Whole-Cell Display of Phosphotransferase in Escherichia coli for High-Efficiency Extracellular ATP Production
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Chemical Reagents
2.2. Construction of Recombinant Plasmids for Cell Surface Display
2.3. Secretory Expression of the Target Protein
2.4. Cell Fractionation
2.5. Fluorescence Microscopy
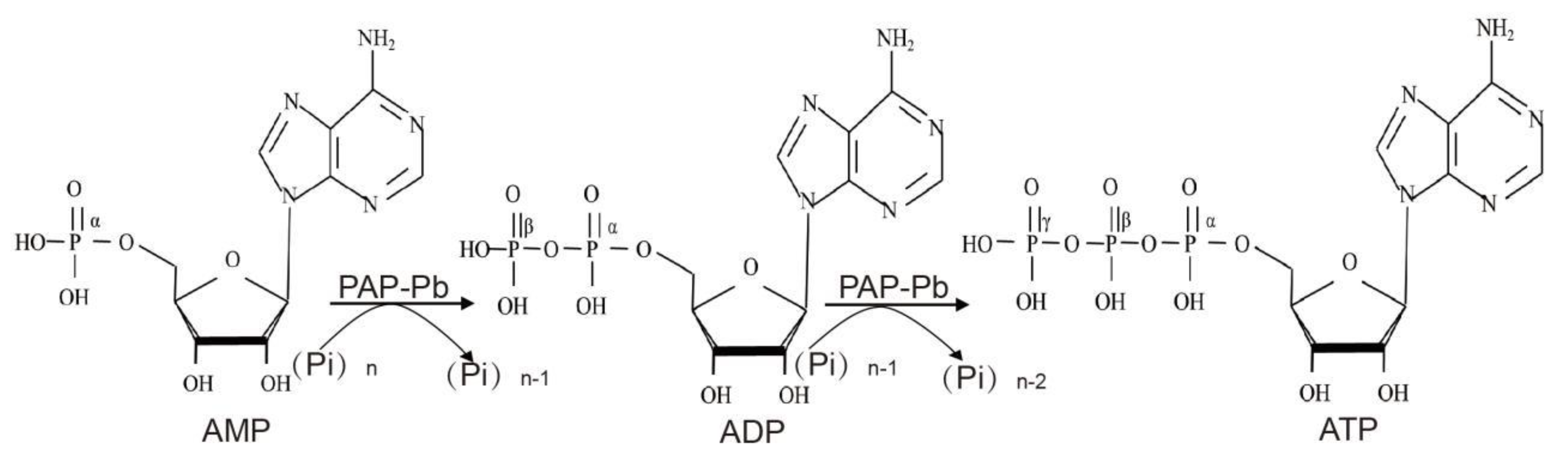
2.6. Biosynthesis of ATP
2.7. Detection and Analysis by High-Performance Liquid Chromatography (HPLC)
2.8. Effect of pH and Mg2+ Concentration ATP Synthesis
2.9. Effect of Polyphosphate Concentration on ATP Synthesis
2.10. Effect of the Cell State on the Production of ATP
2.11. Effects of Temperature and Stability of Whole-Cell Catalysts
2.12. Reuse of Whole-Cell Catalyst
3. Results
3.1. Expression of the Recombinant Proteins
3.2. Biosynthesis of ATP with Fusion Enzyme Displayed on the Surface of E. coli
3.3. Effects of pH and the Concentration of Mg2+
3.4. Effects of the Concentration of Polyphosphate
3.5. Effects of the Cell State on the Production of ATP
3.6. Effects of Temperature and Stability of Whole-Cell Catalysts
3.7. Reuse of Whole-Cell Catalyst
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andexer, J.N.; Richter, M. Emerging enzymes for ATP regeneration in biocatalytic processes. ChemBioChem 2015, 16, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Y.P.J. Enzymatic regeneration and conservation of ATP: Challenges and opportunities. Crit. Rev. Biotechnol. 2021, 41, 16–33. [Google Scholar] [CrossRef]
- Suzuki, S.; Hara, R.; Kino, K. Production of aminoacyl prolines using the adenylation domain of nonribosomal peptide synthetase with class III polyphosphate kinase 2-mediated ATP regeneration. J. Biosci. Bioeng. 2018, 125, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Beis, K. Structural basis for the mechanism of ABC transporters. Biochem. Soc. Trans. 2015, 43, 889–893. [Google Scholar] [CrossRef] [Green Version]
- Otrin, L.; Kleineberg, C.; Caire da Silva, L.; Landfester, K.; Ivanov, I.; Wang, M.; Bednarz, C.; Sundmacher, K.; Vidakovic-Koch, T. Artificial Organelles for Energy Regeneration. Adv. Biosyst. 2019, 3, e1800323. [Google Scholar] [CrossRef]
- Rajendran, M.; Dane, E.; Conley, J.; Tantama, M. Imaging Adenosine Triphosphate (ATP). Biol. Bull. 2016, 231, 73–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calhoun, K.A.; Swartz, J.R. Energy systems for ATP regeneration in cell-free protein synthesis reactions. Methods Mol. Biol. (Clifton N.J.) 2007, 375, 3–17. [Google Scholar] [CrossRef]
- Lian, J.; Ma, Y.; Cai, J.; Wu, M.; Wang, J.; Wang, X.; Xu, Z. High-level expression of soluble subunit b of F1F0 ATP synthase in Escherichia coli cell-free system. Appl. Microbiol. Biotechnol. 2009, 85, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Liao, Y.; Kong, W.Z.; Wang, S.H. ATP dynamic regeneration strategy for enhancing co-production of glutathione and S-adenosylmethionine in Escherichia coli. Biotechnol. Lett. 2020, 42, 2581–2587. [Google Scholar] [CrossRef]
- Chambers, R.W.; Moffatt, J.G.; Khorana, H.G. Nucleoside Polyphosphates. IV.1 A New Synthesis of Guanosine 5’-Phosphate. J. Am. Chem. Soc. 1957, 79, 3752–3755. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resnick, S.M.; Zehnder, A.J. In vitro ATP regeneration from polyphosphate and AMP by polyphosphate:AMP phosphotransferase and adenylate kinase from Acinetobacter johnsonii 210A. Appl. Environ. Microbiol. 2000, 66, 2045–2051. [Google Scholar] [CrossRef] [Green Version]
- Kimura, Y.; Kamatani, S. Catalytic activity profile of polyP:AMP phosphotransferase from Myxococcus xanthus. J. Biosci. Bioeng. 2021, 131, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Fuhner, V.; Heine, P.A.; Zilkens, K.J.C.; Meier, D.; Roth, K.D.R.; Moreira, G.; Hust, M.; Russo, G. Epitope Mapping via Phage Display from Single-Gene Libraries. Methods Mol. Biol. 2019, 1904, 353–375. [Google Scholar] [CrossRef] [PubMed]
- Pédelacq, J.D.; Cabantous, S.; Tran, T.; Terwilliger, T.C.; Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006, 24, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Yu, S.; Qin, F.; Ning, W.; Ma, X.; Tian, K.; Li, Z.; Zhou, K. A secretion-based dual fluorescence assay for high-throughput screening of alcohol dehydrogenases. Biotechnol. Bioeng. 2021, 118, 1624–1635. [Google Scholar] [CrossRef]
- Yao, Y.; Ding, Q.; Ou, L. Biosynthesis of (deoxy)guanosine-5’-triphosphate by GMP kinase and acetate kinase fixed on the surface of E. coli. Enzym. Microb. Technol. 2019, 122, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ou, L.; Ding, Q. Enzymatic Synthesis of Nucleoside Triphosphates and Deoxynucleoside Triphosphates by Surface-Displayed Kinases. Appl. Biochem. Biotechnol. 2020, 190, 1271–1288. [Google Scholar] [CrossRef]
- Salema, V.; Fernández, L. Escherichia coli surface display for the selection of nanobodies. Microb. Biotechnol. 2017, 10, 1468–1484. [Google Scholar] [CrossRef]
- Lee, S.Y.; Choi, J.H.; Xu, Z. Microbial cell-surface display. Trends Biotechnol. 2003, 21, 45–52. [Google Scholar] [CrossRef]
- Schuurmann, J.; Quehl, P.; Festel, G.; Jose, J. Bacterial whole-cell biocatalysts by surface display of enzymes: Toward industrial application. Appl. Microbiol. Biotechnol. 2014, 98, 8031–8046. [Google Scholar] [CrossRef]
- Ding, J.; Liu, Y.; Gao, Y.; Zhang, C.; Wang, Y.; Xu, B.; Yang, Y.; Wu, Q.; Huang, Z. Biodegradation of λ-cyhalothrin through cell surface display of bacterial carboxylesterase. Chemosphere 2021, 289, 133130. [Google Scholar] [CrossRef] [PubMed]
- Le, T.; Park, S. Development of efficient microbial cell factory for whole-cell bioconversion of L-threonine to 2-hydroxybutyric acid. Bioresour. Technol. 2022, 344, 126090. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Du, H.; Zheng, Y.; Sun, J.; Shen, Y.; Lin, J.; Wei, D. Design and engineering of whole-cell biocatalyst for efficient synthesis of (R)-citronellal. Microb. Biotechnol. 2021. [Google Scholar] [CrossRef]
- Nowrouzi, B.; Rios-Solis, L. Redox metabolism for improving whole-cell P450-catalysed terpenoid biosynthesis. Crit. Rev. Biotechnol. 2021, 1–25. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Y.; Ai, M.; Jia, X. Surface display of carbonic anhydrase on Escherichia coli for CO(2) capture and mineralization. Synth. Syst. Biotechnol. 2022, 7, 460–473. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, R.; Zhu, D.; Wang, W.; Yi, L.; Ma, L. Non-peptide guided auto-secretion of recombinant proteins by super-folder green fluorescent protein in Escherichia coli. Sci. Rep. 2017, 7, 6990. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Wang, B.; Wang, F.; Yang, Z.; Gao, D.; Zhang, C.; Ma, L.; Yu, X. Soluble expression of single-chain variable fragment (scFv) in Escherichia coli using superfolder green fluorescent protein as fusion partner. Appl. Microbiol. Biotechnol. 2019, 103, 6071–6079. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.; Liu, Q.; Sun, J.; Secundo, F.; Mao, X. Construction of a Super-Folder Fluorescent Protein-Guided Secretory Expression System for the Production of Phospholipase D in Bacillus subtilis. J. Agric. Food Chem. 2021, 69, 6842–6849. [Google Scholar] [CrossRef]
- Twair, A.; Kassem, I.; Murad, H.; Abbady, A.Q. Secretion of Recombinant Human Annexin V in Fusion with the Super Folder GFP for Labelling Phosphatidylserine-Exposing Membranes. J. Membr. Biol. 2021, 254, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Dong, C.; Wang, X.; Liu, Y.; Ma, L. One-step production of bioactive human lipopolysaccharide binding protein from LPS-eliminated E. coli. Protein Expr. Purif. 2019, 157, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.; Chu, X.; Bao, Z.; Liang, Y.; Wang, X.; Yang, J.; Xian, M.; Sun, Y.; Nian, R. Enhanced anticoagulant activity of hirudin-i analogue co-expressed with arylsulfotransferase in periplasm of E. coli BL21(DE3). J. Biotechnol. 2020, 323, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Quan, S.; Hiniker, A.; Collet, J.F.; Bardwell, J.C. Isolation of bacteria envelope proteins. Methods Mol. Biol. 2013, 966, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Li, X.; Yang, J.; Li, Z.; Hou, J.; Rao, B.; Hu, Y.; Ma, L.; Wang, Y. Cost-Effective Production of ATP and S-Adenosylmethionine Using Engineered Multidomain Scaffold Proteins. Biomolecules 2021, 11. [Google Scholar] [CrossRef]
- Somasundaram, S.; Jeong, J.; Kumaravel, A.; Hong, S.H. Whole-cell display of Pyrococcus horikoshii glutamate decarboxylase in Escherichia coli for high-titer extracellular gamma-aminobutyric acid production. J. Ind. Microbiol. Biotechnol. 2021, 48. [Google Scholar] [CrossRef]
- Sato, M.; Masuda, Y.; Kirimura, K.; Kino, K. Thermostable ATP regeneration system using polyphosphate kinase from Thermosynechococcus elongatus BP-1 for D-amino acid dipeptide synthesis. J. Biosci. Bioeng. 2007, 103, 179–184. [Google Scholar] [CrossRef]








| Gene | Primer | Sequence(5′-3′) |
|---|---|---|
| sfGFP-PAP | SP1 | GGCGGCGGCGGCAGCCATATGTTTGAAAGCGCAGAAATTG |
| SP2 | GCTGCCGCCGCCGCCTTTATACAGTTCATCCATGCCCAGAT | |
| PAP | P1 | CATATGTTTGAAAGCGCAGAAATTGG |
| P2 | GCTTTCAAACATATGATGATGATGATGATGGTGCATATGTATATCTC | |
| sfGFP | S1 | TGAGATCCGGCTGCTAACAA |
| S2 | AGCAGCCGGATCTCATTTATACAGTTCATCCATGCCCAG |
| Strain/Plasmid | Description | Reference |
|---|---|---|
| pET-23a | Vector for expression proteins, T7 promoter, Ampr | This study |
| pPAP-Pb | pET-23a encoding pap-pb, Ampr | This study |
| psfGFP | pET-23a encoding sfgfp, Ampr | This study |
| psfGFP-PAP-Pb | pET-23a encoding sfgfp- pap-pb, Ampr | This study |
| Strain | ||
| E-PAP-Pb | E. coli BL21(DE3) (pPAP-Pb) | This study |
| E-sfGFP | E. coli BL21(DE3) (psfGFP) | This study |
| E-sfGFP-PAP-Pb | E. coli BL21(DE3) (psfGFP-PAP-Pb) | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Yang, G.; Xie, X.; Yan, G.; Wang, F.; Chen, W.; Ma, L. Whole-Cell Display of Phosphotransferase in Escherichia coli for High-Efficiency Extracellular ATP Production. Biomolecules 2022, 12, 139. https://doi.org/10.3390/biom12010139
Zhao S, Yang G, Xie X, Yan G, Wang F, Chen W, Ma L. Whole-Cell Display of Phosphotransferase in Escherichia coli for High-Efficiency Extracellular ATP Production. Biomolecules. 2022; 12(1):139. https://doi.org/10.3390/biom12010139
Chicago/Turabian StyleZhao, Shuai, Guoli Yang, Xiaochen Xie, Guangbo Yan, Fei Wang, Wanping Chen, and Lixin Ma. 2022. "Whole-Cell Display of Phosphotransferase in Escherichia coli for High-Efficiency Extracellular ATP Production" Biomolecules 12, no. 1: 139. https://doi.org/10.3390/biom12010139
APA StyleZhao, S., Yang, G., Xie, X., Yan, G., Wang, F., Chen, W., & Ma, L. (2022). Whole-Cell Display of Phosphotransferase in Escherichia coli for High-Efficiency Extracellular ATP Production. Biomolecules, 12(1), 139. https://doi.org/10.3390/biom12010139






