Phosphorylated Osteopontin Secreted from Cancer Cells Induces Cancer Cell Motility
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Quantitative PCR (qPCR)
- SPP1-Fwd: 5′-CCCCACAGTAGACACATATGATG-3′;
- SPP1-Rev: 5′-TTCAACTCCTCGCTTTCCAT-3′;
- HPRT1-Fwd: 5′-TTGTTGTAGGATATGCCCTTGA-3′;
- HPRT1-Rev: 5′-GCGATGTCAATAGGACTCCAG-3′.
2.3. Western Blot
2.4. Preparation of CM and Purification of Recombinant OPN
2.5. Cell Migration and Invasion Assays
2.6. Plasmids and Transfectants
2.7. Phos-Tag Enzyme Linked Immunosorbent Assay (ELISA)
2.8. Statistics and Reproducibility
3. Results
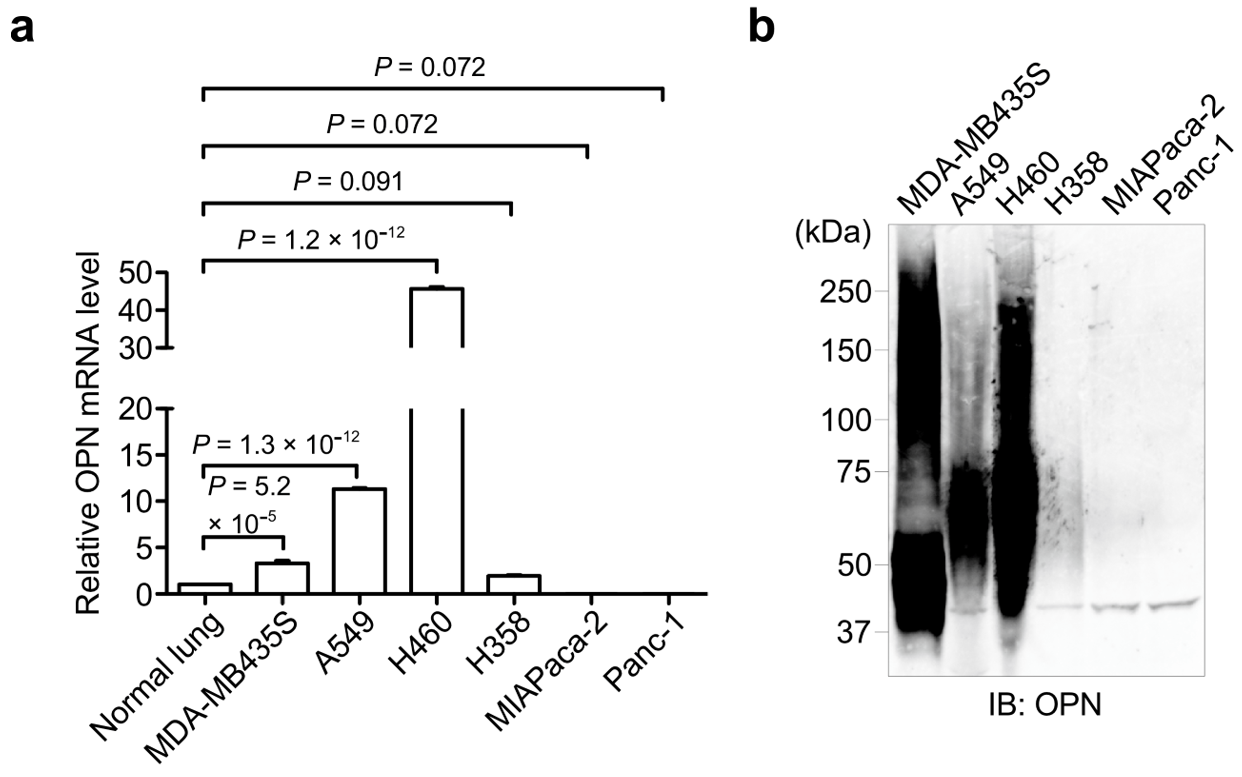
3.1. OPN Expression in Cancer Cell Lines
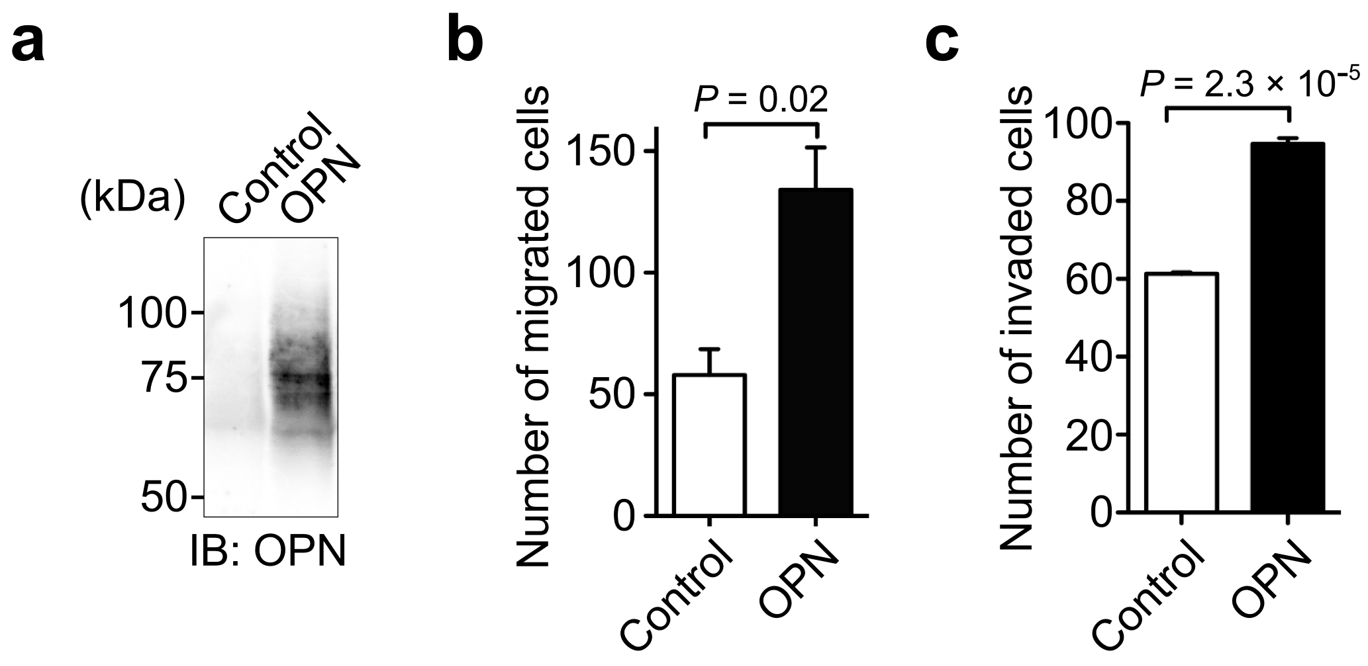
3.2. Identification of OPN as a Cell Motility Inducing Factor in Culture Media of Cancer Cells
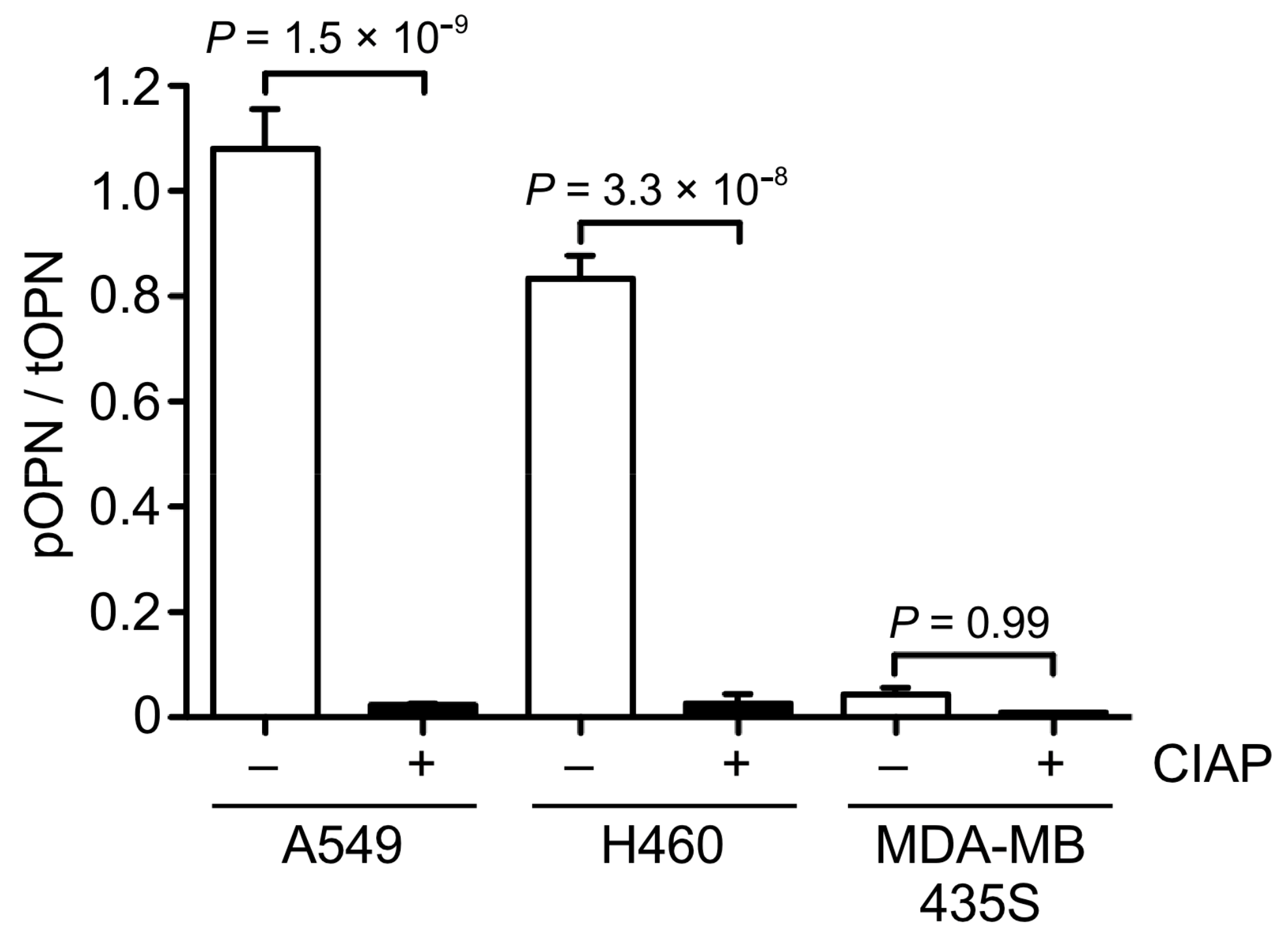
3.3. OPN Phosphorylation in Cancer Cells
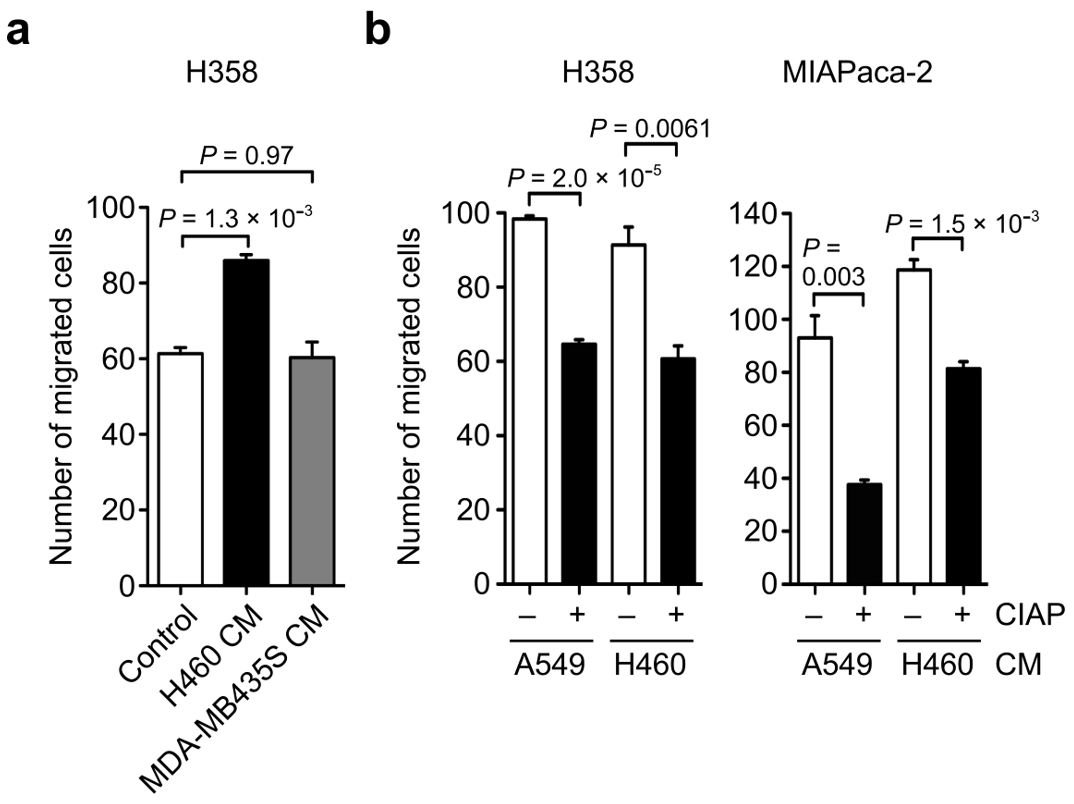
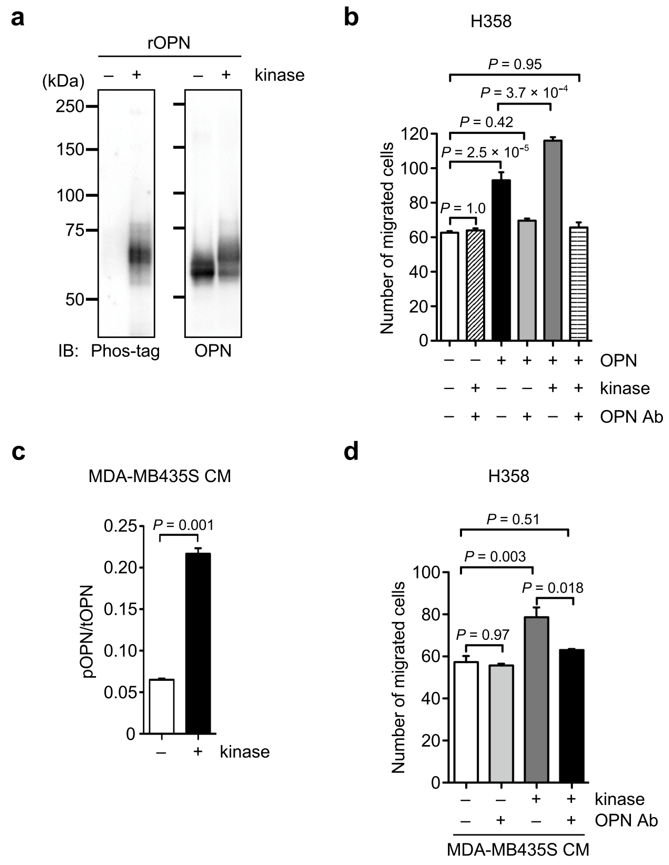
3.4. Effect of OPN Phosphorylation on OPN-Induced Cancer Cell Motility
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509, 582–587. [Google Scholar] [CrossRef]
- Lee, M.J.; Yaffe, M.B. Protein Regulation in Signal Transduction. Cold Spring Harb. Perspect. Biol. 2016, 8, a005918. [Google Scholar] [CrossRef]
- Yalak, G.; Olsen, B.R. Proteomic database mining opens up avenues utilizing extracellular protein phosphorylation for novel therapeutic applications. J. Transl. Med. 2015, 13, 125. [Google Scholar] [CrossRef]
- Yalak, G.; Vogel, V. Extracellular phosphorylation and phosphorylated proteins: Not just curiosities but physiologically important. Sci. Signal. 2012, 5, re7. [Google Scholar] [CrossRef]
- Tagliabracci, V.S.; Engel, J.L.; Wen, J.; Wiley, S.E.; Worby, C.A.; Kinch, L.N.; Xiao, J.; Grishin, N.V.; Dixon, J.E. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science 2012, 336, 1150–1153. [Google Scholar] [CrossRef]
- Bordoli, M.R.; Yum, J.; Breitkopf, S.B.; Thon, J.N.; Italiano, J.E., Jr.; Xiao, J.; Worby, C.; Wong, S.K.; Lin, G.; Edenius, M.; et al. A secreted tyrosine kinase acts in the extracellular environment. Cell 2014, 158, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Babiker, A.A.; Ronquist, G.; Nilsson, B.; Ekdahl, K.N. Overexpression of ecto-protein kinases in prostasomes of metastatic cell origin. Prostate 2006, 66, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Zawadzka, A.M.; Schilling, B.; Cusack, M.P.; Sahu, A.K.; Drake, P.; Fisher, S.J.; Benz, C.C.; Gibson, B.W. Phosphoprotein secretome of tumor cells as a source of candidates for breast cancer biomarkers in plasma. Mol. Cell Proteomics 2014, 13, 1034–1049. [Google Scholar] [CrossRef] [PubMed]
- Senger, D.R.; Wirth, D.F.; Hynes, R.O. Transformed mammalian cells secrete specific proteins and phosphoproteins. Cell 1979, 16, 885–893. [Google Scholar] [CrossRef]
- Senger, D.R.; Wirth, D.F.; Hynes, R.O. Transformation-specific secreted phosophoproteins. Nature 1980, 286, 619–621. [Google Scholar] [CrossRef] [PubMed]
- Senger, D.R.; Perruzzi, C.A.; Gracey, C.F.; Papadopoulos, A.; Tenen, D.G. Secreted phosphoproteins associated with neoplastic transformation: Close homology with plasma proteins cleaved during blood coagulation. Cancer Res. 1988, 48, 5770–5774. [Google Scholar] [PubMed]
- Subraman, V.; Thiyagarajan, M.; Malathi, N.; Rajan, S.T. OPN -Revisited. J. Clin. Diagn. Res. 2015, 9, ZE10–ZE13. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Wong, J.P.C.; Kwok, H.F. Osteopontin—A promising biomarker for cancer therapy. J. Cancer 2017, 8, 2173–2183. [Google Scholar] [CrossRef]
- Shevde, L.A.; Samant, R.S. Role of osteopontin in the pathophysiology of cancer. Matrix Biol. 2014, 37, 131–141. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, Q.; Alam, A.; Cui, J.; Suen, K.C.; Soo, A.P.; Eguchi, S.; Gu, J.; Ma, D. The role of osteopontin in the progression of solid organ tumour. Cell Death Dis. 2018, 9, 356. [Google Scholar] [CrossRef] [PubMed]
- Kariya, Y.; Kanno, M.; Matsumoto-Morita, K.; Konno, M.; Yamaguchi, Y.; Hashimoto, Y. Osteopontin O-glycosylation contributes to its phosphorylation and cell-adhesion properties. Biochem. J. 2014, 463, 93–102. [Google Scholar] [CrossRef]
- Christensen, B.; Nielsen, M.S.; Haselmann, K.F.; Petersen, T.E.; Sorensen, E.S. Post-translationally modified residues of native human osteopontin are located in clusters: Identification of 36 phosphorylation and five O-glycosylation sites and their biological implications. Biochem. J. 2005, 390, 285–292. [Google Scholar] [CrossRef]
- Li, H.; Shen, H.; Yan, G.; Zhang, Y.; Liu, M.; Fang, P.; Yu, H.; Yang, P. Site-specific structural characterization of O-glycosylation and identification of phosphorylation sites of recombinant osteopontin. Biochim. Biophys. Acta 2015, 1854, 581–591. [Google Scholar] [CrossRef]
- Oyama, M.; Kariya, Y.; Kariya, Y.; Matsumoto, K.; Kanno, M.; Yamaguchi, Y.; Hashimoto, Y. Biological role of site-specific O-glycosylation in cell adhesion activity and phosphorylation of osteopontin. Biochem. J. 2018, 475, 1583–1595. [Google Scholar] [CrossRef]
- Zhu, X.; Luo, C.; Ferrier, J.M.; Sodek, J. Evidence of ectokinase-mediated phosphorylation of osteopontin and bone sialoprotein by osteoblasts during bone formation in vitro. Biochem. J. 1997, 323, 637–643. [Google Scholar] [CrossRef]
- Tagliabracci, V.S.; Wiley, S.E.; Guo, X.; Kinch, L.N.; Durrant, E.; Wen, J.; Xiao, J.; Cui, J.; Nguyen, K.B.; Engel, J.L.; et al. A Single Kinase Generates the Majority of the Secreted Phosphoproteome. Cell 2015, 161, 1619–1632. [Google Scholar] [CrossRef] [PubMed]
- Safran, J.B.; Butler, W.T.; Farach-Carson, M.C. Modulation of osteopontin post-translational state by 1, 25-(OH)2-vitamin D3. Dependence on Ca2+ influx. J. Biol. Chem. 1998, 273, 29935–29941. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nemir, M.; DeVouge, M.W.; Mukherjee, B.B. Normal rat kidney cells secrete both phosphorylated and nonphosphorylated forms of osteopontin showing different physiological properties. J. Biol. Chem. 1989, 264, 18202–18208. [Google Scholar] [CrossRef]
- Ek-Rylander, B.; Flores, M.; Wendel, M.; Heinegard, D.; Andersson, G. Dephosphorylation of osteopontin and bone sialoprotein by osteoclastic tartrate-resistant acid phosphatase. Modulation of osteoclast adhesion in vitro. J. Biol. Chem. 1994, 269, 14853–14856. [Google Scholar] [CrossRef]
- Christensen, B.; Klaning, E.; Nielsen, M.S.; Andersen, M.H.; Sorensen, E.S. C-terminal Modification of Osteopontin Inhibits Interaction with the alphaVbeta3-Integrin. J. Biol. Chem. 2012, 287, 3788–3797. [Google Scholar] [CrossRef]
- Katayama, Y.; House, C.M.; Udagawa, N.; Kazama, J.J.; McFarland, R.J.; Martin, T.J.; Findlay, D.M. Casein kinase 2 phosphorylation of recombinant rat osteopontin enhances adhesion of osteoclasts but not osteoblasts. J. Cell Physiol. 1998, 176, 179–187. [Google Scholar] [CrossRef]
- Schytte, G.N.; Christensen, B.; Bregenov, I.; Kjøge, K.; Scavenius, C.; Petersen, S.V.; Enghild, J.J.; Sørensen, E.S. FAM20C phosphorylation of the RGDSVVYGLR motif in osteopontin inhibits interaction with the αvβ3 integrin. J. Cell Biochem. 2020, 121, 4809–4818. [Google Scholar] [CrossRef] [PubMed]
- Al-Shami, R.; Sorensen, E.S.; Ek-Rylander, B.; Andersson, G.; Carson, D.D.; Farach-Carson, M.C. Phosphorylated osteopontin promotes migration of human choriocarcinoma cells via a p70 S6 kinase-dependent pathway. J. Cell Biochem. 2005, 94, 1218–1233. [Google Scholar] [CrossRef]
- Pellegatti, P.; Raffaghello, L.; Bianchi, G.; Piccardi, F.; Pistoia, V.; Di Virgilio, F. Increased level of extracellular ATP at tumor sites: In vivo imaging with plasma membrane luciferase. PLoS ONE 2008, 3, e2599. [Google Scholar] [CrossRef]
- Britton, D.; Zen, Y.; Quaglia, A.; Selzer, S.; Mitra, V.; Lobetaner, C.; Jung, S.; Bohm, G.; Schmid, P.; Prefot, P.; et al. Quantification of pancreatic cancer proteome and phosphorylome: Indicates molecular events likely contributing to cancer and activity of drug targets. PLoS ONE 2014, 9, e90948. [Google Scholar] [CrossRef]
- Schweppe, D.K.; Rigas, J.R.; Gerber, S.A. Quantitative phosphoproteomic profiling of human non-small cell lung cancer tumors. J. Proteomics 2013, 91, 286–296. [Google Scholar] [CrossRef]
- Liu, X.; Zhan, Y.; Xu, W.; Liu, X.; Geng, Y.; Liu, L.; Da, J.; Wang, J.; Zhang, X.; Jin, H.; et al. Prognostic and immunological role of Fam20C in pan-cancer. Biosci. Rep. 2021, 41, BSR20201920. [Google Scholar] [CrossRef]
- Du, S.; Guan, S.; Zhu, C.; Guo, Q.; Cao, J.; Guan, G.; Cheng, W.; Cheng, P.; Wu, A. Secretory Pathway Kinase FAM20C, a Marker for Glioma Invasion and Malignancy, Predicts Poor Prognosis of Glioma. Onco. Targets Ther. 2020, 13, 11755–11768. [Google Scholar] [CrossRef]
- Schytte, G.N.; Christensen, B.; Bregenov, I.; Sørensen, E.S. Ras-transformation reduce FAM20C expression and osteopontin phosphorylation. Biosci. Rep. 2020, 40, BSR20194378. [Google Scholar] [CrossRef] [PubMed]
- Kazanecki, C.C.; Uzwiak, D.J.; Denhardt, D.T. Control of osteopontin signaling and function by post-translational phosphorylation and protein folding. J. Cell Biochem. 2007, 102, 912–924. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Hanashima, S.; Yagi, H.; Takahashi, Y.; Sasakawa, H.; Kurimoto, E.; Iguchi, T.; Kon, S.; Uede, T.; Kato, K. NMR characterization of intramolecular interaction of osteopontin, an intrinsically disordered protein with cryptic integrin-binding motifs. Biochem. Biophys. Res. Commun. 2010, 393, 487–491. [Google Scholar] [CrossRef]
- Platzer, G.; Schedlbauer, A.; Chemelli, A.; Ozdowy, P.; Coudevylle, N.; Auer, R.; Kontaxis, G.; Hartl, M.; Miles, A.J.; Wallace, B.A.; et al. The metastasis-associated extracellular matrix protein osteopontin forms transient structure in ligand interaction sites. Biochemistry 2011, 50, 6113–6124. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Bah, A.; Vernon, R.M.; Siddiqui, Z.; Krzeminski, M.; Muhandiram, R.; Zhao, C.; Sonenberg, N.; Kay, L.E.; Forman-Kay, J.D. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 2015, 519, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Bah, A.; Forman-Kay, J.D. Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. J. Biol. Chem. 2016, 291, 6696–6705. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kariya, Y.; Oyama, M.; Kariya, Y.; Hashimoto, Y. Phosphorylated Osteopontin Secreted from Cancer Cells Induces Cancer Cell Motility. Biomolecules 2021, 11, 1323. https://doi.org/10.3390/biom11091323
Kariya Y, Oyama M, Kariya Y, Hashimoto Y. Phosphorylated Osteopontin Secreted from Cancer Cells Induces Cancer Cell Motility. Biomolecules. 2021; 11(9):1323. https://doi.org/10.3390/biom11091323
Chicago/Turabian StyleKariya, Yoshinobu, Midori Oyama, Yukiko Kariya, and Yasuhiro Hashimoto. 2021. "Phosphorylated Osteopontin Secreted from Cancer Cells Induces Cancer Cell Motility" Biomolecules 11, no. 9: 1323. https://doi.org/10.3390/biom11091323
APA StyleKariya, Y., Oyama, M., Kariya, Y., & Hashimoto, Y. (2021). Phosphorylated Osteopontin Secreted from Cancer Cells Induces Cancer Cell Motility. Biomolecules, 11(9), 1323. https://doi.org/10.3390/biom11091323




