Inhibition of UBA5 Expression and Induction of Autophagy in Breast Cancer Cells by Usenamine A
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis
2.2. Enrichment Analysis of Gene Ontology (GO)
2.3. Cell Culture
2.4. Cell Viability Assay
2.5. Microscopic Analysis
2.6. Colony Formation Assay
2.7. Staining Assay Using 5-Ethynyl-2′-Deoxyuridine (EdU)
2.8. Cell Cycle Distribution Analysis
2.9. Cell Apoptosis Analysis
2.10. Transwell Assay
2.11. Plasmids
2.12. Western Blot
2.13. RFP-LC3B Puncta Assay
2.14. Molecular Docking
2.15. Isolation of Usenamine A
2.16. Statistical Analysis
3. Results
3.1. UBA5 Is Highly Expressed in Breast Cancer and Correlates with Poor Prognosis
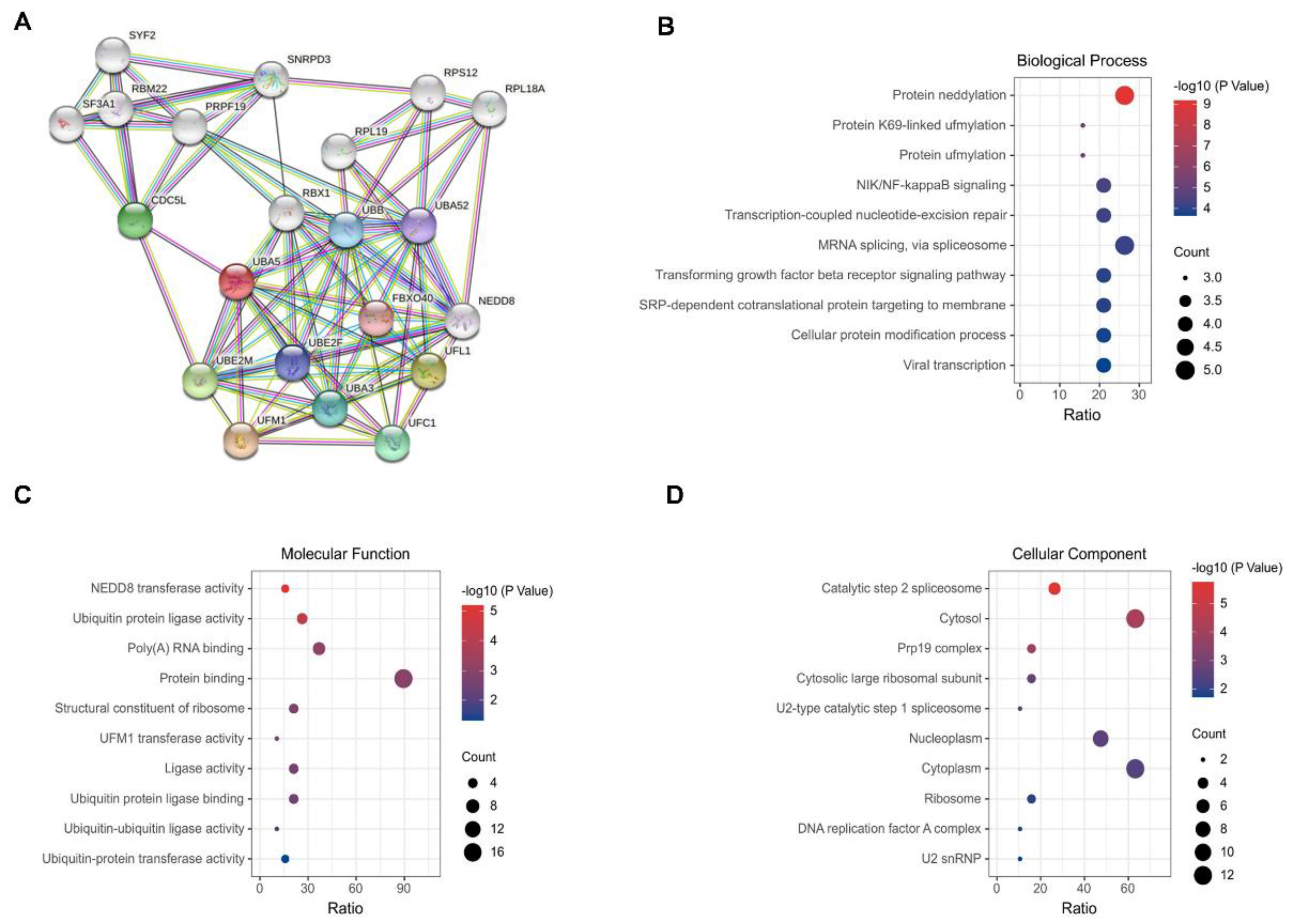
3.2. Construction of a PPI Network and GO Analysis
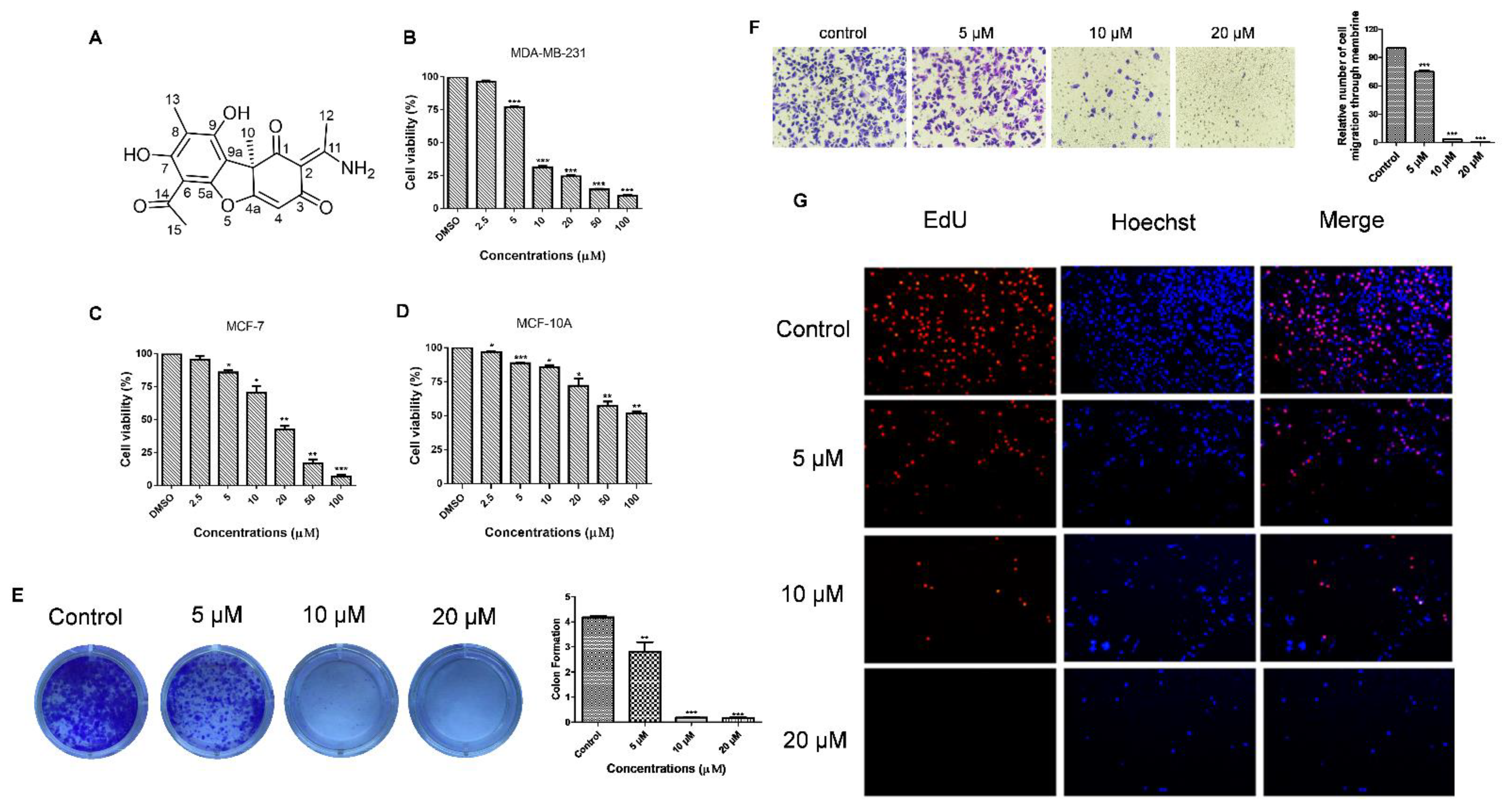
3.3. Usenamine A Inhibits the Proliferation and Invasion of MDA-MB-231 Cells
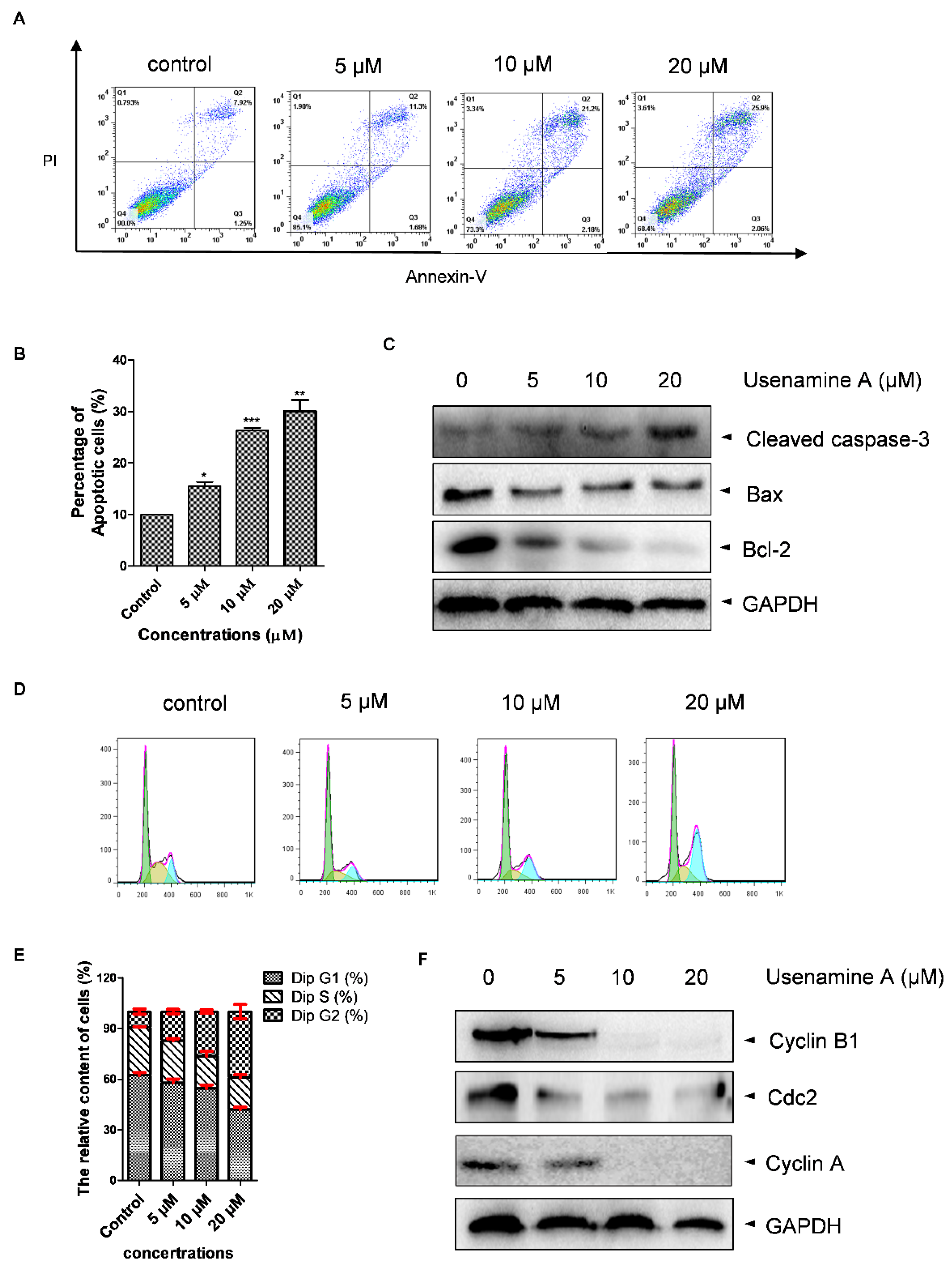
3.4. Usenamine A Induces Cell Apoptosis and G2/M Phase Arrest in MDA-MB-231 Cells
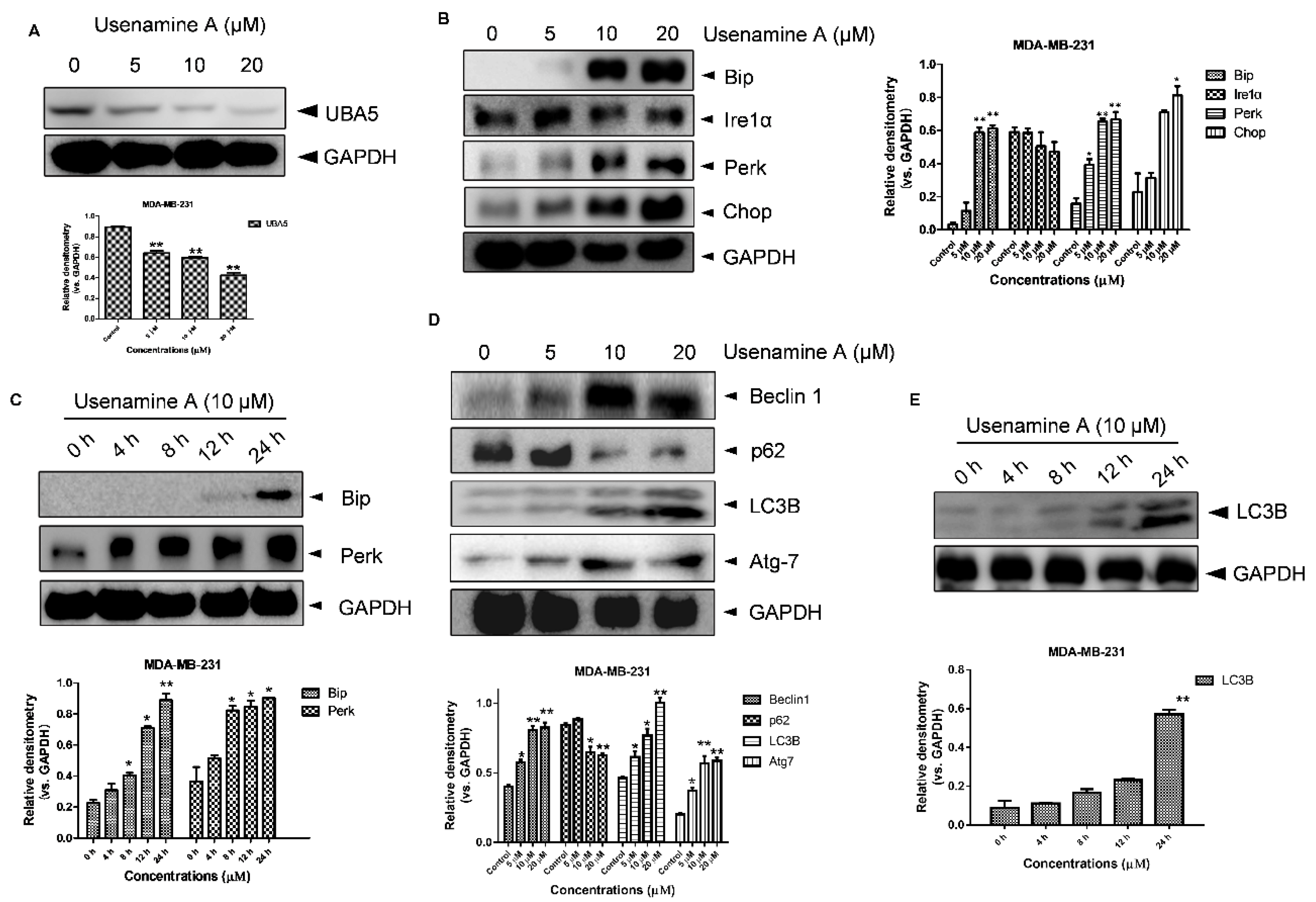
3.5. Usenamine A Activates ER Stress and Induces Autophagy in MDA-MB-231 Cells
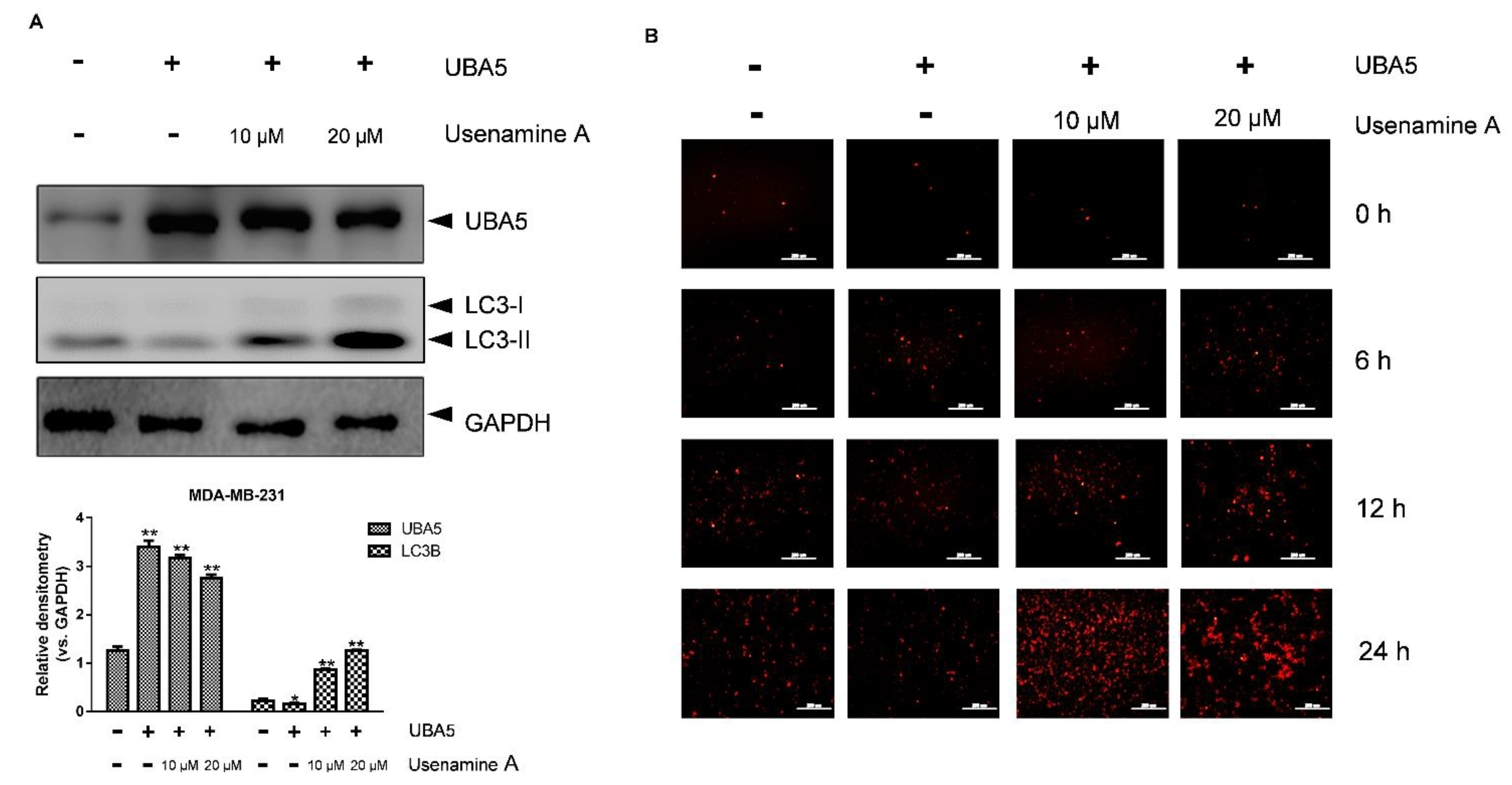
3.6. Overexpression of UBA5 Can Reduce Autophagy in MDA-MB-231 Cells
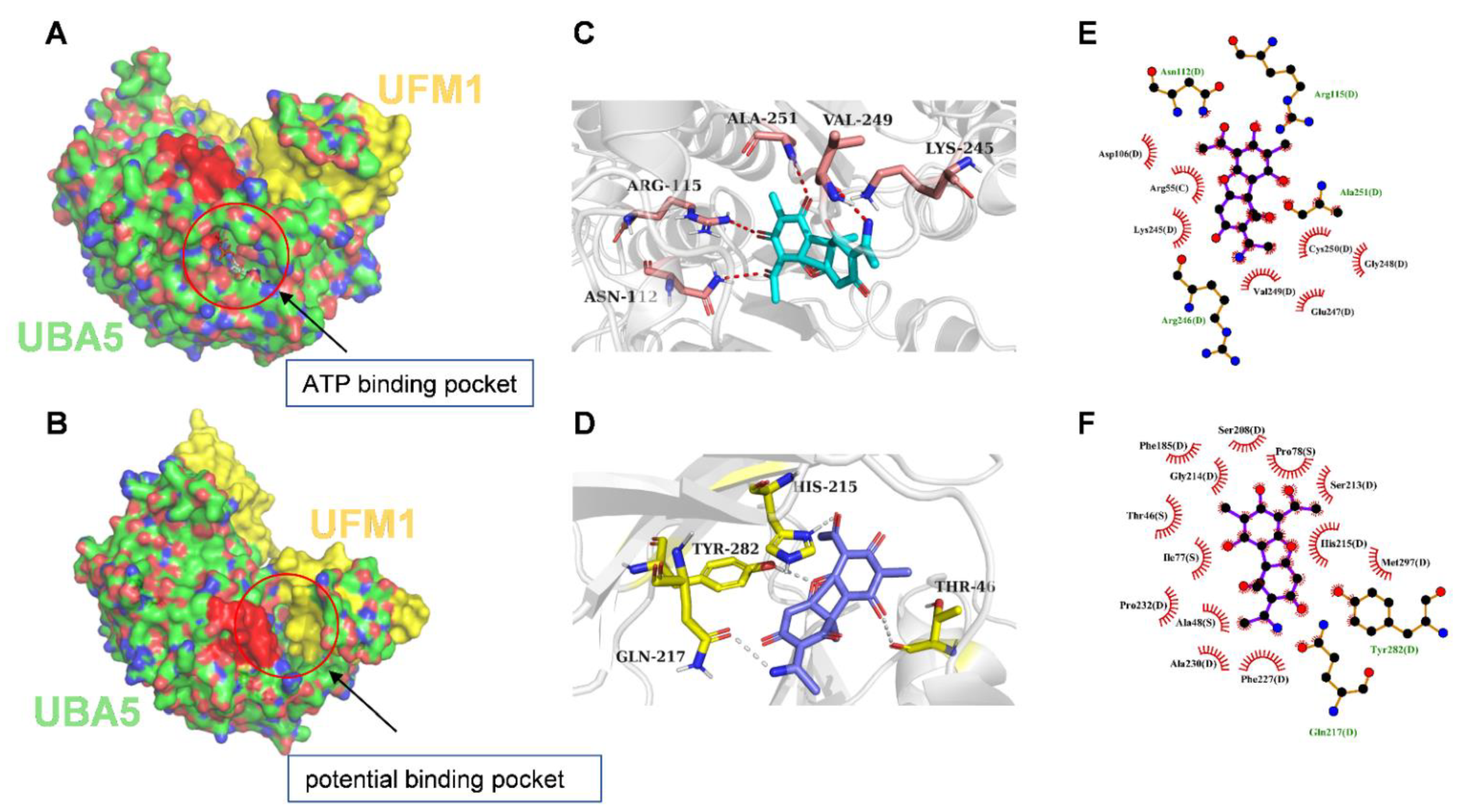
3.7. Molecular Docking of UBA5 to Usenamine A
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Patten, D.K.; Corleone, G.; Győrffy, B.; Perone, Y.; Slaven, N.; Barozzi, I.; Erdős, E.; Saiakhova, A.; Goddard, K.; Vingiani, A.; et al. Enhancer mapping uncovers phenotypic heterogeneity and evolution in patients with luminal breast cancer. Nat. Med. 2018, 24, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.W.; Huang, C.Y.; Tam, K.W.; Lin, C.Y.; Huang, L.C.; Lin, C.L.; Hsieh, W.S.; Chi, W.M.; Chang, Y.J.; Wei, P.L.; et al. The Application of Non-Invasive Apoptosis Detection Sensor (NIADS) on Histone Deacetylation Inhibitor (HDACi)-Induced Breast Cancer Cell Death. Int J. Mol. Sci. 2018, 19, 452. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.P.; Xu, T.Q.; Liu, B.L.; Lei, X.P.; Hambrook, J.R.; Zhang, D.M.; Zhou, G.X. Sasanquasaponin ΙΙΙ from Schima crenata Korth induces autophagy through Akt/mTOR/p70S6K pathway and promotes apoptosis in human melanoma A375 cells. Phytomedicine 2019, 58, 152769. [Google Scholar] [CrossRef]
- Wong, R.S. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011, 30, 87. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, N.; Liu, K.; Zhou, G.; Gan, J.; Wang, Z.; Shi, T.; He, W.; Wang, L.; Guo, T.; et al. Autophagy mediated CoCrMo particle-induced peri-implant osteolysis by promoting osteoblast apoptosis. Autophagy 2015, 11, 2358–2369. [Google Scholar] [CrossRef]
- Gerakis, Y.; Quintero, M.; Li, H.; Hetz, C. The UFMylation System in Proteostasis and Beyond. Trends Cell Biol. 2019, 29, 974–986. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, K.; Moura, R.F.; Granvik, M.; Igoillo-Esteve, M.; Hohmeier, H.E.; Hendrickx, N.; Newgard, C.B.; Waelkens, E.; Cnop, M.; Schuit, F. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLoS ONE 2011, 6, e18517. [Google Scholar]
- Zhu, H.; Bhatt, B.; Sivaprakasam, S.; Cai, Y.; Liu, S.; Kodeboyina, S.K.; Patel, N.; Savage, N.M.; Sharma, A.; Kaufman, R.J.; et al. Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR. Nat. Commun 2019, 10, 1084. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.M.; Park, J.H.; Jeon, Y.J.; Chung, C.H. Ubiquitin-fold modifier 1 acts as a positive regulator of breast cancer. Front. Endocrinol. 2015, 6, 36. [Google Scholar] [CrossRef]
- Yoo, H.M.; Kang, S.H.; Kim, J.Y.; Lee, J.E.; Seong, M.W.; Lee, S.W.; Ka, S.H.; Sou, Y.S.; Komatsu, M.; Tanaka, K.; et al. Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol. Cell 2014, 56, 261–274. [Google Scholar] [CrossRef]
- Roberts, A.M.; Miyamoto, D.K.; Huffman, T.R.; Bateman, L.A.; Ives, A.N.; Akopian, D.; Heslin, M.J.; Contreras, C.M.; Rape, M.; Skibola, C.F.; et al. Chemoproteomic Screening of Covalent Ligands Reveals UBA5 As a Novel Pancreatic Cancer Target. ACS Chem. Biol. 2017, 12, 899–904. [Google Scholar] [CrossRef]
- Da Silva, S.R.; Paiva, S.L.; Bancerz, M.; Geletu, M.; Lewis, A.M.; Chen, J.; Cai, Y.; Lukkarila, J.L.; Li, H.; Gunning, P.T. A selective inhibitor of the UFM1-activating enzyme, UBA5. Bioorg. Med. Chem. Lett. 2016, 26, 4542–4547. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. J. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- The Gene Expression Profiling Interactive Analysis. Available online: https://www.gepia.cancer-pku.cn/ (accessed on 9 September 2021).
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (N. Y.) 2017, 19, 649–658. [Google Scholar] [CrossRef]
- UALCAN. Available online: https://www.ualcan.path.uab.edu/ (accessed on 9 September 2021).
- Szklarczyk, D.; Gable, A.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.; Morris, J.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. J. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- STRING. Available online: https://www.string-db.org/ (accessed on 9 September 2021).
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Tao, Y.; Shan, L.; Xu, X.; Jiang, H.; Chen, R.; Qian, Z.; Yang, Z.; Liang, B.; Zheng, H.; Cai, F.; et al. Huaier Augmented the Chemotherapeutic Sensitivity of Oxaliplatin via Downregulation of YAP in Hepatocellular Carcinoma. J. Cancer 2018, 9, 3962–3970. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- The protein Data Bank. Available online: https://www.rcsb.org/ (accessed on 9 September 2021).
- Soudah, N.; Padala, P.; Hassouna, F.; Kumar, M.; Mashahreh, B.; Lebedev, A.A.; Isupov, M.N.; Cohen-Kfir, E.; Wiener, R. An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation. J. Mol. Biol. 2019, 431, 463–478. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- He, L.F.; Xu, H.W.; Chen, M.; Xian, Z.R.; Wen, X.F.; Chen, M.N.; Du, C.W.; Huang, W.H.; Wu, J.D.; Zhang, G.J. Activated-PAK4 predicts worse prognosis in breast cancer and promotes tumorigenesis through activation of PI3K/AKT signaling. Oncotarget 2017, 8, 17573–17585. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.R.; Lingeman, E.; Luong, T.; Ahmed, S.; Muhar, M.; Nguyen, T.; Olzmann, J.A.; Corn, J.E. A Genome-wide ER-phagy Screen Highlights Key Roles of Mitochondrial Metabolism and ER-Resident UFMylation. Cell 2020, 180, 1160–1177.e20. [Google Scholar] [CrossRef] [PubMed]
- Gavin, J.M.; Hoar, K.; Xu, Q.; Ma, J.; Lin, Y.; Chen, J.; Chen, W.; Bruzzese, F.J.; Harrison, S.; Mallender, W.D.; et al. Mechanistic study of Uba5 enzyme and the Ufm1 conjugation pathway. J. Biol. Chem. 2014, 289, 22648–22658. [Google Scholar]
- Cai, Y.; Pi, W.; Sivaprakasam, S.; Zhu, X.; Zhang, M.; Chen, J.; Makala, L.; Lu, C.; Wu, J.; Teng, Y.; et al. UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development. PLoS Genet. 2015, 11, e1005643. [Google Scholar] [CrossRef] [PubMed]
- Heath-Engel, H.M.; Chang, N.C.; Shore, G.C. The endoplasmic reticulum in apoptosis and autophagy: Role of the BCL-2 protein family. Oncogene 2008, 27, 6419–6433. [Google Scholar] [CrossRef] [PubMed]
- Clarke, R.; Cook, K.L.; Hu, R.; Facey, C.O.; Tavassoly, I.; Schwartz, J.L.; Baumann, W.T.; Tyson, J.J.; Xuan, J.; Wang, Y.; et al. Endoplasmic reticulum stress, the unfolded protein response, autophagy, and the integrated regulation of breast cancer cell fate. Cancer Res. 2012, 72, 1321–1331. [Google Scholar]
- Wang, R.; Deng, X.; Yoshioka, Y.; Vougiouklakis, T.; Park, J.H.; Suzuki, T.; Dohmae, N.; Ueda, K.; Hamamoto, R.; Nakamura, Y. Effects of SMYD2-mediated EML4-ALK methylation on the signaling pathway and growth in non-small-cell lung cancer cells. Cancer Sci. 2017, 108, 1203–1209. [Google Scholar] [CrossRef] [PubMed]
- Moll, L.; Roitenberg, N.; Bejerano-Sagie, M.; Boocholez, H.; Carvalhal Marques, F.; Volovik, Y.; Elami, T.; Siddiqui, A.A.; Grushko, D.; Biram, A.; et al. The insulin/IGF signaling cascade modulates SUMOylation to regulate aging and proteostasis in Caenorhabditis elegans. Elife 2018, 7, e38635. [Google Scholar] [CrossRef] [PubMed]
- Veggiani, G.; Gerpe, M.C.R.; Sidhu, S.S.; Zhang, W. Emerging drug development technologies targeting ubiquitination for cancer therapeutics. Pharmacol. Ther. 2019, 199, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Xu, X. UFMylation: A Unique & Fashionable Modification for Life. Genom. Proteom. Bioinform. 2016, 14, 140–146. [Google Scholar]
- Xi, P.; Ding, D.; Zhou, J.; Wang, M.; Cong, Y.S. DDRGK1 regulates NF-κB activity by modulating IκBα stability. PLoS ONE 2013, 8, e64231. [Google Scholar]
- Liu, J.; Guan, D.; Dong, M.; Yang, J.; Wei, H.; Liang, Q.; Song, L.; Xu, L.; Bai, J.; Liu, C.; et al. UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nat. Cell Biol. 2020, 22, 1056–1063. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Shen, Q.; Li, J. Traditional Chinese medicine targeting apoptotic mechanisms for esophageal cancer therapy. Acta Pharmacol. Sin. 2016, 37, 295–302. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, C.; Li, X.; Zhang, Y. Effects of pterostilbene on treating hyperprolactinemia and related mechanisms. Am. J. Transl. Res. 2016, 8, 3049–3055. [Google Scholar] [PubMed]
- Senavirathna, L.K.; Huang, C.; Yang, X.; Munteanu, M.C.; Sathiaseelan, R.; Xu, D.; Henke, C.A.; Liu, L. Hypoxia induces pulmonary fibroblast proliferation through NFAT signaling. Sci. Rep. 2018, 8, 2709. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, M.; Wu, J.; Lei, G.; Li, H. Transcriptional regulation of the Ufm1 conjugation system in response to disturbance of the endoplasmic reticulum homeostasis and inhibition of vesicle trafficking. PLoS ONE 2012, 7, e48587. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Song, L.; Zeng, L.; Yi, W.; Liu, T.; Chen, H.; Wang, M.; Ju, Z.; Cong, Y.S. A critical role of DDRGK1 in endoplasmic reticulum homoeostasis via regulation of IRE1α stability. Nat. Commun. 2017, 8, 14186. [Google Scholar] [CrossRef]
- Cao, Y.; Li, R.; Shen, M.; Li, C.; Zou, Y.; Jiang, Q.; Liu, S.; Lu, C.; Li, H.; Liu, H.; et al. DDRGK1, a crucial player of ufmylation system, is indispensable for autophagic degradation by regulating lysosomal function. Cell Death Dis. 2021, 12, 416. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, J.; Li, K.; Deng, L.; Wang, H. Combination of an Autophagy Inducer and an Autophagy Inhibitor: A Smarter Strategy Emerging in Cancer Therapy. Front. Pharmacol. 2020, 11, 408. [Google Scholar] [CrossRef]
- Wu, Y.T.; Tan, H.L.; Shui, G.; Bauvy, C.; Huang, Q.; Wenk, M.R.; Ong, C.N.; Codogno, P.; Shen, H.M. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 2010, 285, 10850–10861. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T. How to interpret LC3 immunoblotting. Autophagy 2007, 3, 542–545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Teaching the basics of autophagy and mitophagy to redox biologists--mechanisms and experimental approaches. Redox Biol. 2015, 4, 242–259. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in mammalian autophagy research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Kelly, C.; Challacombe, S.J.; Donaldson, A.N.; Bhogal, B.S.; Setterfield, J.F. Serum and salivary IgG and IgA antibodies to desmoglein 3 in mucosal pemphigus vulgaris. Br. J. Dermatol. 2016, 175, 113–121. [Google Scholar] [CrossRef]
- Vidal, R.L.; Matus, S.; Bargsted, L.; Hetz, C. Targeting autophagy in neurodegenerative diseases. Trends Pharmacol. Sci. 2014, 35, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.P.; Debnath, J. Therapeutic implications of autophagy-mediated cell survival in gastrointestinal stromal tumor after treatment with imatinib mesylate. Autophagy 2010, 6, 1190–1191. [Google Scholar] [CrossRef][Green Version]
- Yang, L.; Xie, P.; Wu, J.; Yu, J.; Li, X.; Ma, H.; Yu, T.; Wang, H.; Ye, J.; Wang, J.; et al. Deferoxamine Treatment Combined With Sevoflurane Postconditioning Attenuates Myocardial Ischemia-Reperfusion Injury by Restoring HIF-1/BNIP3-Mediated Mitochondrial Autophagy in GK Rats. Front. Pharmacol. 2020, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Tan, J.; Miao, Y.; Li, M.; Zhang, Q. Crosstalk of autophagy and apoptosis: Involvement of the dual role of autophagy under ER stress. J. Cell. Physiol. 2017, 232, 2977–2984. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, B.; Li, Z.; Qiu, Y.; Cho, N.; Yoo, H.M. Inhibition of UBA5 Expression and Induction of Autophagy in Breast Cancer Cells by Usenamine A. Biomolecules 2021, 11, 1348. https://doi.org/10.3390/biom11091348
Fang B, Li Z, Qiu Y, Cho N, Yoo HM. Inhibition of UBA5 Expression and Induction of Autophagy in Breast Cancer Cells by Usenamine A. Biomolecules. 2021; 11(9):1348. https://doi.org/10.3390/biom11091348
Chicago/Turabian StyleFang, Bo, Zijun Li, Yinda Qiu, Namki Cho, and Hee Min Yoo. 2021. "Inhibition of UBA5 Expression and Induction of Autophagy in Breast Cancer Cells by Usenamine A" Biomolecules 11, no. 9: 1348. https://doi.org/10.3390/biom11091348
APA StyleFang, B., Li, Z., Qiu, Y., Cho, N., & Yoo, H. M. (2021). Inhibition of UBA5 Expression and Induction of Autophagy in Breast Cancer Cells by Usenamine A. Biomolecules, 11(9), 1348. https://doi.org/10.3390/biom11091348






