Revealing the Mutation Patterns of Drug-Resistant Reverse Transcriptase Variants of Human Immunodeficiency Virus through Proteochemometric Modeling
Abstract
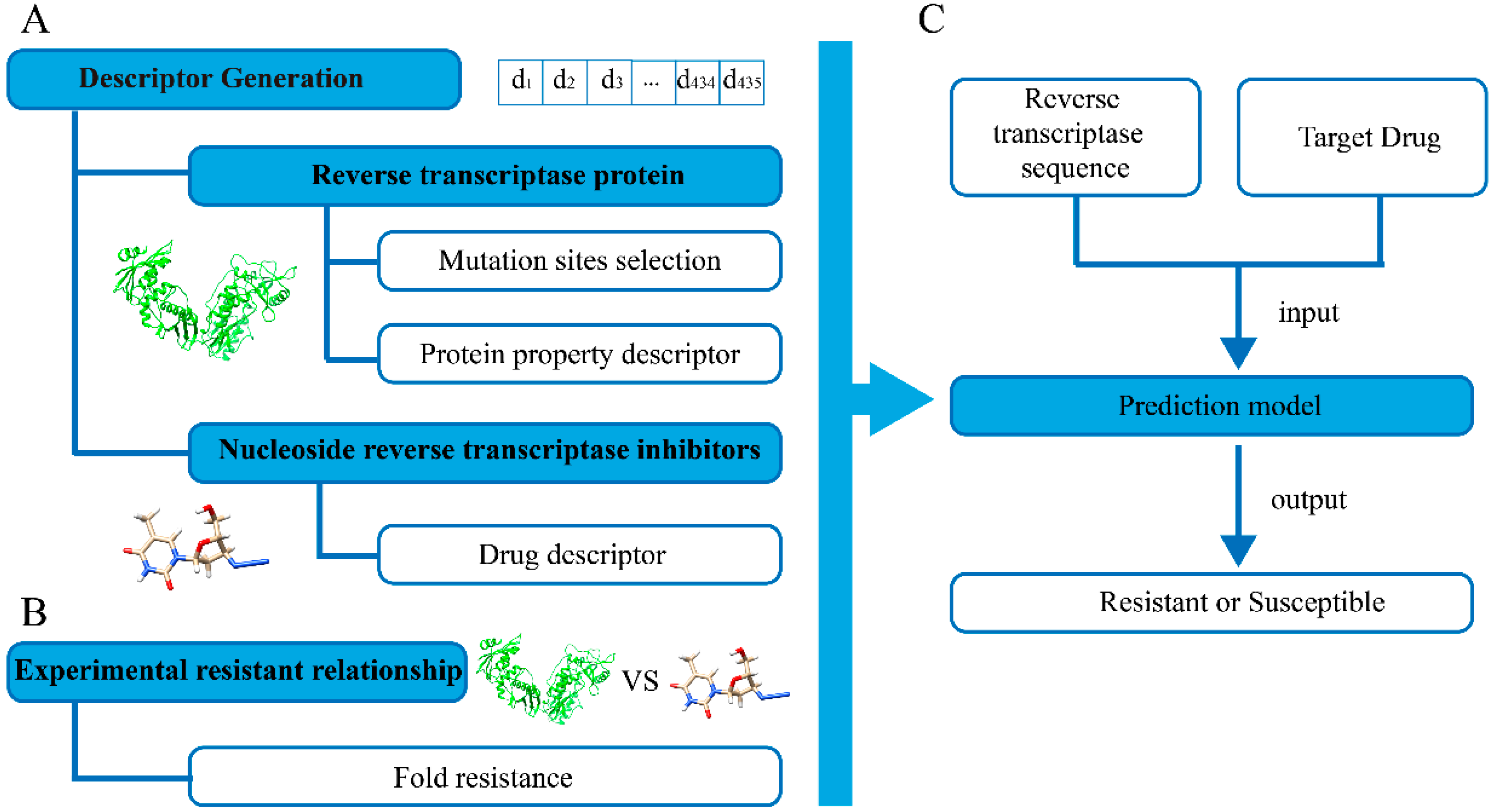
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Protein Structure Modeling
2.3. Mutation Sites Selection
2.4. Ligand Binding Site Prediction
2.5. Descriptor Generation
- (1).
- Z1: lipophilicity scale. Negative Z1 refers to lipophilic residues, and positive Z1 correlates to hydrophilic ones.
- (2).
- Z2: steric bulk, molecular weight and van der Waals volume.
- (3).
- Z3: description of polarity.
- (4).
- Z4 and Z5: combined properties, including electronegativity, electrophilicity, and hardness.
2.6. Model Construction
2.7. Model Evaluation
2.8. Calculating Feature Importance
2.9. Detecting Mutation Patterns in Experimental Pairs
- (1)
- Calculate the residue distribution on the individual target sites. For each target site, the residue frequencies in both experimentally determined drug-susceptible proteins and drug-resistant proteins were calculated.
- (2)
- Deriving the mutation patterns of the target sites. The amino acids on the dominant sites (Section 2.8) were joined as peptide fragments. Then, the distribution of each joint fragment was counted to form the mutation pattern.
2.10. Evaluation of Mutation Patterns
3. Results
3.1. Spatial Location of Screened Mutation Sites
3.2. Model Performance on Drug Susceptibility Prediction
3.3. Detecting Important Features for HIV-1 Drug Resistance
3.4. Mutation Patterns of Joint Fragment on Target Sites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/hiv-aids (accessed on 20 June 2021).
- Opar, A. New HIV drug classes on the horizon. Nat. Rev. Drug Discov. 2007, 6, 258–259. [Google Scholar] [CrossRef]
- Tarasova, O.; Poroikov, V. HIV Resistance Prediction to Reverse Transcriptase Inhibitors: Focus on Open Data. Molecules 2018, 23, 956. [Google Scholar] [CrossRef] [Green Version]
- Shafer, R.W.; Jung, D.R.; Betts, B.J.; Xi, Y.; Gonzales, M.J. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2000, 28, 346–348. [Google Scholar] [CrossRef] [Green Version]
- Santos, L.H.; Ferreira, R.S.; Caffarena, E.R. Computational drug design strategies applied to the modelling of human immunodeficiency virus-1 reverse transcriptase inhibitors. Mem. Inst. Oswaldo Cruz 2015, 110, 847–864. [Google Scholar] [CrossRef] [Green Version]
- Kantor, R.; Machekano, R.; Gonzales, M.J.; Dupnik, K.; Schapiro, J.M.; Shafer, R.W. Human Immunodeficiency Virus Reverse Transcriptase and Protease Sequence Database: An expanded data model integrating natural language text and sequence analysis programs. Nucleic Acids Res. 2001, 29, 296–299. [Google Scholar] [CrossRef] [Green Version]
- Shafer, R.W. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin. Microbiol. Rev. 2002, 15, 247–277. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, M.S.; Brun-Vezinet, F.; D’Aquila, R.T.; Hammer, S.M.; Johnson, V.A.; Kuritzkes, D.R.; Loveday, C.; Mellors, J.W.; Clotet, B.; Conway, B.; et al. Antiretroviral drug resistance testing in adult HIV-1 infection—Recommendations of an International AIDS Society-USA panel. JAMA-J. Am. Med. Assoc. 2000, 283, 2417–2426. [Google Scholar] [CrossRef]
- Beerenwinkel, N.; Schmidt, B.; Walter, H.; Kaiser, R.; Lengauer, T.; Hoffmann, D.; Korn, K.; Selbig, J. Diversity and complexity of HIV-1 drug resistance: A bioinformatics approach to predicting phenotype from genotype. Proc. Natl. Acad. Sci. USA 2002, 99, 8271–8276. [Google Scholar] [CrossRef] [Green Version]
- Beerenwinkel, N.; Daumer, M.; Oette, M.; Korn, K.; Hoffmann, D.; Kaiser, R.; Lengauer, T.; Selbig, J.; Walter, H. Geno2pheno: Estimating phenotypic drug resistance from HIV-1 genotypes. Nucleic Acids Res. 2003, 31, 3850–3855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhee, S.Y.; Taylor, J.; Wadhera, G.; Ben-Hur, A.; Brutlag, D.L.; Shafer, R.W. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc. Natl. Acad. Sci. USA 2006, 103, 17355–17360. [Google Scholar] [CrossRef] [Green Version]
- Tarasova, O.; Biziukova, N.; Filimonov, D.; Poroikov, V. A Computational Approach for the Prediction of HIV Resistance Based on Amino Acid and Nucleotide Descriptors. Molecules 2018, 23, 2751. [Google Scholar] [CrossRef] [Green Version]
- Paneth, A.; Plonka, W.; Paneth, P. What do docking and QSAR tell us about the design of HIV-1 reverse transcriptase nonnucleoside inhibitors? J. Mol. Model. 2017, 23, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiri, F.; Pirhadi, S.; Rahmani, A. Identification of new potential HIV-1 reverse transcriptase inhibitors by QSAR modeling and structure-based virtual screening. J. Recept. Sig. Transd. 2018, 38, 37–47. [Google Scholar] [CrossRef]
- Ramon, E.; Belanche-Munoz, L.; Perez-Enciso, M. HIV drug resistance prediction with weighted categorical kernel functions. BMC Bioinform. 2019, 20, 410. [Google Scholar] [CrossRef]
- Brand, L.; Yang, X.; Liu, K.; Elbeleidy, S.; Wang, H.; Zhang, H.; Nie, F. Learning Robust Multilabel Sample Specific Distances for Identifying HIV-1 Drug Resistance. J. Comput. Biol. 2020, 27, 655–672. [Google Scholar] [CrossRef]
- Leidner, F.; Yilmaz, N.K.; Schiffer, C.A. Target-Specific Prediction of Ligand Affinity with Structure-Based Interaction Fingerprints. J. Chem. Inf. Model. 2019, 59, 3679–3691. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Gonzales, M.J.; Kantor, R.; Betts, B.J.; Ravela, J.; Shafer, R.W. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003, 31, 298–303. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Rhee, S.Y.; Taylor, J.; Shafer, R.W. Comparison of the precision and sensitivity of the antivirogram and PhenoSense HIV drug susceptibility assays. Jaids-J. Acq. Imm. Def. 2005, 38, 439–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Zhou, Y.; Tanaka, I.; Yao, M. Roll: A new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics 2010, 26, 46–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandberg, M.; Eriksson, L.; Jonsson, J.; Sjostrom, M.; Wold, S. New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. J. Med. Chem. 1998, 41, 2481–2491. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- van Westen, G.J.P.; Hendriks, A.; Wegner, J.K.; IJzerman, A.P.; van Vlijmen, H.W.T.; Bender, A. Significantly Improved HIV Inhibitor Efficacy Prediction Employing Proteochemometric Models Generated From Antivirogram Data. PLoS Comput. Biol. 2013, 9, e1002899. [Google Scholar] [CrossRef]






| AUC | Accuracy | F-Score | Precision | Recall | |
|---|---|---|---|---|---|
| Random Forest | 0.921 ± 0.060 * | 0.827 ± 0.073 | 0.822 ± 0.087 | 0.815 ± 0.070 | 0.777 ± 0.206 |
| Logistic Regression | 0.871 ± 0.076 | 0.768 ± 0.094 | 0.758 ± 0.112 | 0.752 ± 0.104 | 0.750 ± 0.195 |
| Decision Tree | 0.791 ± 0.073 | 0.788 ± 0.069 | 0.793 ± 0.068 | 0.766 ± 0.070 | 0.772 ± 0.142 |
| Naïve Bayes | 0.813 ± 0.136 | 0.712 ± 0.099 | 0.685 ± 0.133 | 0.743 ± 0.121 | 0.596 ± 0.287 |
| Supporting Vector Machine | 0.896 ± 0.068 | 0.772 ± 0.098 | 0.758 ± 0.119 | 0.780 ± 0.107 | 0.717 ± 0.241 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, J.; Tian, X.; Liu, J.; Qin, Y.; Zhu, J.; Xu, D.; Qiu, T. Revealing the Mutation Patterns of Drug-Resistant Reverse Transcriptase Variants of Human Immunodeficiency Virus through Proteochemometric Modeling. Biomolecules 2021, 11, 1302. https://doi.org/10.3390/biom11091302
Qiu J, Tian X, Liu J, Qin Y, Zhu J, Xu D, Qiu T. Revealing the Mutation Patterns of Drug-Resistant Reverse Transcriptase Variants of Human Immunodeficiency Virus through Proteochemometric Modeling. Biomolecules. 2021; 11(9):1302. https://doi.org/10.3390/biom11091302
Chicago/Turabian StyleQiu, Jingxuan, Xinxin Tian, Jiangru Liu, Yulong Qin, Junjie Zhu, Dongpo Xu, and Tianyi Qiu. 2021. "Revealing the Mutation Patterns of Drug-Resistant Reverse Transcriptase Variants of Human Immunodeficiency Virus through Proteochemometric Modeling" Biomolecules 11, no. 9: 1302. https://doi.org/10.3390/biom11091302






