Shock-Induced Damage Mechanism of Perineuronal Nets
Abstract
:1. Introduction
- The link protein (LP) and core protein (CP) of proteoglycan (PG),
- Tenascin-R (TR) and CP,
- The LP and hyaluronan (HA),
2. Methodology
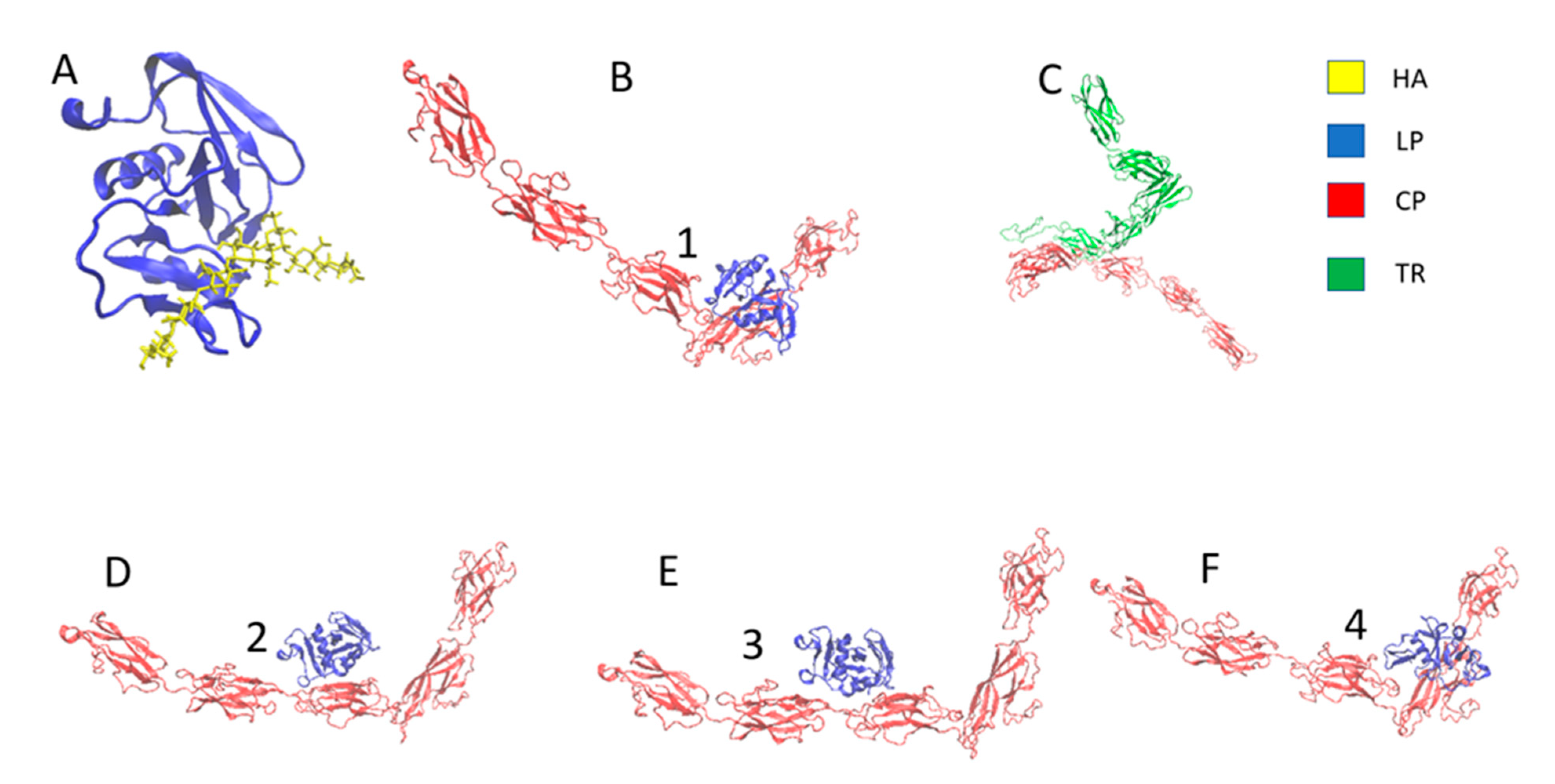
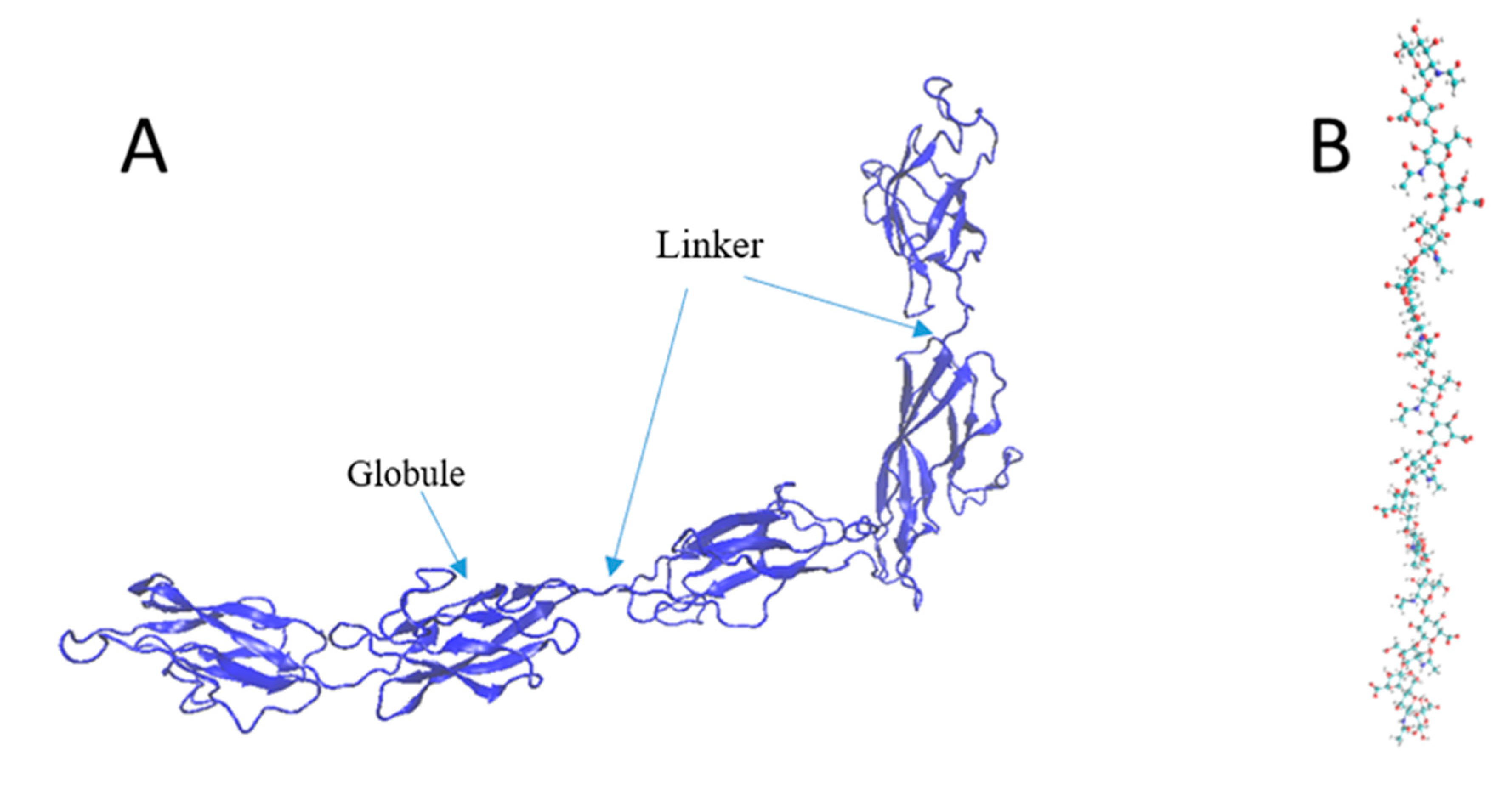
2.1. Modeling of the PNN Structure
2.2. Interfacial Strength of PNN Components
2.3. Mechanical Property Evaluation of PNN Components
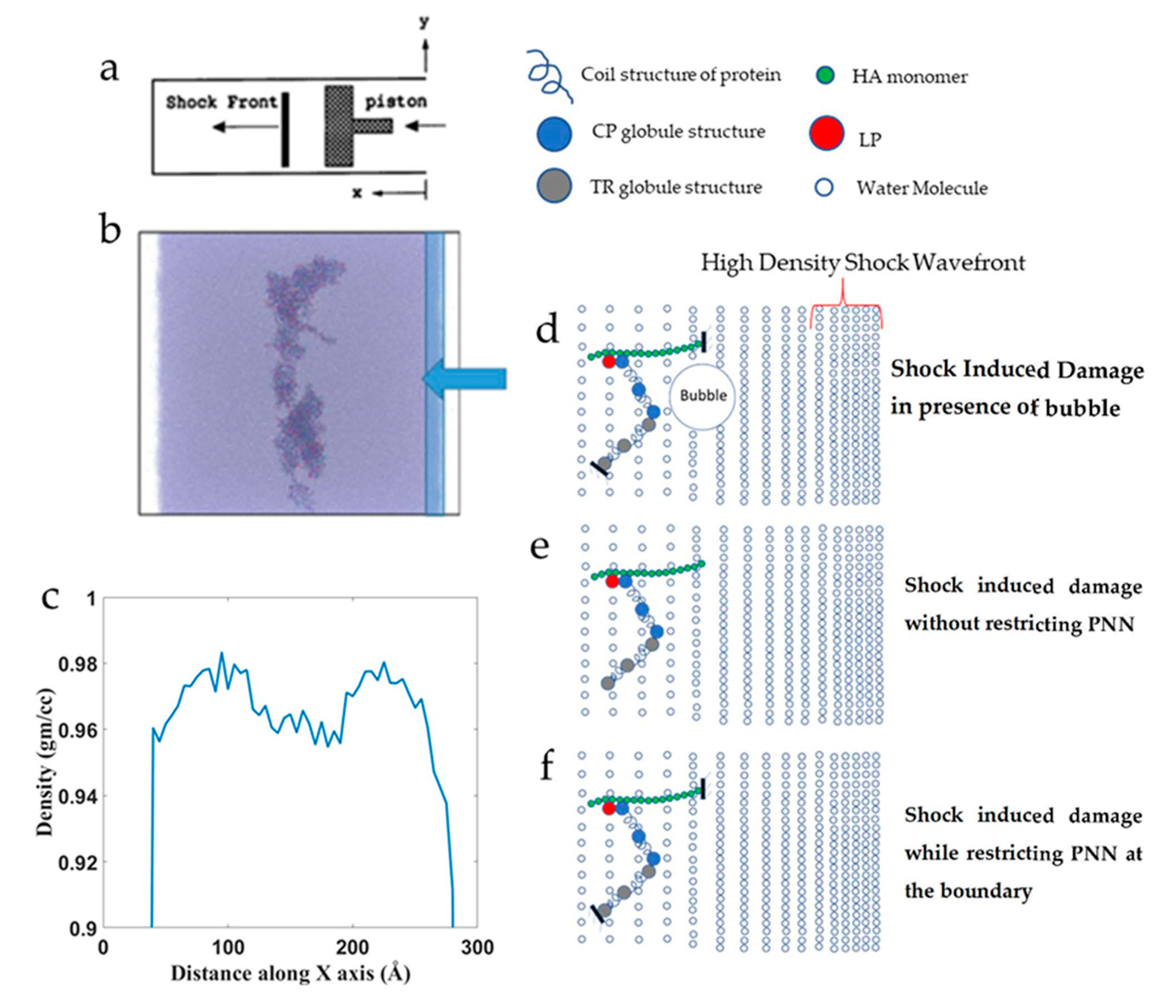
2.4. Shock Simulation
3. Results and Discussions
3.1. Mechanical Strength of the PNN Components
3.2. Interfacial Strength
3.3. Shock Simulation
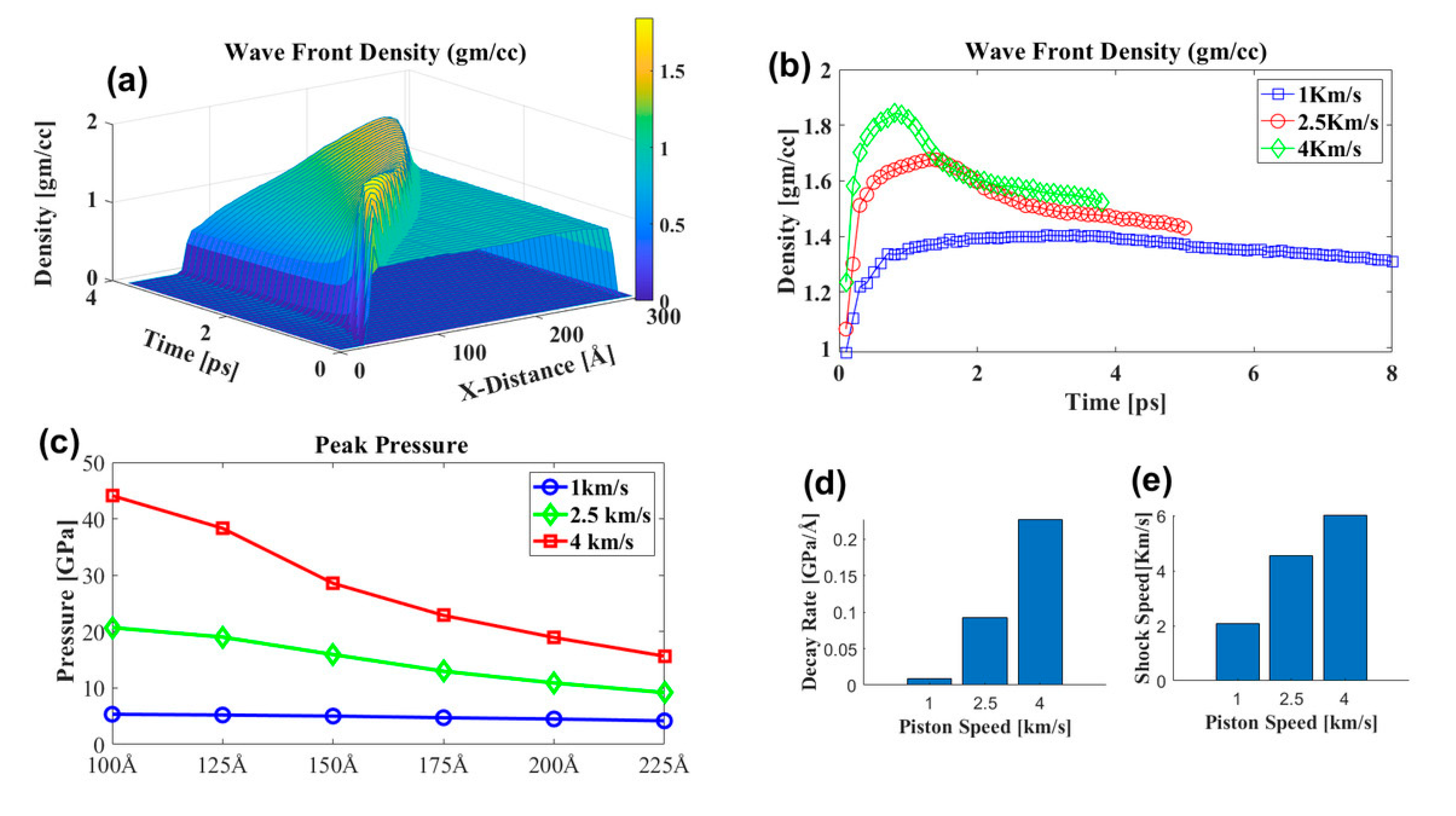
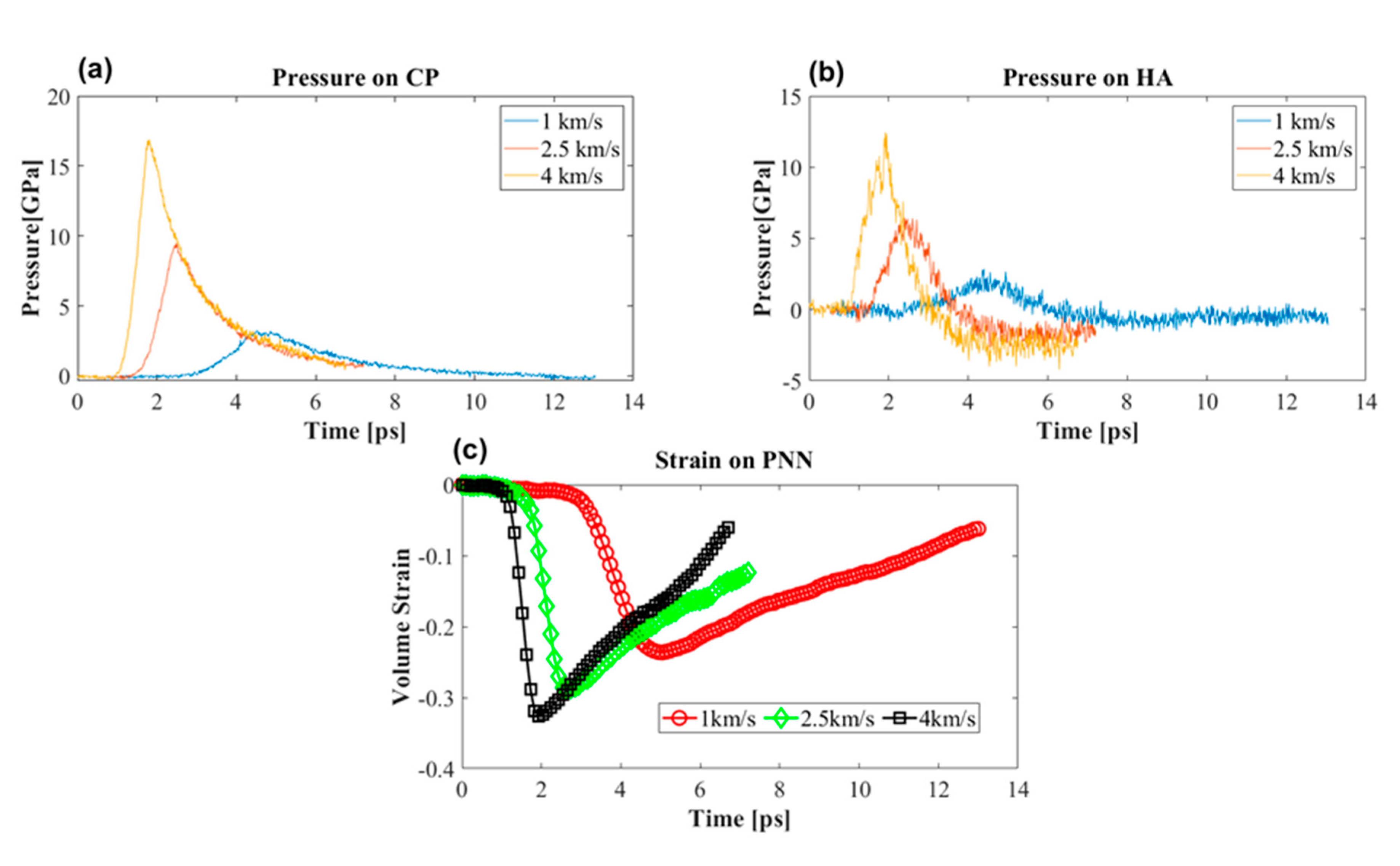
3.3.1. Effect of Shock Speed
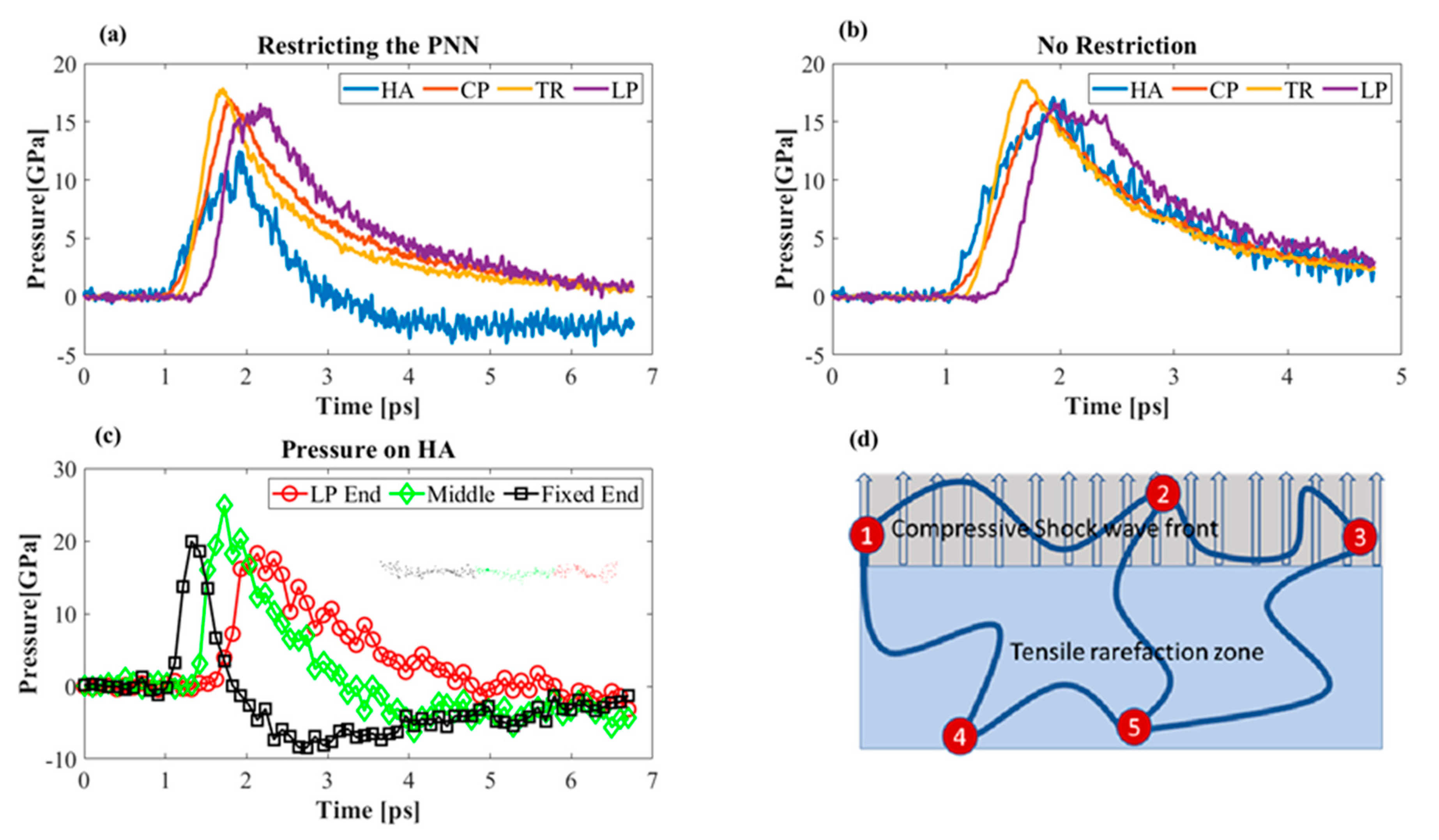
3.3.2. Effect of Cavitation Bubble
3.3.3. Effect of Boundary Conditions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- McKee, A.C.; Robinson, M.E. Military-related traumatic brain injury and neurodegeneration. Alzheimers Dement. 2014, 10, S242–S253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franck, C. Microcavitation: The key to modeling blast traumatic brain injury? Future Med. 2017, 2, CNC47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, C.; Center, S.; Stewart, C. Blast Injuries True Weapons of Mass Destruction. Work 2009, 918, 660–2828. [Google Scholar]
- Qu, T.; Zhou, M. Implosion of composite cylinders due to underwater impulsive loads. In Dynamic Response and Failure of Composite Materials and Structures; Woodhead Publishing: Sawston, UK, 2017; pp. 239–262. [Google Scholar]
- Özer, M.T.; Coşkun, K.; Öğünç, G.İ.; Eryılmaz, M.; Yiğit, T.; Kozak, O.; Apaydın, K. The disguised face of blast injuries: Shock waves. Turkish J. Trauma Emerg. Surg. 2010, 16, 395–400. [Google Scholar]
- Al Mahmud, K.A.H.; Hasan, F.; Khan, M.I.; Adnan, A. On the Molecular Level cavitation in Soft Gelatin Hydrogel. Sci. Rep. 2020, 10, 9635. [Google Scholar] [CrossRef]
- Khan, M.I.; Hasan, F.; Al Mahmud, K.A.H.; Adnan, A. Recent Computational Approaches on Mechanical Behavior of Axonal Cytoskeletal Components of Neuron: A Brief Review. Multiscale Sci. Eng. 2020, 2, 199–213. [Google Scholar] [CrossRef]
- Wu, Y.-T.; Adnan, A. Damage and Failure of Axonal Microtubule under Extreme High Strain Rate: An In-Silico Molecular Dynamics Study. Sci. Rep. 2018, 8, 12260. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Hasan, F.; Al Mahmud, K.A.H.; Adnan, A. Domain focused and residue focused phosphorylation effect on tau protein: A molecular dynamics simulation study. J. Mech. Behav. Biomed. Mater. 2020, 113, 104149. [Google Scholar] [CrossRef] [PubMed]
- Bosiacki, M.; Gąssowska-Dobrowolska, M.; Kojder, K.; Fabiańska, M.; Jeżewski, D.; Gutowska, I.; Lubkowska, A. Perineuronal nets and their role in synaptic homeostasis. Int. J. Mol. Sci. 2019, 20, 4108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, T.H.; Binder, D.K.; Ethell, I.M.; Razak, K.A. The perineuronal ‘safety’net? Perineuronal net abnormalities in neurological disorders. Front. Mol. Neurosci. 2018, 11, 270. [Google Scholar] [CrossRef]
- Agarwal, G.; Krishnan, K.V.; Prasad, S.B.; Bhaduri, A.; Jayaraman, G. Biosynthesis of Hyaluronic acid polymer: Dissecting the role of sub structural elements of hyaluronan synthase. Sci. Rep. 2019, 9, 12510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraser, J.R.E.; Laurent, T.C.; Laurent, U.B.G. Hyaluronan: Its nature, distribution, functions and turnover. J. Intern. Med. 1997, 242, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Frischknecht, R.; Seidenbecher, C.I. The crosstalk of hyaluronan-based extracellular matrix and synapses. Neuron Glia Biol. 2008, 4, 249–257. [Google Scholar] [CrossRef]
- Kwok, J.C.F.; Dick, G.; Wang, D.; Fawcett, J.W. Extracellular matrix and perineuronal nets in CNS repair. Dev. Neurobiol. 2011, 71, 1073–1089. [Google Scholar] [CrossRef] [PubMed]
- Carulli, D. Perineuronal Nets: A Mechanism to Control Brain Plasticity. The Scientists, 1 April 2018. Available online: https://www.the-scientist.com/cover-story/perineuronal-nets-a-mechanism-to-control-brain-plasticity-29877 (accessed on 9 September 2019).
- Schofield, P.W.; Logroscino, G.; Andrews, H.F.; Albert, S.; Stern, Y. An association between head circumference and Alzheimer’s disease in a population-based study of aging and dementia. Neurology 1997, 49, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.-T.; Adnan, A. Effect of shock-induced cavitation bubble collapse on the damage in the simulated perineuronal net of the brain. Sci. Rep. 2017, 7, 5323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, H.; Xie, Y.; Dong, Q.; Kimata, K.; Nishida, Y.; Ishiguro, N.; Zhuo, L. Roles of hyaluronan in cardiovascular and nervous system disorders. J. Zhejiang Univ. B 2019, 20, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Oohashi, T.; Edamatsu, M.; Bekku, Y.; Carulli, D. The hyaluronan and proteoglycan link proteins: Organizers of the brain extracellular matrix and key molecules for neuronal function and plasticity. Exp. Neurol. 2015, 274, 134–144. [Google Scholar] [CrossRef]
- Morawski, M.; Dityatev, A.; Hartlage-Rübsamen, M.; Blosa, M.; Holzer, M.; Flach, K.; Pavlica, S.; Dityateva, G.; Grosche, J.; Brückner, G. Tenascin-R promotes assembly of the extracellular matrix of perineuronal nets via clustering of aggrecan. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20140046. [Google Scholar] [CrossRef]
- Wu, Y.-T.; Adnan, A. Effects of Bubble Size and Gas Density on the Shock-induced Collapse of Nanoscale Cavitation Bubble. Multiscale Sci. Eng. 2020, 2, 127–134. [Google Scholar] [CrossRef]
- Kfoury, R.; Marzban, B.; Makki, E.; Greenfield, M.L.; Yuan, H. Effect of pressure profile of shock waves on lipid membrane deformation. PLoS ONE 2019, 14, e0212566. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Lin, X.; Zhang, Z.; Gu, N. Impact of shock-induced lipid nanobubble collapse on a phospholipid membrane. J. Phys. Chem. C 2016, 120, 18803–18810. [Google Scholar] [CrossRef]
- Koshiyama, K.; Kodama, T.; Yano, T.; Fujikawa, S. Molecular dynamics simulation of structural changes of lipid bilayers induced by shock waves: Effects of incident angles. Biochim. Biophys. Acta-Biomembr. 2008, 1778, 1423–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Wu, C.; Hao, H.; Liu, Z. Post-blast capacity of ultra-high performance concrete columns. Eng. Struct. 2017, 134, 289–302. [Google Scholar] [CrossRef]
- Huber, B.R.; Meabon, J.S.; Martin, T.J.; Mourad, P.D.; Bennett, R.; Kraemer, B.C.; Cernak, I.; Petrie, E.C.; Emery, M.J.; Swenson, E.R. Blast exposure causes early and persistent aberrant phospho-and cleaved-tau expression in a murine model of mild blast-induced traumatic brain injury. J. Alzheimers Dis. 2013, 37, 309–323. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Li, X.; Xiao, S.; Deng, H.; Huang, B.; Zhu, W.; Hu, W. Molecular dynamics simulations of shock loading of nearly fully dense granular Ni–Al composites. Phys. Chem. Chem. Phys. 2019, 21, 20252–20261. [Google Scholar] [CrossRef]
- Lau, E.Y.; Berkowitz, M.L.; Schwegler, E. Shock wave-induced damage of a protein by void collapse. Biophys. J. 2016, 110, 147–156. [Google Scholar] [CrossRef] [Green Version]
- Fröhlich, M.G.; Sewell, T.D.; Thompson, D.L. Molecular dynamics simulations of shock waves in hydroxyl-terminated polybutadiene melts: Mechanical and structural responses. J. Chem. Phys. 2014, 140, 24902. [Google Scholar] [CrossRef]
- Eisenhaber, F.; Persson, B.; Argos, P. Protein structure prediction: Recognition of primary, secondary, and tertiary structural features from amino acid sequence. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 1–94. [Google Scholar] [CrossRef]
- Khan, M.I.; Gilpin, K.; Hasan, F.; Mahmud, K.A.H.A.; Adnan, A. Effect of strain rate on single tau, dimerized tau and tau-microtubule interface: A molecular dynamics simulation study. Biomolecules 2021, 11, 1308. [Google Scholar] [CrossRef]
- Creighton, T.E. Protein folding. Biochem. J. 1990, 270, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Hasan, F.; Mahmud, K.A.; Adnan, A. Viscoelastic Response of Neurofilaments: An Atomistic Simulation Approach. Biomolecules 2021, 11, 540. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins Struct. Funct. Bioinform. 2013, 81, 2159–2166. [Google Scholar] [CrossRef] [Green Version]
- Jo, S.; Song, K.C.; Desaire, H.; MacKerell, A.D., Jr.; Im, W. Glycan Reader: Automated sugar identification and simulation preparation for carbohydrates and glycoproteins. J. Comput. Chem. 2011, 32, 3135–3141. [Google Scholar] [CrossRef] [Green Version]
- Haxaire, K.; Braccini, I.; Milas, M.; Rinaudo, M.; Pérez, S. Conformational behavior of hyaluronan in relation to its physical properties as probed by molecular modeling. Glycobiology 2000, 10, 587–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerji, S.; Wright, A.J.; Noble, M.; Mahoney, D.J.; Campbell, I.D.; Day, A.J.; Jackson, D.G. Structures of the Cd44–hyaluronan complex provide insight into a fundamental carbohydrate-protein interaction. Nat. Struct. Mol. Biol. 2007, 14, 234–239. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Isralewitz, B.; Baudry, J.; Gullingsrud, J.; Kosztin, D.; Schulten, K. Steered molecular dynamics investigations of protein function. J. Mol. Graph. Model. 2001, 19, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Rief, M.; Grubmüller, H. Force spectroscopy of single biomolecules. ChemPhysChem 2002, 3, 255–261. [Google Scholar] [CrossRef] [Green Version]
- Ravelo, R.; Germann, T.C.; Guerrero, O.; An, Q.; Holian, B.L. Shock-induced plasticity in tantalum single crystals: Interatomic potentials and large-scale molecular-dynamics simulations. Phys. Rev. B 2013, 88, 134101. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, U.; Goliaei, A.; Berkowitz, M.L. Mechanism of membrane poration by shock wave induced nanobubble collapse: A molecular dynamics study. J. Phys. Chem. B 2015, 119, 6225–6234. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Stukowski, A. Visualization and analysis of atomistic simulation data with OVITO–the Open Visualization Tool. Model. Simul. Mater. Sci. Eng. 2009, 18, 15012. [Google Scholar] [CrossRef]
- Tanemura, M.; Ogawa, T.; Ogita, N. A new algorithm for three-dimensional Voronoi tessellation. J. Comput. Phys. 1983, 51, 191–207. [Google Scholar] [CrossRef]
- Rycroft, C. Voro++: A Three-Dimensional Voronoi Cell Library in C++; Lawrence Berkeley National Lab.(LBNL): Berkeley, CA, USA, 2009. [Google Scholar]
- Thompson, A.P.; Plimpton, S.J.; Mattson, W. General formulation of pressure and stress tensor for arbitrary many-body interaction potentials under periodic boundary conditions. J. Chem. Phys. 2009, 131, 154107. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, Y.; Kobayashi, A.; Kawakami, M. Mechanical unfolding studies of protein molecules. Biophysics 2012, 8, 51–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuendet, M.A.; Michielin, O. Protein-protein interaction investigated by steered molecular dynamics: The TCR-pMHC complex. Biophys. J. 2008, 95, 3575–3590. [Google Scholar] [CrossRef] [Green Version]
- Bayas, M.V.; Schulten, K.; Leckband, D. Forced detachment of the CD2-CD58 complex. Biophys. J. 2003, 84, 2223–2233. [Google Scholar] [CrossRef] [Green Version]
- Chahal, R.; Adnan, A.; Reifsnider, K.; Raihan, R.; Ting Wu, Y.; Vadlamudi, V.; Elenchezhian, M.R.P. Molecular Dynamics for the Prediction of the Interfacial Shear Stress and Interface Dielectric Properties of Carbon Fiber Epoxy Composites. In Proceedings of the American Society for Composites—Thirty-Third Technical Conference on Composite Materials; DEStech Publication, Inc.: Lancaster, PA, USA, 2018. [Google Scholar]
- Bosshard, H.R.; Marti, D.N.; Jelesarov, I. Protein stabilization by salt bridges: Concepts, experimental approaches and clarification of some misunderstandings. J. Mol. Recognit. 2004, 17, 1–16. [Google Scholar] [CrossRef]
- Guetta, O.; Varfman, B.H.; Rittel, D. Shock attenuation characteristics of methylcellulose hydrogels: Phenomenological modeling. J. Mech. Phys. Solids 2021, 146, 104220. [Google Scholar] [CrossRef]
- Bremond, N.; Arora, M.; Dammer, S.M.; Lohse, D. Interaction of cavitation bubbles on a wall. Phys. Fluids 2006, 18, 121505. [Google Scholar] [CrossRef]
- Cooper, P.W. Explosives Engineering; John Wiley & Sons: Hoboken, NJ, USA, 2018; ISBN 1119537134. [Google Scholar]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmud, K.A.H.A.; Hasan, F.; Khan, M.I.; Adnan, A. Shock-Induced Damage Mechanism of Perineuronal Nets. Biomolecules 2022, 12, 10. https://doi.org/10.3390/biom12010010
Mahmud KAHA, Hasan F, Khan MI, Adnan A. Shock-Induced Damage Mechanism of Perineuronal Nets. Biomolecules. 2022; 12(1):10. https://doi.org/10.3390/biom12010010
Chicago/Turabian StyleMahmud, Khandakar Abu Hasan Al, Fuad Hasan, Md Ishak Khan, and Ashfaq Adnan. 2022. "Shock-Induced Damage Mechanism of Perineuronal Nets" Biomolecules 12, no. 1: 10. https://doi.org/10.3390/biom12010010
APA StyleMahmud, K. A. H. A., Hasan, F., Khan, M. I., & Adnan, A. (2022). Shock-Induced Damage Mechanism of Perineuronal Nets. Biomolecules, 12(1), 10. https://doi.org/10.3390/biom12010010





