Chakravarthy, S.; Kumar, S. Systems biological approach on neurological disorders: A novel molecular connectivity to aging and psychiatric diseases. BMC Syst. Biol. 2011, 5, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malan-Müller, S.; de Souza, V.B.C.; Daniels, W.M.U.; Seedat, S.; Robinson, M.D.; Hemmings, S.M.J. Shedding Light on the Transcriptomic Dark Matter in Biological Psychiatry: Role of Long Noncoding RNAs in D-cycloserine-Induced Fear Extinction in Posttraumatic Stress Disorder. OMICS J. Integr. Biol. 2020, 24, 352–369. [Google Scholar] [CrossRef]
- Qiu, L.; Liu, X. Identification of key genes involved in myocardial infarction. Eur. J. Med. Res. 2019, 24, 22. [Google Scholar] [CrossRef] [Green Version]
- Lin, H. Identification of Potential coregenes in Sevoflurance induced Myocardial Energy Metabolismin Patients Undergoing Off-pump Coronary Artery Bypass Graft Surgery using Bioinformatics analysis. Res. Sq. 2019, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Chekouo, T.; Safo, S.E. Bayesian Integrative Analysis and Prediction with Application to Atherosclerosis Cardiovascular Disease. arXiv 2020, arXiv:2005.11586. [Google Scholar]
- Winer, D.A.; Winer, S.; Shen, L.; Wadia, P.P.; Yantha, J.; Paltser, G.; Tsui, H.; Wu, P.; Davidson, M.G.; Alonso, M.N.; et al. B cells promote insulin resistance through modulation of T cells and production of pathogenic IgG antibodies. Nat. Med. 2011, 17, 610–617. [Google Scholar] [CrossRef]
- Xia, B.; Li, Y.; Zhou, J.; Tian, B.; Feng, L. Identification of potential pathogenic genes associated with osteoporosis. Bone Jt. Res. 2017, 6, 640–648. [Google Scholar] [CrossRef] [Green Version]
- Thankam, F.G.; Boosani, C.S.; Dilisio, M.F.; Agrawal, D.K. MicroRNAs associated with inflammation in shoulder tendinopathy and glenohumeral arthritis. Mol. Cell. Biochem. 2018, 437, 81–97. [Google Scholar] [CrossRef]
- Wang, J.C.; Ramaswami, G.; Geschwind, D.H. Gene co-expression network analysis in human spinal cord highlights mechanisms underlying amyotrophic lateral sclerosis susceptibility. bioRxiv 2020, 11, 1–14. [Google Scholar]
- Lv, L.; Zhang, D.; Hua, P.; Yang, S. The glial-specific hypermethylated 3′ untranslated region of histone deacetylase 1 may modulates several signal pathways in Alzheimer’s disease. Life Sci. 2021, 265, 118760. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Voineagu, I.; Paşca, S.P.; Won, H.; Chandran, V.; Horvath, S.; Dolmetsch, R.E.; Geschwind, D.H. Alteration in basal and depolarization induced transcriptional network in iPSC derived neurons from Timothy syndrome. Genome Med. 2014, 6, 75. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Fujisawa, N.; Tashiro, Y.; Takanabe, N.; Sugiyama, A.; Tashiro, F. The Role of Transcriptional Corepressor Nif3l1 in Early Stage of Neural Differentiation via Cooperation with Trip15/CSN2. J. Biol. Chem. 2003, 278, 10752–10762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duzyj, C.M.; Paidas, M.J.; Jebailey, L.; Huang, J.; Barnea, E.R. PreImplantation factor (PIF*) promotes embryotrophic and neuroprotective decidual genes: Effect negated by epidermal growth factor. J. Neurodev. Disord. 2014, 6, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyama, H. Implication of Trip15/CSN2 in early stage of neuronal differentiation of P19 embryonal carcinoma cells. Dev. Brain Res. 2003, 140, 45–56. [Google Scholar] [CrossRef]
- Boswell, W.T.; Boswell, M.; Walter, D.J.; Navarro, K.L.; Chang, J.; Lu, Y.; Savage, M.G.; Shen, J.; Walter, R.B. Exposure to 4100 K fluorescent light elicits sex specific transcriptional responses in Xiphophorus maculatus skin. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2018, 208, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Zuccotti, M.; Merico, V.; Sacchi, L.; Bellone, M.; Brink, T.C.; Bellazzi, R.; Stefanelli, M.; Redi, C.; Garagna, S.; Adjaye, J. Maternal Oct-4 is a potential key regulator of the developmental competence of mouse oocytes. BMC Dev. Biol. 2008, 8, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skottman, H.; Mikkola, M.; Lundin, K.; Olsson, C.; Strömberg, A.-M.; Tuuri, T.; Otonkoski, T.; Hovatta, O.; Lahesmaa, R. Gene Expression Signatures of Seven Individual Human Embryonic Stem Cell Lines. Stem Cells 2005, 23, 1343–1356. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yao, X.; Bachvarov, D.; Saifudeen, Z.; El-Dahr, S.S. Genome-wide analysis of gestational gene-environment interactions in the developing kidney. Physiol. Genom. 2014, 46, 655–670. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.; Bi, Y.; Wang, H.; Dong, S.; Li, K.; Li, J. Gene Expression Profiling of Clostridium botulinum under Heat Shock Stress. BioMed Res. Int. 2013, 2013, 760904. [Google Scholar] [CrossRef] [Green Version]
- Selby, K.; Mascher, G.; Somervuo, P.; Lindström, M.; Korkeala, H. Heat shock and prolonged heat stress attenuate neurotoxin and sporulation gene expression in group I Clostridium botulinum strain ATCC 3502. PLoS ONE 2017, 12, e0176944. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.L.; Roux, C.M.; Olson, M.W.; Luong, T.T.; Lee, C.Y.; Olson, R.; Dunman, P.M. Characterizing the effects of inorganic acid and alkaline shock on the Staphylococcus aureus transcriptome and messenger RNA turnover. FEMS Immunol. Med. Microbiol. 2010, 60, 208–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belvin, B.R.; Gui, Q.; Hutcherson, J.A.; Lewis, J.P. The Porphyromonas gingivalis hybrid cluster protein Hcp is required for growth with nitrite and survival with host cells. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aurass, P.; Pless, B.; Rydzewski, K.; Holland, G.; Bannert, N.; Flieger, A. bdhA-patD Operon as a Virulence Determinant, Revealed by a Novel Large-Scale Approach for Identification of Legionella pneumophila Mutants Defective for Amoeba Infection. Appl. Environ. Microbiol. 2009, 75, 4506–4515. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Caro, F.; Robins, W.; Mekalanos, J.J. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 2018, 359, 210–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangaiah, D.; Labandeira-Rey, M.; Zhang, X.; Fortney, K.R.; Ellinger, S.; Zwickl, B.; Baker, B.; Liu, Y.; Janowicz, D.M.; Katz, B.P.; et al. Haemophilus ducreyi Hfq Contributes to Virulence Gene Regulation as Cells Enter Stationary Phase. mBio 2014, 5, e01081-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labandeira-Rey, M.; Mock, J.R.; Hansen, E.J. Regulation of Expression of the Haemophilus ducreyi LspB and LspA2 Proteins by CpxR. Infect. Immun. 2009, 77, 3402–3411. [Google Scholar] [CrossRef] [Green Version]
- Spinola, S.M.; Fortney, K.R.; Baker, B.; Janowicz, D.M.; Zwickl, B.; Katz, B.P.; Blick, R.J.; Munson, R.S. Activation of the CpxRA System by Deletion of cpxA Impairs the Ability of Haemophilus ducreyi To Infect Humans. Infect. Immun. 2010, 78, 3898–3904. [Google Scholar] [CrossRef] [Green Version]
- Rahmani-Badi, A.; Sepehr, S.; Fallahi, H.; Heidari-Keshel, S. Erratum: Exposure of E. coli to DNA-Methylating Agents Impairs Biofilm Formation and Invasion of Eukaryotic Cells via Down Regulation of the N-Acetylneuraminate Lyase NanA. Front. Microbiol. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription Profiling-Based Identification of Staphylococcus aureus Genes Regulated by the agr and/or sarA Loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, L.E.; Tsang, J.; Mrázek, J.; Hoover, T.R. The zinc-ribbon domain of Helicobacter pylori HP0958: Requirement for RpoN accumulation and possible roles of homologs in other bacteria. Microb. Inform. Exp. 2011, 1, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pomposiello, P.J.; Bennik, M.H.J.; Demple, B. Genome-Wide Transcriptional Profiling of the Escherichia coli Responses to Superoxide Stress and Sodium Salicylate. J. Bacteriol. 2001, 183, 3890–3902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, C.; Andersen, B.; Arshid, S.; Larsen, M.R.; Albergaria, H.; Lametsch, R.; Arneborg, N. Proteomics insights into the responses of Saccharomyces cerevisiae during mixed-culture alcoholic fermentation with Lachancea thermotolerans. FEMS Microbiol. Ecol. 2019, 95, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Shulami, S.; Shenker, O.; Langut, Y.; Lavid, N.; Gat, O.; Zaide, G.; Zehavi, A.; Sonenshein, A.L.; Shoham, Y. Multiple Regulatory Mechanisms Control the Expression of the Geobacillus stearothermophilus Gene for Extracellular Xylanase. J. Biol. Chem. 2014, 289, 25957–25975. [Google Scholar] [CrossRef] [Green Version]
- Ogura, M.; Sato, T.; Abe, K. Bacillus subtilis YlxR, Which Is Involved in Glucose-Responsive Metabolic Changes, Regulates Expression of tsaD for Protein Quality Control of Pyruvate Dehydrogenase. Front. Microbiol. 2019, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.-C.; Huang, C.-H.; Yang, C.S.; Kuan, S.-M.; Lin, C.-T.; Chou, S.-H.; Chen, Y. Crystal Structure of a Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii Reveals a Novel Quaternary Assembly in the Nif3 Family. BioMed Res. Int. 2014, 2014, 171263. [Google Scholar] [CrossRef]
- Tomoike, F.; Wakamatsu, T.; Nakagawa, N.; Kuramitsu, S.; Masui, R. Crystal structure of the conserved hypothetical protein TTHA1606 from Thermus thermophilus HB8. Proteins Struct. Funct. Bioinforma. 2009, 76, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Fujishiro, T.; Ermler, U.; Shima, S. A possible iron delivery function of the dinuclear iron center of HcgD in [Fe]-hydrogenase cofactor biosynthesis. FEBS Lett. 2014, 588, 2789–2793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lie, T.J.; Costa, K.C.; Pak, D.; Sakesan, V.; Leigh, J.A. Phenotypic evidence that the function of the [Fe]-hydrogenase Hmd in Methanococcus maripaludis requires seven hcg ( hmd co-occurring genes) but not hmdII. FEMS Microbiol. Lett. 2013, 343, 156–160. [Google Scholar] [CrossRef] [Green Version]
- Godsey, M.H.; Minasov, G.; Shuvalova, L.; Brunzelle, J.S.; Vorontsov, I.I.; Collart, F.R.; Anderson, W.F. The 2.2 Å resolution crystal structure of Bacillus cereus Nif3-family protein YqfO reveals a conserved dimetal-binding motif and a regulatory domain. Protein Sci. 2007, 16, 1285–1293. [Google Scholar] [CrossRef] [PubMed]
- Lamba, J.K.; Feldberg, T.; Ghosh, T.M.; Bhise, N.; Fridley, B. Abstract 2214: Genome-wide association analysis identified genetic markers associated with triptolide cellular sensitivity using HapMap LCLs as model system. In Proceedings of the Experimental and Molecular Therapeutics; American Association for Cancer Research: Philadelphia, PA, USA, 2013; Volume 73, p. 2214. [Google Scholar]
- Malik, A.; Pande, K.; Kumar, A.; Vemula, A.; Chandramohan, M.R.V. Finding Pathogenic nsSNP’s and their structural effect on COPS2 using Molecular Dynamic Approach. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kuan, S.-M.; Chen, H.-C.; Huang, C.-H.; Chang, C.-H.; Chen, S.-C.; Yang, C.S.; Chen, Y. Crystallization and preliminary X-ray diffraction analysis of the Nif3-family protein MJ0927 from Methanocaldococcus jannaschii. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 80–82. [Google Scholar] [CrossRef] [Green Version]
- Saikatendu, K.S.; Zhang, X.; Kinch, L.; Leybourne, M.; Grishin, N.V.; Zhang, H. Structure of a conserved hypothetical protein SA1388 from S. aureus reveals a capped hexameric toroid with two PII domain lids and a dinuclear metal center. BMC Struct. Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Constantine, K.L.; Krystek, S.R.; Healy, M.D.; Doyle, M.L.; Siemers, N.O.; Thanassi, J.; Yan, N.; Xie, D.; Goldfarb, V.; Yanchunas, J.; et al. Structural and functional characterization of CFE88: Evidence that a conserved and essential bacterial protein is a methyltransferase. Protein Sci. 2009, 14, 1472–1484. [Google Scholar] [CrossRef] [Green Version]
- Qijing, G.; Zhang, Y. NIF3 Superfamily protein. Chin. J. Cell Biol. 2007, 29, 816–820. [Google Scholar]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Li, Q.; Yang, H.; Yan, L.; Yang, L.; Yu, L. Overexpression of human CUTA isoform 2 enhances the cytotoxicity of copper to HeLa cells. Acta Biochim. Pol. 2008, 55, 411–415. [Google Scholar] [CrossRef]
- Gupta, S.D.; Lee, B.T.O.; Camakaris, J.; Wu, H.C. Identification of cutC and cutF (nlpE) genes involved in copper tolerance in Escherichia coli. J. Bacteriol. 1995, 177, 4207–4215. [Google Scholar] [CrossRef] [Green Version]
- Fong, S.T.; Camakaris, J.; Lee, B.T. Molecular genetics of a chromosomal locus involved in copper tolerance in Escherichia coli K-12. Mol. Microbiol. 1995, 15, 1127–1137. [Google Scholar] [CrossRef]
- Tanaka, Y.; Tsumoto, K.; Nakanishi, T.; Yasutake, Y.; Sakai, N.; Yao, M.; Tanaka, I.; Kumagai, I. Structural implications for heavy metal-induced reversible assembly and aggregation of a protein: The case of Pyrococcus horikoshii CutA. FEBS Lett. 2004, 556, 167–174. [Google Scholar] [CrossRef] [Green Version]
- Odermatt, A.; Solioz, M. Two trans-acting metalloregulatory proteins controlling expression of the copper-ATPases of Enterococcus hirae. J. Biol. Chem. 1995, 270, 4349–4354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rensing, C.; Franke, S. Copper Homeostasis in Escherichia coli and Other Enterobacteriaceae. EcoSal Plus 2007, 2, ecosalplus.5.4.4.1. [Google Scholar] [CrossRef]
- Bagautdinov, B. The structures of the CutA1 proteins from Thermus thermophilus and Pyrococcus horikoshii: Characterization of metal-binding sites and metal-induced assembly. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 404–413. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2256–2268. [Google Scholar] [CrossRef]
- Siltberg-Liberles, J.; Martinez, A. Searching distant homologs of the regulatory ACT domain in phenylalanine hydroxylase. Amino Acids 2009, 36, 235–249. [Google Scholar] [CrossRef]
- Arnesano, F.; Banci, L.; Benvenuti, M.; Bertini, I.; Calderone, V.; Mangani, S.; Viezzoli, M.S. The Evolutionarily Conserved Trimeric Structure of CutA1 Proteins Suggests a Role in Signal Transduction. J. Biol. Chem. 2003, 278, 45999–46006. [Google Scholar] [CrossRef] [Green Version]
- Forchhammer, K.; Lüddecke, J. Sensory properties of the PII signalling protein family. FEBS J. 2016, 283, 425–437. [Google Scholar] [CrossRef]
- Selim, K.A.; Tremiño, L.; Marco-Marín, C.; Alva, V.; Espinosa, J.; Contreras, A.; Hartmann, M.D.; Forchhammer, K.; Rubio, V. Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role. FEBS J. 2021, 288, 1142–1162. [Google Scholar] [CrossRef]
- Selim, K.A.; Haffner, M. Heavy Metal Stress Alters the Response of the Unicellular Cyanobacterium Synechococcus elongatus PCC 7942 to Nitrogen Starvation. Life 2020, 10, 275. [Google Scholar] [CrossRef]
- Koga, R.; Matsumoto, A.; Kouzuma, A.; Watanabe, K. Identification of an extracytoplasmic function sigma factor that facilitates c-type cytochrome maturation and current generation under electrolyte-flow conditions in Shewanella oneidensis MR-1. Environ. Microbiol. 2020, 22, 3671–3684. [Google Scholar] [CrossRef] [PubMed]
- Manina, G.; Bellinzoni, M.; Pasca, M.R.; Neres, J.; Milano, A.; De Jesus Lopes Ribeiro, A.L.; Buroni, S.; Škovierová, H.; Dianišková, P.; Mikušová, K.; et al. Biological and structural characterization of the Mycobacterium smegmatis nitroreductase NfnB, and its role in benzothiazinone resistance. Mol. Microbiol. 2010, 77, 1172–1185. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Chen, I.M.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Grechkin, Y.; Ratner, A.; Anderson, I.; Lykidis, A.; Mavromatis, K.; et al. The integrated microbial genomes system: An expanding comparative analysis resource. Nucleic Acids Res. 2009, 38, 382–390. [Google Scholar] [CrossRef] [Green Version]
- Grigoriev, I.V.; Nordberg, H.; Shabalov, I.; Aerts, A.; Cantor, M.; Goodstein, D.; Kuo, A.; Minovitsky, S.; Nikitin, R.; Ohm, R.A.; et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012, 40, D26–D32. [Google Scholar] [CrossRef] [PubMed]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014, 42, 26–31. [Google Scholar] [CrossRef]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nature 2009, 460, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Haakonsen, D.L.; Sanderlin, A.G.; Liu, Y.J.; Shen, L.; Zhuang, N.; Laub, M.T.; Zhang, Y. Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA. Nucleic Acids Res. 2018, 46, 3245–3256. [Google Scholar] [CrossRef]
- Stamford, N.P.; Lilley, P.E.; Dixon, N.E. Enriched sources of Escherichia coli replication proteins. The dnaG primase is a zinc metalloprotein. Biochim. Biophys. Acta 1992, 1132, 17–25. [Google Scholar] [CrossRef]
- Czubat, B.; Minias, A.; Brzostek, A.; Żaczek, A.; Struś, K.; Zakrzewska-Czerwińska, J.; Dziadek, J. Functional Disassociation Between the Protein Domains of MSMEG_4305 of Mycolicibacterium smegmatis (Mycobacterium smegmatis) in vivo. Front. Microbiol. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Nowotny, M.; Yang, W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006, 25, 1924–1933. [Google Scholar] [CrossRef]
- Niyomporn, B.; Dahl, J.L.; Strominger, J.L. Biosynthesis of the peptidoglycan of bacterial cell walls. IX. Purification and properties of glycyl transfer ribonucleic acid synthetase from Staphylococcus aureus. J. Biol. Chem. 1968, 243, 773–778. [Google Scholar] [CrossRef]
- Pelosi, L.; Vo, C.-D.-T.; Abby, S.S.; Loiseau, L.; Rascalou, B.; Hajj Chehade, M.; Faivre, B.; Goussé, M.; Chenal, C.; Touati, N.; et al. Ubiquinone Biosynthesis over the Entire O2 Range: Characterization of a Conserved O2-Independent Pathway. mBio 2019, 10, e01319-19. [Google Scholar] [CrossRef] [Green Version]
- Kato, T.; Takahashi, N.; Kuramitsu, H.K. Sequence analysis and characterization of the Porphyromonas gingivalis prtC gene, which expresses a novel collagenase activity. J. Bacteriol. 1992, 174, 3889–3895. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, R.P.; Ahern, H.; Xing, D.; Thayer, M.M.; Tainer, J.A. Structure and function of Escherichia coli endonuclease III. Ann. N. Y. Acad. Sci. 1994, 726, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.A.; Karim, N.; Worku, M.; Moore, S.A.; Penn, C.W.; O’Toole, P.W. HP0958 is an essential motility gene in Helicobacter pylori. FEMS Microbiol. Lett. 2005, 248, 47–55. [Google Scholar] [CrossRef]
- Kumar, A.; Karthikeyan, S. Crystal structure of the MSMEG_4306 gene product from Mycobacterium smegmatis. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2018, 74, 166–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barta, M.L.; Battaile, K.P.; Lovell, S.; Hefty, P.S. Hypothetical protein CT398 (CdsZ) interacts with σ54 (RpoN)-holoenzyme and the type III secretion export apparatus in Chlamydia trachomatis. Protein Sci. 2015, 24, 1617–1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rees, W.D.; Lorenzo-Leal, A.C.; Steiner, T.S.; Bach, H. Mycobacterium avium Subspecies paratuberculosis Infects and Replicates within Human Monocyte-Derived Dendritic Cells. Microorganisms 2020, 8, 994. [Google Scholar] [CrossRef]
- Kim, W.S.; Shin, M.-K.; Shin, S.J. MAP1981c, a Putative Nucleic Acid-Binding Protein, Produced by Mycobacterium avium subsp. paratuberculosis, Induces Maturation of Dendritic Cells and Th1-Polarization. Front. Cell. Infect. Microbiol. 2018, 8. [Google Scholar] [CrossRef]
- Sassetti, C.M.; Boyd, D.H.; Rubin, E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003, 48, 77–84. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Yanai, I.; Hunter, C.P. Comparison of diverse developmental transcriptomes reveals that coexpression of gene neighbors is not evolutionarily conserved. Genome Res. 2009, 19, 2214–2220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheftel, A.D.; Wilbrecht, C.; Stehling, O.; Niggemeyer, B.; Elsässer, H.P.; Mühlenhoff, U.; Lill, R. The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol. Biol. Cell 2012, 23, 1157–1166. [Google Scholar] [CrossRef]
- Cai, K.; Markley, J. NMR as a Tool to Investigate the Processes of Mitochondrial and Cytosolic Iron-Sulfur Cluster Biosynthesis. Molecules 2018, 23, 2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katzemeier, G.; Schmid, C.; Kellermann, J.; Lottspeich, F.; Bacher, A. Biosynthesis of Tetrahydrofolate. Sequence of GTP Cyclohydrolase I from Escherichia coli. Biol. Chem. Hoppe. Seyler. 1991, 372, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Cossins, E.A.; Chen, L. Folates and one-carbon metabolism in plants and fungi. Phytochemistry 1997, 45, 437–452. [Google Scholar] [CrossRef]
- Burg, A.W.; Brown, G.M. The biosynthesis of folic acid. 8. Purification and properties of the enzyme that catalyzes the production of formate from carbon atom 8 of guanosine triphosphate. J. Biol. Chem. 1968, 243, 2349–2358. [Google Scholar] [CrossRef]
- Thöny, B.; Auerbach, G.; Blau, N. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 2000, 347, 1–16. [Google Scholar] [CrossRef]
- Phillips, G.; El Yacoubi, B.; Lyons, B.; Alvarez, S.; Iwata-Reuyl, D.; De Crécy-Lagard, V. Biosynthesis of 7-deazaguanosine-modified tRNA nucleosides: A new role for GTP cyclohydrolase I. J. Bacteriol. 2008, 190, 7876–7884. [Google Scholar] [CrossRef] [Green Version]
- El Yacoubi, B.; Bonnett, S.; Anderson, J.N.; Swairjo, M.A.; Iwata-Reuyl, D.; De Crécy-Lagard, V. Discovery of a new prokaryotic type I GTP cyclohydrolase family. J. Biol. Chem. 2006, 281, 37586–37593. [Google Scholar] [CrossRef] [Green Version]
- Paranagama, N.; Bonnett, S.A.; Alvarez, J.; Luthra, A.; Stec, B.; Gustafson, A.; Iwata-Reuyl, D.; Swairjo, M.A. Mechanism and catalytic strategy of the prokaryotic-specific GTP cyclohydrolase-IB. Biochem. J. 2017, 474, 1017–1039. [Google Scholar] [CrossRef] [Green Version]
- Sankaran, B.; Bonnett, S.A.; Shah, K.; Gabriel, S.; Reddy, R.; Schimmel, P.; Rodionov, D.A.; De Crécy-Lagard, V.; Helmann, J.D.; Iwata-Reuyl, D.; et al. Zinc-independent folate biosynthesis: Genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB. J. Bacteriol. 2009, 191, 6936–6949. [Google Scholar] [CrossRef] [Green Version]
- de Crécy-Lagard, V.; El Yacoubi, B.; de la Garza, R.D.; Noiriel, A.; Hanson, A.D. Comparative genomics of bacterial and plant folate synthesis and salvage: Predictions and validations. BMC Genom. 2007, 8, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Gorelova, V.; Bastien, O.; De Clerck, O.; Lespinats, S.; Rébeillé, F.; Van Der Straeten, D. Evolution of folate biosynthesis and metabolism across algae and land plant lineages. Sci. Rep. 2019, 9, 5731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerdes, S.Y.; Scholle, M.D.; Campbell, J.W.; Balázsi, G.; Ravasz, E.; Daugherty, M.D.; Somera, A.L.; Kyrpides, N.C.; Anderson, I.; Gelfand, M.S.; et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 2003, 185, 5673–5684. [Google Scholar] [CrossRef] [Green Version]
- Salama, N.R.; Shepherd, B.; Falkow, S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 2004, 186, 7926–7935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahba, A.J.; Friedkin, M. The Enzymatic Synthesis of Thymidylate. J. Biol. Chem. 1962, 237, 3794–3801. [Google Scholar] [CrossRef]
- Rebelo, J.; Auerbach, G.; Bader, G.; Bracher, A.; Nar, H.; Hösl, C.; Schramek, N.; Kaiser, J.; Bacher, A.; Huber, R.; et al. Biosynthesis of Pteridines. Reaction Mechanism of GTP Cyclohydrolase I. J. Mol. Biol. 2003, 326, 503–516. [Google Scholar] [CrossRef]
- Philpott, C.C.; Jadhav, S. The ins and outs of iron: Escorting iron through the mammalian cytosol. Free Radic. Biol. Med. 2019, 133, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.R.; Wang, J.; Weiss, A.; Skaar, E.P.; Capdevila, D.A.; Giedroc, D.P. Mechanistic Insights into the Metal-Dependent Activation of Zn II -Dependent Metallochaperones. Inorg. Chem. 2019, 58, 13661–13672. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, K.A.; Jordan, M.R.; Giedroc, D.P. COG0523 proteins: A functionally diverse family of transition metal-regulated G3E P-loop GTP hydrolases from bacteria to man. Metallomics 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Chandrangsu, P.; Huang, X.; Gaballa, A.; Helmann, J.D. Bacillus subtilis FolE is sustained by the ZagA zinc metallochaperone and the alarmone ZTP under conditions of zinc deficiency. Mol. Microbiol. 2019, 112, 751–765. [Google Scholar] [CrossRef] [PubMed]

| Name | Organisms | Phenotype, Biological Relevance | Reference |
|---|---|---|---|
| YqfO/BC_4286 | Bacillus cereus | Inserted domain similar to PII-like/CutA1 family proteins; present in select bacterial clades; domain may regulate catalytic activity | [134] |
| YqfO/BSU_25170 | Bacillus subtilis subsp. subtilis str. 168 | With YlxR, coregulates tsaEBD (t6A synthesis [62]); disruption impairs tsaEDB regulation, loss of glucose-induction of sigX via PDHc expression dysregulation | [129] |
| BmNIF3l | Bombyx mori | Translocates to nucleus from cytoplasm upon ATRA tx; higher transcript levels in differentiating tissues; no expression detected in the egg stage | [73] |
| YbgI/b0710 | Escherichia coli | Structure, homohexameric toroid; monomers possess dinuclear metal ion-binding site; putatively involved in DNA repair | [26] |
| No survival impairment upon mutant UV tx; polar localization during cell division (co-localized with PstB, TktA); GlmS putative interaction partner; mutant sensitive to antibiotics affecting cell wall synthesis | [32] | ||
| XynX | Geobacillus stearothermophilus | Negatively regulates expression of xynA (encodes a secreted xylanase); may be negatively regulated by xylR | [128] |
| NIF3L1/ALS2CR1/CALS-7/MDS015/My018 | Homo sapiens | Ubiquitously expressed during embryonic development; strong over-expression in spermatogonia-derived, teratocarcinoma cell lines; Isolated, characterized; cytosolic subcellular localization; highly conserved N-, C-terminal regions; shares inserted region of its murine homolog (CutA1-like) | [24] |
| NIF3L1 interacts with splice variant, NIF3L1 BP1 (THOC7), cytosolic colocalization; C-terminal leucine zipper-like domain of variant mediates interaction; not indicated in repression in NIH3T3 cells; binding partner, NIF3L1 BP1, demonstrates additional passive presence in the nucleus | [25] | ||
| Retinoic acid-induced binding, cooperative translocation with Trip15/CSN2 from the cytosol to the nucleus (early neuronal development, silences differentiation suppressor Oct-3/4); ubiquitous expression, important in neuronal development | [107] | ||
| Detected in brain, spinal cord, and lymphocytes; observed as two distinct transcripts with similar patterns of expression; highest levels of both transcripts in heart, skeletal muscle, testis; smaller transcript was expressed at a higher level than the other; no deletions, polymorphisms linked to ALS patients relative to controls; 1 of 6 candidates eliminated for a causative link to ALS2 | [30] | ||
| 1 of 4 hypermethylated, significant differential expression shared between two cancellous bone specimen groups: osteoarthritis, osteoporosis | [75] | ||
| With 14-3-3, co-regulates transcriptional of Wbscr14 by preventing its nuclear localization via complex formation (Wbscr14 participates in the complex-mediated transcription of lipogenic enzymes, promoting fat accumulation) | [31] | ||
| Included in a 7.5-Mb interstitial deletion on 2q32.3–33.1 (28 genes) inpatient diagnosed with SATB2-Associated 2q32-q33 microdeletion syndrome | [76] | ||
| Significantly associated with triptolide chemosensitivity in lymphoblast cell lines | [135] | ||
| COPS2 point mutations consistent with previously defined NIF3L1-COPS2 co-repression interaction model (limited; pathogenesis associated COPS2 mutations: S120C, N144S, Y159H, R173C) | [136] | ||
| HP0959 | Helicobacter pylori | GTP-binding, hydrolysis in vitro, biologically irrelevant pH, temperature | [34] |
| HcgD/MJ0927 | Methanocaldococcus jannaschii | Proposed iron chaperone required for FeGP cofactor biosynthesis Homohexameric via 2 interfaced homotrimeric units; binds to ssDNA/dsDNA | [132] [130,137] |
| Nif3l1/1110030G24Rik | Mus musculus | Isolated, characterized; ubiquitous expression across tissues; cytosolic localization; highly conserved N-, C-terminal regions; shares inserted region of the human homolog | [24] |
| Retinoic acid-induced binding, cooperative translocation with Trip15/CSN2 from the cytosol to the nucleus (early neuronal development, results in the silence of the differentiation suppressor Oct-3/4); ubiquitous tissue expression, important in neuronal development | [107] | ||
| WP_046236688 WP_032702676 PP_1038 VT47_06255 WP_017124074 WP_054077596 | Pseudomonas sp. | (“YqfO03”) small, secreted protein; demonstrated high potency as nematicide against C. elegans, M. incognita; free-standing YqfO domain-containing protein (no NIF3/DUF34 domains) is a member of the NIF3 protein family | [74] |
| Nif3/YGL221C | Saccharomyces cerevisiae | Determined to have dual/multiple localizations (cytosolic, mitochondrial) | [72] |
| SA1388 | Staphylococcus aureus | The central domain of NIF3 homolog has high structural similarity to CutA1 (family linked to cation tolerance, homeostasis) | [138] |
| SP1609 | Streptococcus pneumoniae | Described as a member of the same orthologous group (COG2384) as TrmK, RpoD protein families via structural alignment (incorrect*) | [139] |
| TTHA1606 | Thermus thermophilus HB8 | Binds to ssDNA (very weakly, in vitro) | [131] |
| NIF3-like protein superfamily | NA | (electronic translation) describes family members of model organisms (Eukaryota, Bacteria), structures published prior to 2007 | [140] |
| Name | Organisms | Ligands | PII Domain | PDB | Phenotype | Reference |
|---|---|---|---|---|---|---|
| YbgI | Escherichia coli | (2)Fe3+ | No | 1NMO | NA | [26] |
| (2)Mg2+ | No | 1NMP | ||||
| HcgD/MJ0927 | Methanocaldococcus jannaschii | (1)Cl−, (2)Fe3+ | No | 3WSD | Weaker Fe1 site under oxidized conditions in vitro | [132] |
| (2)Fe2+, (1)PO43− | No | 3WSE | ||||
| (1)Fe3+, (1)citrate | No | 3WSF | ||||
| (1)Fe2+, (1)citrate | No | 3WSG | ||||
| (1)Fe3+, (1)SO42− | No | 3WSH | ||||
| (1)Fe2+, (1)PO43− | No | 3WSI | ||||
| NA | No | 4IWG | Binds to ssDNA, dsDNA in vitro | [130,137] | ||
| NA | No | 4IWM | ||||
| SA1388 | Staphylococcus aureus | (2)Zn2+, (1)B3P | Yes | 3LNL | Cavity diameter = 38 Å; opening edge length = 20 Å (triangular opening) | [138] |
| (2)Zn2+ | Yes | 2NYD | ||||
| SP1609 | Streptococcus pneumoniae | NA | No | 2FYW | NA | PDB only |
| TTHA1606 | Thermus thermophilus | NA | No | 2YYB | Binds ssDNA not dsDNA in vitro | [131] |
| Sthe_0840 | Sphaerobacter thermophilus | (7)Cl− *, (14)FMT *, (1)ACT * | No | 3RXY | NA | PDB only |
| YqfO | Bacillus cereus | (2)Zn2+, (1)HEPES, (1)TRS | Yes | 2GX8 | NA | [134] |
| Rank | COG | Name/Description | Metal(s) | References (PMID, EC Number) |
|---|---|---|---|---|
| 1 | COG0327 | Putative GTP cyclohydrolase 1 type 2, NIF3 family | Fe2+/Fe3+, Zn2+, Mg2+ | [26], [132], [138], [26.88.147.156], [26.89.148.157] |
| 2 | COG1579 | Predicted nucleic acid-binding protein DR0291, contains C4-type Zn-ribbon domain | Zn2+ | [125] |
| 3 | COG0568 | DNA-directed RNA polymerase, sigma subunit (sigma70/sigma32) | Zn2+, Mg2+ | [162], [2.7.7.6] |
| 4 | COG0358 | DNA primase (bacterial type) | Zn2+, Mg2+, Mn2+ | [163], [2.7.7.101] |
| 5 | COG0457 a | Tetratricopeptide (TPR) repeat | NA | None listed |
| 6 | COG2384 | tRNA A22 N1-methylase | NA | [2.1.1.217] |
| 7 | COG0079 | Histidinol-phosphate/aromatic aminotransferase or cobyric acid decarboxylase | NA; Co (cobalamin) | [164], [2.6.1.9] |
| 8 | COG0240 | Glycerol-3-phosphate dehydrogenase | NA | [1.1.1.94] |
| 9 | COG0328 | Ribonuclease HI (RnhA) | Mg2+, Mn2+, Co2+, Ni2+ | [165], [3.1.26.4] |
| 10 | COG0500 b | SAM-dependent methyltransferase | NA | [2.1.1.242] |
| 11 | COG0513 c | Superfamily II DNA and RNA helicase (SrmB/RhlB) | Mg2+, Mn2+ | [3.6.4.13] |
| 12 | COG0596 | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase MenH and related esterases, alpha/beta hydrolase fold (MhpC) | NA | [3.7.1.14] |
| 13 | COG0655 | Multimeric flavodoxin WrbA, includes NAD(P)H:quinone oxidoreductase | Most req. Fe-S cluster; subtypes without Fe-S clusters | [1.6.5.2], [1.6.5.6] |
| 14 | COG0752 | Glycyl-tRNA synthetase, alpha subunit | Mg2+, Mn2+, Co2+ | [166], [6.1.1.14] |
| 15 | COG0826 | 23S rRNA C2501 and tRNA U34 5’-hydroxylation protein RlhA/YrrN/YrrO, U32 peptidase family; ubiquinone biosynthesis protein, UbiU/YhbU | Fe-S cluster/Fe, Ca2+ | [167,168] |
| 16 | COG1028 | NAD(P)-dependent dehydrogenase, short-chain alcohol dehydrogenase family | Co2+, Fe/Fe2+, Mg2+, Mn2+, Zn/Zn2+ | [1.1.1.2] |
| 17 | COG1897 | Homoserine O-succinyltransferase | NA | [2.3.1.31], [2.3.1.46] |
| 18 | COG0177 d | Endonuclease III (Nth) | Fe-S cluster, Ca2+, Co2+, Fe/Fe2+, Mg2+, Mn2+, Ni2+, Zn2+ | [169], [4.2.99.18] |
| 19 | COG0477 d | MFS family permease (includes anhydromuropeptide permease AmpG, ProP) | NA | None listed |
| 20 | COG0494 e | 8-oxo-dGTP pyrophosphatase MutT and related house-cleaning NTP pyrophosphohydrolases, NUDIX family | Co2+, Mg2+, Mn2+, Zn2+ | [3.6.1.13] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reed, C.J.; Hutinet, G.; de Crécy-Lagard, V. Comparative Genomic Analysis of the DUF34 Protein Family Suggests Role as a Metal Ion Chaperone or Insertase. Biomolecules 2021, 11, 1282. https://doi.org/10.3390/biom11091282
Reed CJ, Hutinet G, de Crécy-Lagard V. Comparative Genomic Analysis of the DUF34 Protein Family Suggests Role as a Metal Ion Chaperone or Insertase. Biomolecules. 2021; 11(9):1282. https://doi.org/10.3390/biom11091282
Chicago/Turabian StyleReed, Colbie J., Geoffrey Hutinet, and Valérie de Crécy-Lagard. 2021. "Comparative Genomic Analysis of the DUF34 Protein Family Suggests Role as a Metal Ion Chaperone or Insertase" Biomolecules 11, no. 9: 1282. https://doi.org/10.3390/biom11091282






