Abstract
Background: HSP90B1, a member of the heat-shock protein 90 family, plays a vital role as a molecular chaperone for oncogenes and stimulates tumour growth. However, its role in various cancers remains unexplored. Methods: Using the cancer genome atlas, gene expression omnibus the Human Protein Atlas databases and various other bioinformatic tools, this study investigated the involvement of HSP90B1 in 33 different tumour types. Results: The over-expression of HSP90B1 generally predicted poor overall survival and disease-free survival for patients with tumours, such as adrenocortical carcinoma, bladder urothelial carcinoma, kidney renal papillary cell carcinoma, and lung adenocarcinoma. In this study, HSP90B1 was highly expressed in the majority of tumours. A comparison was made between the phosphorylation of HSP90B1 in normal and primary tumour tissues, and putative functional mechanisms in HSP90B1-mediated oncogenesis were investigated. Additionally, the mutation burden of HSP90B1 in cancer was evaluated along with the survival rate of patients with cancer patients. Conclusion: This first pan-cancer investigation reveals the oncogenic functions of HSP90B1 in various cancers.
1. Introduction
It is essential to identify new pan-cancer-related genes and elucidate their functions to have a better understanding of the incredibly complicated process of carcinogenesis. The development of better diagnoses, treatments, and preventive methods for cancer is closely related to a better understanding of genetic changes in tumours [1]. The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases utilise innovative genomic analysis techniques to obtain a comprehensive understanding of cancer, thereby aiding in generating novel cancer therapies, diagnostics, and preventive strategies [2]. These two databases provide a wealth of publicly available data, providing researchers around the world with invaluable knowledge about the physiological, genetic, and epigenetic profiles of cancer [1]. It also helps researchers screen candidate cancer biomarkers and drug targets, thereby translating cancer genomics into therapeutics and diagnostics and aiding in developing personalised cancer treatments. With the development of the TCGA and GEO databases, there is currently a significant increase in the amount of cancer-related functional genomics datasets of various malignancies, which can be used for an in-depth pan-cancer study [3].
Heat-shock protein 90kDa beta1 (HSP90B1), a stress-inducible molecular chaperone that is a member of the heat-shock protein (HSP) 90 family, is also known as GRP94 and GP96 [4]. Through the maintenance of the endoplasmic reticulum (ER) stress sensors, the preservation of the ER protein folding capability, and the repression of ER-associated proapoptotic machinery, HSP90B1 plays an essential part in regulating the delicate balance between the survival and death of cancer cells [5]. HSP90B1 is crucial for the chaperoning of various proteins, including Toll-like receptors (TLRs) [6], integrin subunits [7], and wnt co-receptor low-density lipoprotein receptor-related protein 6 (LRP6) [8], that have been linked to immune response and cancer development. HSP90B1 mRNA expression has been reported to be elevated in several different cancer tissue types, including breast cancer, oesophageal cancer, glioma tissues, and liver cancer [9,10,11,12]. Various immuno-histochemistry studies report that HSP90B1 protein is significantly overexpressed in a variety of malignancies, including breast, lung, colorectal, oral, oesophageal, and gastric cancers, indicating a connection with cancer development [9,13,14,15,16,17]. Furthermore, HSP90B1 overexpression has been reported to be negatively associated with patient survival in many of the above-mentioned malignancies. For example, patients with breast cancer expressing high levels of HSP90B1 had a significantly lower survival time than those expressing low levels of HSP90B1 [9]. Additionally, HSP90B1 is speculated to be a factor that contributes to bad prognosis in many cancer types, including lung, oesophageal, gastric, and colorectal cancers [13,16,17,18]. Thus, HSP90B1 has a significant relationship with cancer growth and metastases. A previous study reports that administering Her2/neu DNA vaccination to HSP90B1 HER2+ breast cancer-carrying mice improved immune response against tumours, which was indicated by higher IFN-/IL-4 levels and reduced Tregs at the tumour site [19]. Thus, owing to its ability to activate anti-tumour immune responses on its own, extra-cellular HSP90B1 has potential as an antigen in tumour vaccines. However, the examination of HSP90B1 has been limited to a select few forms of cancer, and its significance in other types of tumours remains unknown.
For the first time, to the best of our knowledge, this study aims to comprehensively analyse the role of HSP90B1 in various cancers. To investigate the potential role of the HSP90B1 gene as a molecular mechanism in the development of various cancers, HSP90B1 gene expression, survival status, gene mutations, protein phosphorylation, immune infiltration, and cellular pathways were studied.
2. Results
2.1. Expression of HSP90B1 in Tumours and Normal Tissues
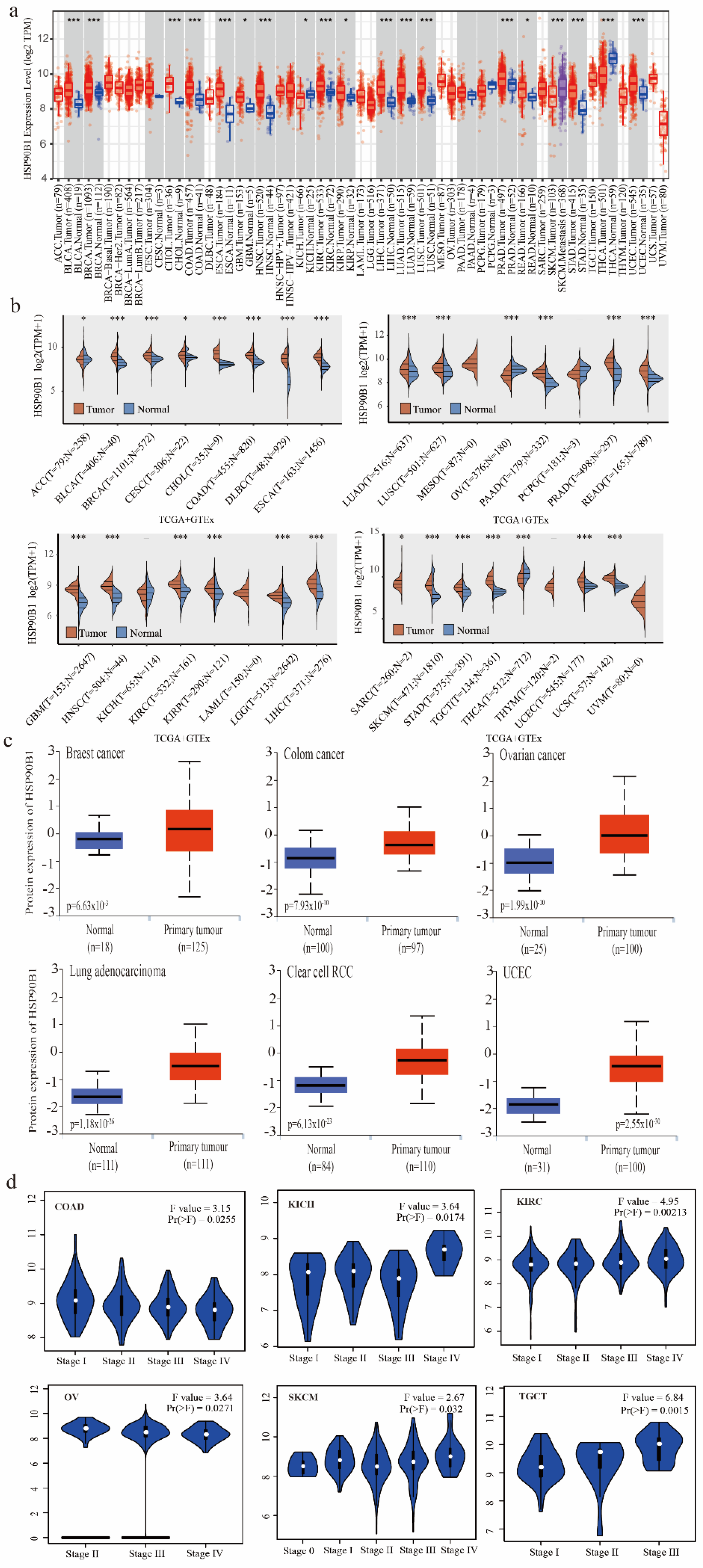
The TIMER method was used to evaluate the expression patterns of HSP90B1 in various cancer types, which were obtained from the TCGA database. HSP90B1 expression was higher in BLCA, BRCA, CHOL, COAD, ESCA, NHSC, KIRC, LIHC, LUAD, LUSC, PRAD, SKCM, STAD, THCA, UCEC (p < 0.001), GBM, KICH, KIRP, and READ (p < 0.05) than in the corresponding non-tumour tissues (Figure 1a). As shown in Figure 1b, data from the TCGA and GTEx databases were used to analyse the expression of HSP90B1 in different cancer and normal tissue types. Following this, we performed an analysis of HSP90B1 protein expression in breast cancer, colon cancer, ovarian cancer, lung adenocarcinoma, clear cell RCC, and UCEC using the CPTAC dataset. The expression of HSP90B1 protein in these tumour tissues was observed to be significantly higher than that in normal tissues (p < 0.01) (Figure 1c). Furthermore, GEPIA2 analysis revealed that HSP90B1 expression levels were significantly correlated to the clinical stage of the following cancer types: COAD (p = 0.0255), KICH (p = 0.0174), KIRC (p = 0.00213), OV (p = 0.00271), SKCM (p = 0.032), and TGCT (p = 0.0015) (Figure 1d). Additionally, immunohistochemical staining of the Human Protein Atlas database showed that HSP90B1 was highly expressed in most malignant tumours (Figure S1). Thus, these findings indicate that HSP90B1 is significantly expressed in the majority of cancers, which offers a foundation for further investigation.
Figure 1.
Expression and pathological staging of HSP90B1 in different tumours. (a). Using TIMER2, we examined the levels of HSP90B1 mRNA expression in various malignancies. (b) In the TCGA and GTEx datasets, HSP90B1 expression was upregulated in most tumours. (c) In the CPTAC dataset, we examined the expression levels of HSP90B1 protein between primary tumour tissues and normal tissues of breast cancer, LUAD, colon cancer, clear cell RCC, UCEC cancer, and ovarian cancer. (d) The pathological stages of HSP90B1 in COAD, KICH, KIRC, OV, SKCM, and TGCT were analysed using data from the TCGA database. Logarithmic analysis used Log2 (TPM+1). *p < 0.05; ***p < 0.001.
2.2. Prognostic Significance of HSP90B1 in Various Tumours
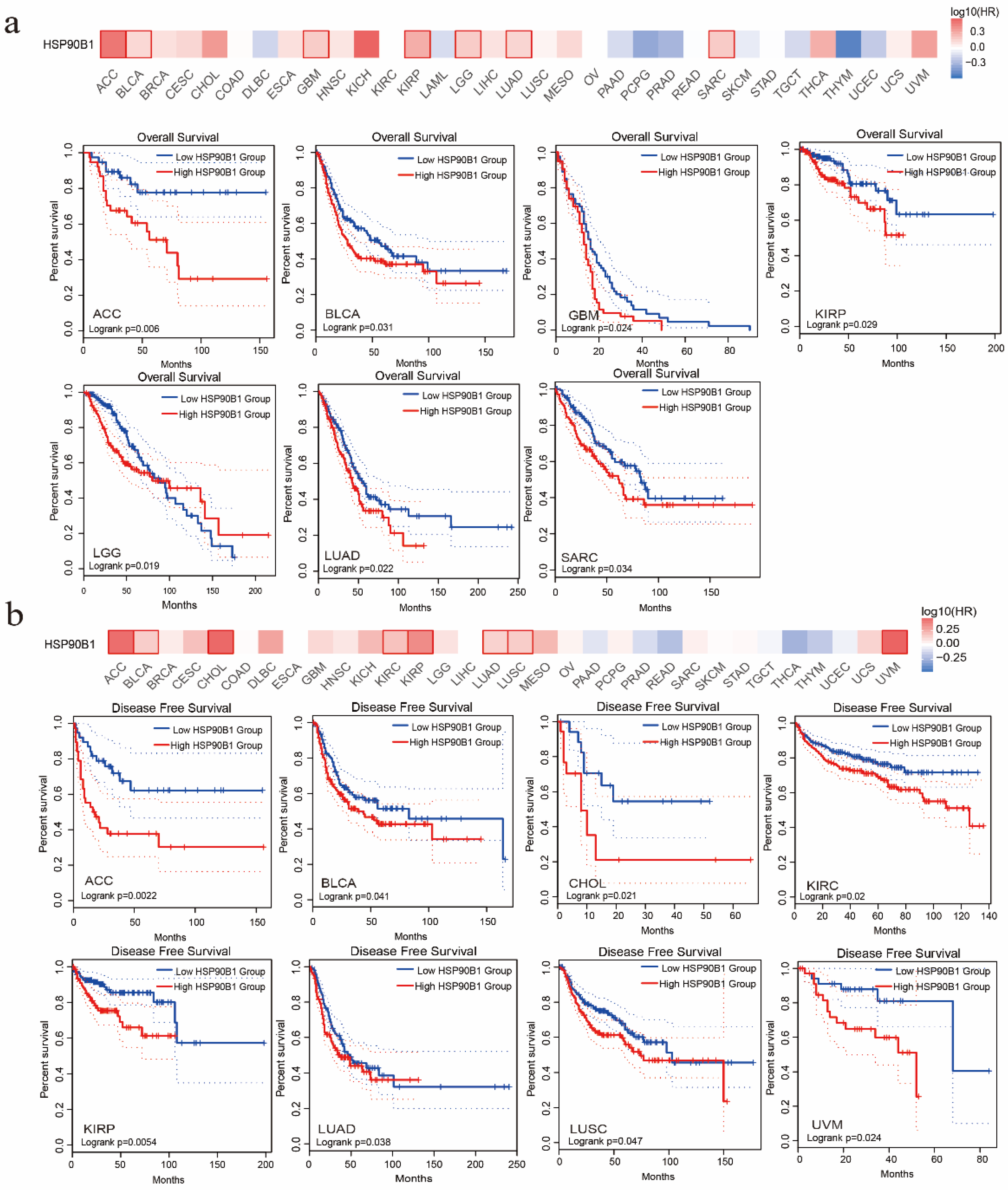
The association between HSP90B1 expression and the patient prognosis was investigated using TCGA datasets. Two groups were formed based on HSP90B1 expression (high- and low-level HSP90B1 expression) to examine the link between HSP90B1 expression and overall patient survival. According to the TCGA datasets, high levels of HSP90B1 expression were linked to poor prognosis for patients with ACC (p = 0.006), BLCA (p = 0.031), GBM (p = 0.024), KIRP (p = 0.029), LGG (p = 0.019), LUAD (p = 0.022), and SARC (p = 0.034) (Figure 2a). DFS analysis (Figure 2b) indicated that the high expression of HSP90B1 was associated with poor prognosis of ACC (p = 0.0022), BLCA (p = 0.041), CHOL (p = 0.021), KIRC (p = 0.02), KIRP (p = 0.0054), LUAD (p = 0.038), LUSC (p = 0.047), and UVM (p = 0.024) tumours. Therefore, tumours with high HSP90B1 expression are speculated to have a bad prognosis.
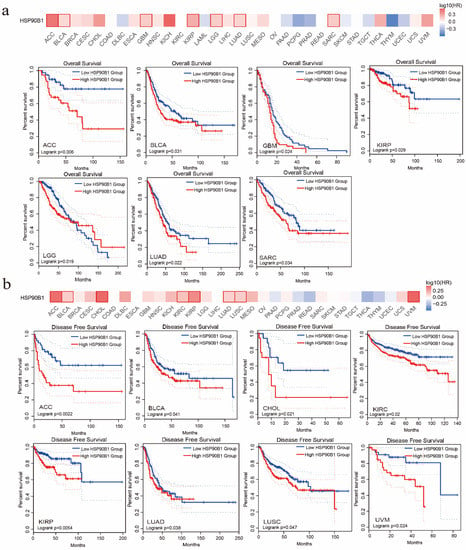
Figure 2.
The prognostic relationship of HSP90B1 expression in various tumours. GEPIA2 was used to examine the impact of HSP90B1 on overall survival (a) and disease-free survival (b) in various tumour types.
2.3. The Genetic Alteration of HSP90B1
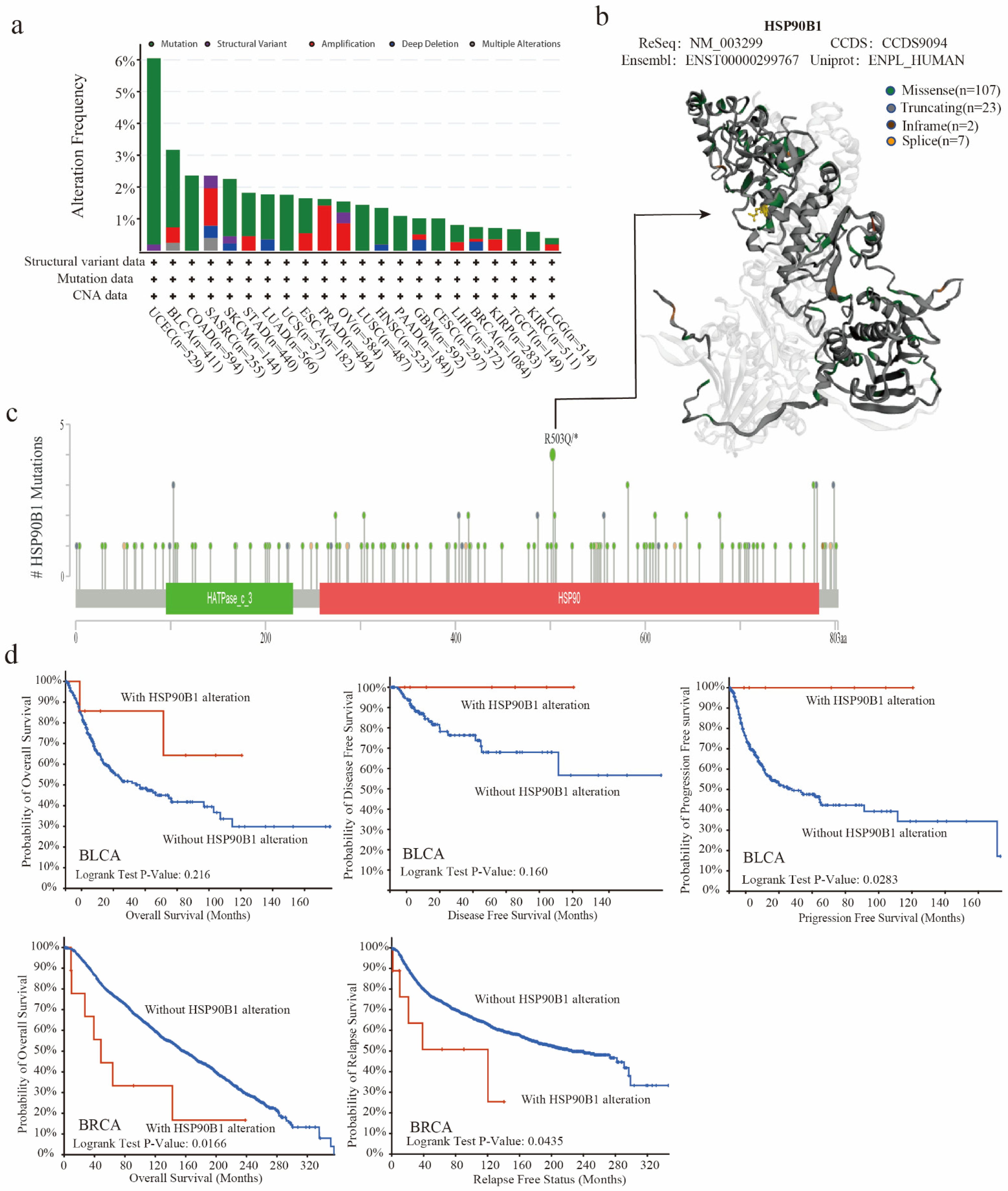
The TCGA database was used to examine the genetic changes of HSP90B1 in various tumour samples. As shown in Figure 3a, the highest frequency of HSP90B1 alterations (>6 percent) was observed in patients with UCEC who had a “mutation” as the primary feature, followed by patients with BLCA. Furthermore, Figure 3b displays 145 mutation sites (including 107 missenses, 23 truncating, 2 in frames, and 7 splices) in the 3D structure map using different colours. HSP90B1 acts as a molecular chaperone to promote protein folding by binding to nascent polypeptides, thereby preventing protein misfolding and aggregation. Mutations in genes disrupt this binding, affecting the function of the molecular chaperones. Gene mutations can also alter the role of molecular chaperones in tumours by affecting their sub-localization. Additionally, chaperones act by regulating their client proteins, with mutations affecting the binding of chaperones to their client proteins. Moreover, the missense mutation in HSP90B1 was observed to be the major type of genetic alteration, with R503Q/* alteration detected in four cases of UCEC (Figure 3c). Furthermore, we hypothesised a possible connection between genetic alterations in HSP90B1 and the clinical survival outcomes of UCEC, BLCA, and breast cancer. Figure 3d shows that HSP90B1-altered patients with BLCA had an improved prognosis in progression-free survival (p = 0.0283) but not in overall survival (p = 0.216) and disease-free survival (p = 0.16) compared with HSP90B1-unaltered cases. In breast cancer, HSP90B1-altered patients showed a significantly worse prognosis in overall survival (p = 0.0166) and relapse-free survival (p = 0.0435). These findings indicate that mutations in the HSP90B1 gene could affect the prognosis of individuals with varying malignancies.
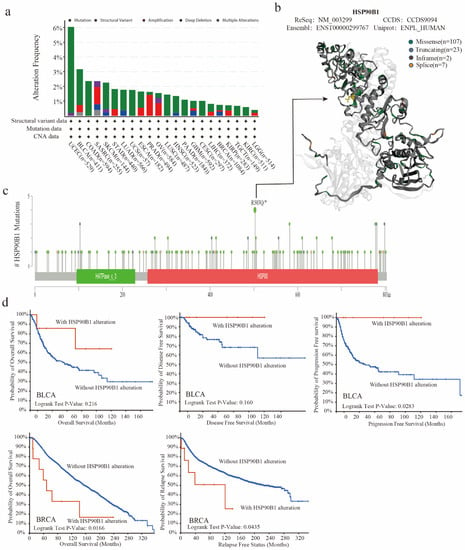
Figure 3.
Mutation characteristics of HSP90B1 in various cancer types. (a) Alteration frequency of mutation types and mutation sites of HSP90B1 in different tumours. (b,c) Different mutation types and the position of the highest mutation site (R503Q/*) in the 3D structure of HSP90B1. (d) Potential correlations between BLCA and breast cancer mutational status, overall survival, disease-free survival, progression-free survival, and recurrence-free survival.
2.4. Phosphorylation of HSP90B1 Protein was Analysed in Different Tumours
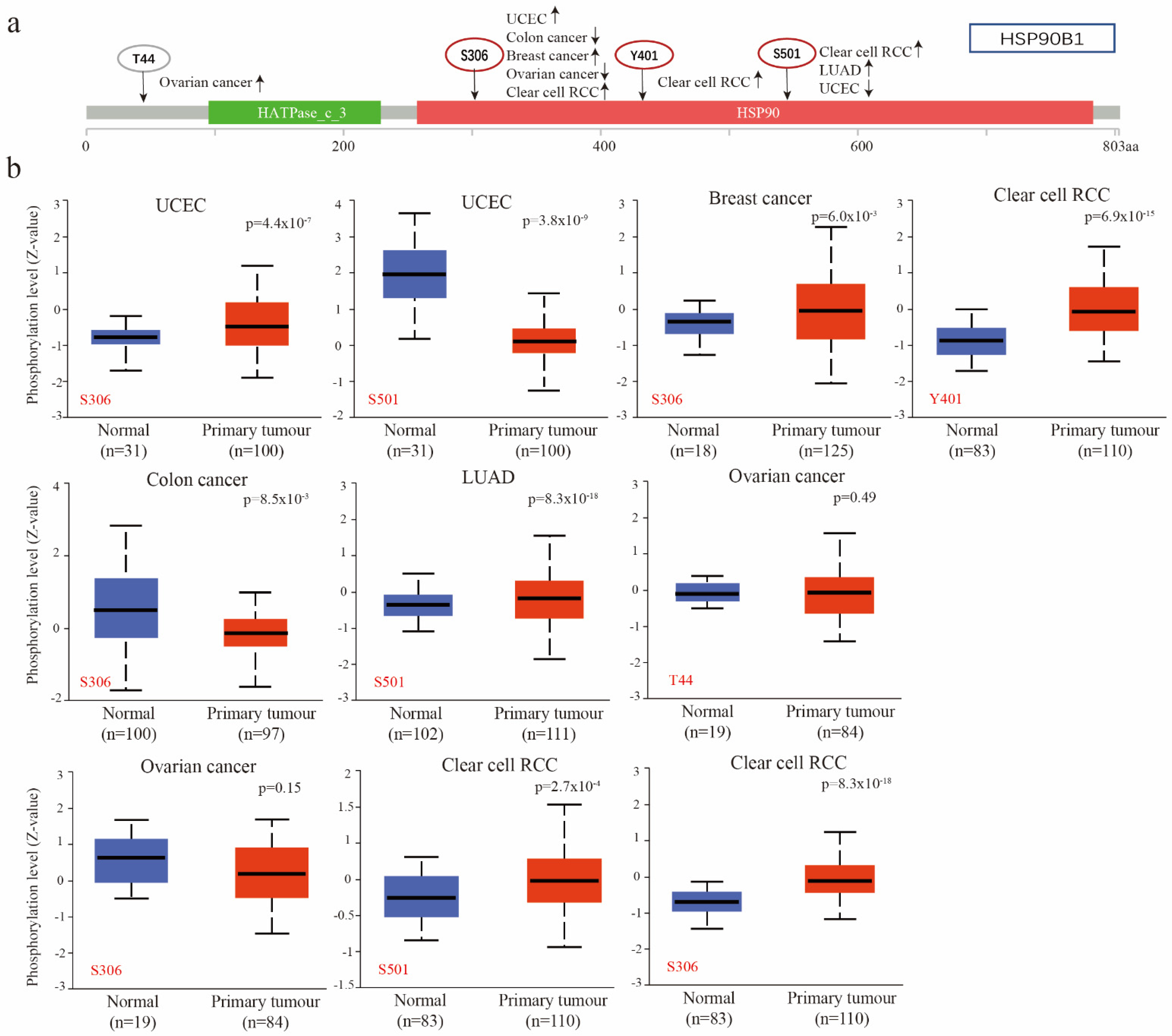
Utilizing the CPTAC database, researchers compared the phosphorylation levels of HSP90B1 in normal and primary tumour tissues of patients with UCEC, breast cancer, clear cell RCC, colon cancer, LUAD, and ovarian cancer, revealing significant differences. Figure 4a provides a summary of the HSP90B1 phosphorylation locations and the changes in expression. Figure 4b highlights the difference in HSP90B1 phosphorylation levels between normal and primary tumour tissues. At Y401 site, we found that the phosphorylation level of HSP90B1 in clear cell renal cell carcinoma tissues was higher than that in normal tissues, but there was no significant difference between ovarian cancer tissues at T44 site and corresponding normal tissues.. At S306, the phosphorylation levels of HSP90B1 were higher in UCEC, breast cancer, and clear cell RCC tissues than in normal tissues; however, the opposite effect was observed in colon cancer. Finally, at S501, the phosphorylation levels of HSP90B1 were higher in LUAD and clear cell RCC tissues than in normal tissues and reversed in UCEC. Additionally, the expression of HSP90B1 methylation levels was evaluated in various cancers, revealing that HSP90B1 methylation levels were higher in CHOL and KIRC tumours than in normal tissues, while the opposite was observed in BLCA, COAD, ESCA, HNSC, KIRC, PRAD, and STAD (Figure S2).
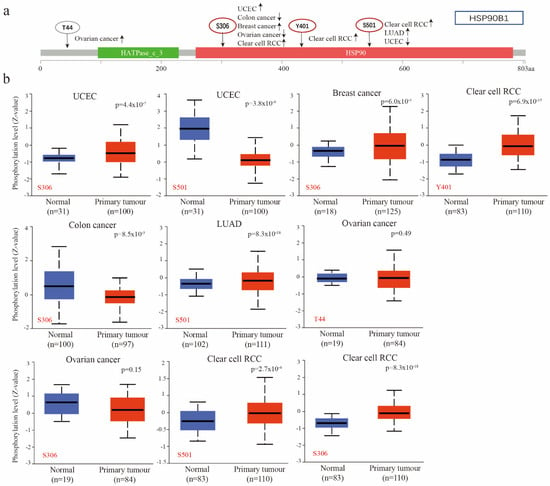
Figure 4.
Phosphorylation of HSP90B1 was analysed in different tumours. (a) Four phosphorylated protein sites (T44, S306, Y401, and S501) of HSP90B1 were confirmed using the cBioPortal tool. (b) We analysed the differences in HSP90B1 phosphorylated proteins between primary tumour and normal tissues in UCEC, breast cancer, colon cancer, LUAD, ovarian cancer, and clear cell RCC.
2.5. Analysis of HSP90B1 Expression and Tumour-Associated Immune Infiltration
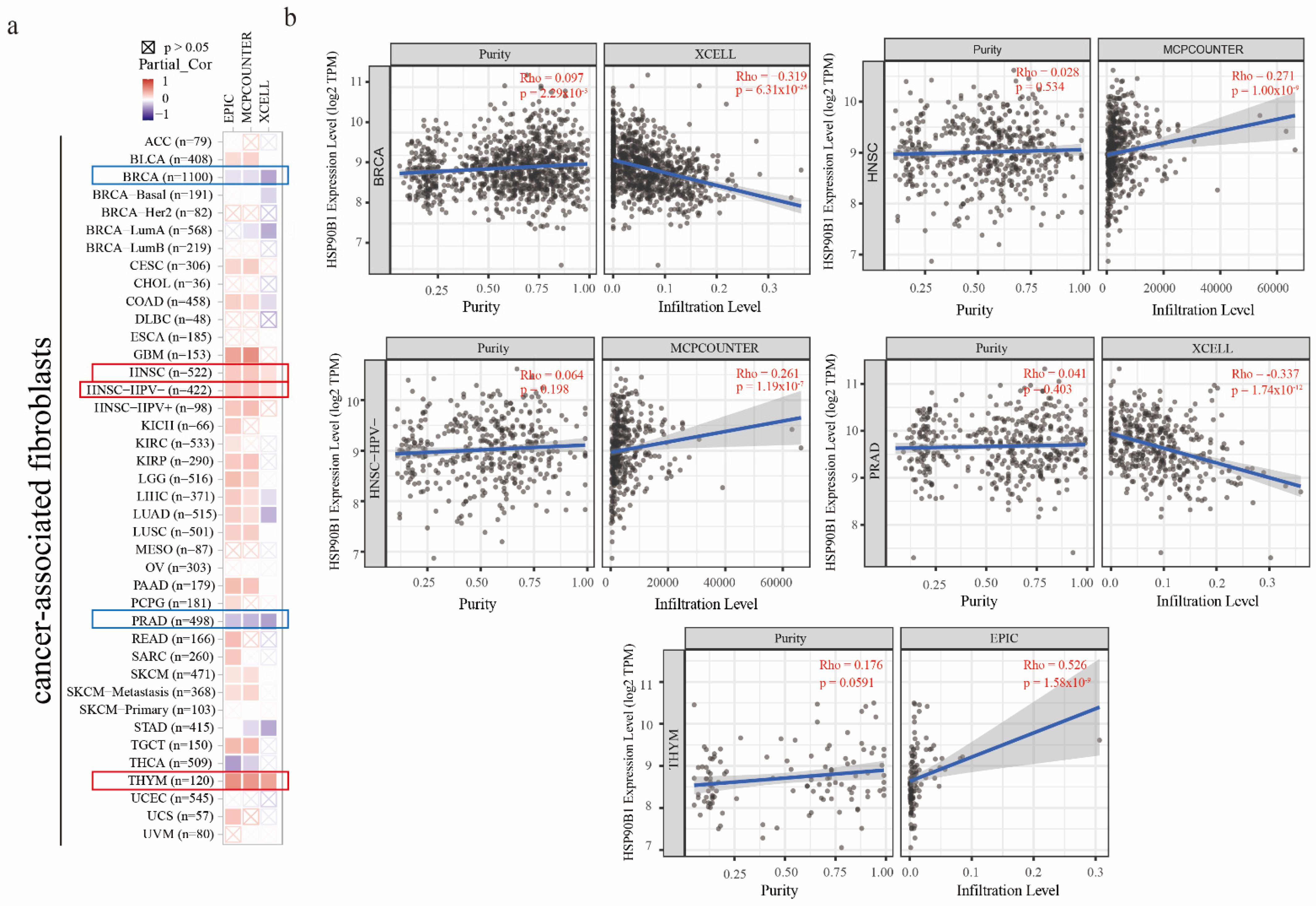
It has been reported that cancer-associated fibroblasts have features that promote tumorigenesis [20]. To investigate the relationship between HSP90B1 expression and cancer-associated fibroblasts, three different algorithms, namely EPIC, MCPCOUNTER, and XCELL, were used. In the TCGA tumours of HNSC, HNSC-HPV(-), and THYM, a statistically positive association between the estimated infiltration value of cancer-associated fibroblasts and HSP90B1 expression was observed. However, a statistically negative correlation was observed between BRCA, PRAD, and HSP90B1 (Figure 5a). Figure 5b displays the scatter plot of the aforementioned tumours, which were generated using an algorithm.
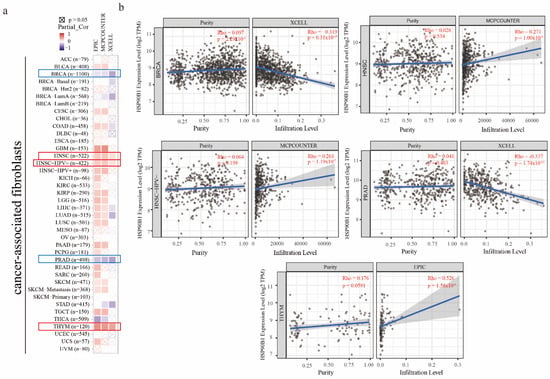
Figure 5.
The relationship between immune infiltration of cancer-associated fibroblasts and HSP90B1 expression. (a) HSP90B1 expression and the invasion of cancer-associated fibroblasts in various forms of cancer were investigated using the EPIC, MCPCOUNTER, and XCELL algorithms. (b) Significant correlation was found between HSP90B and cancer-associated fibroblasts in BRCA, HNSC, HNSC-HPV(-), PRAD, and THYM.
2.6. Enrichment Analysis of HSP90B1-Related Genes
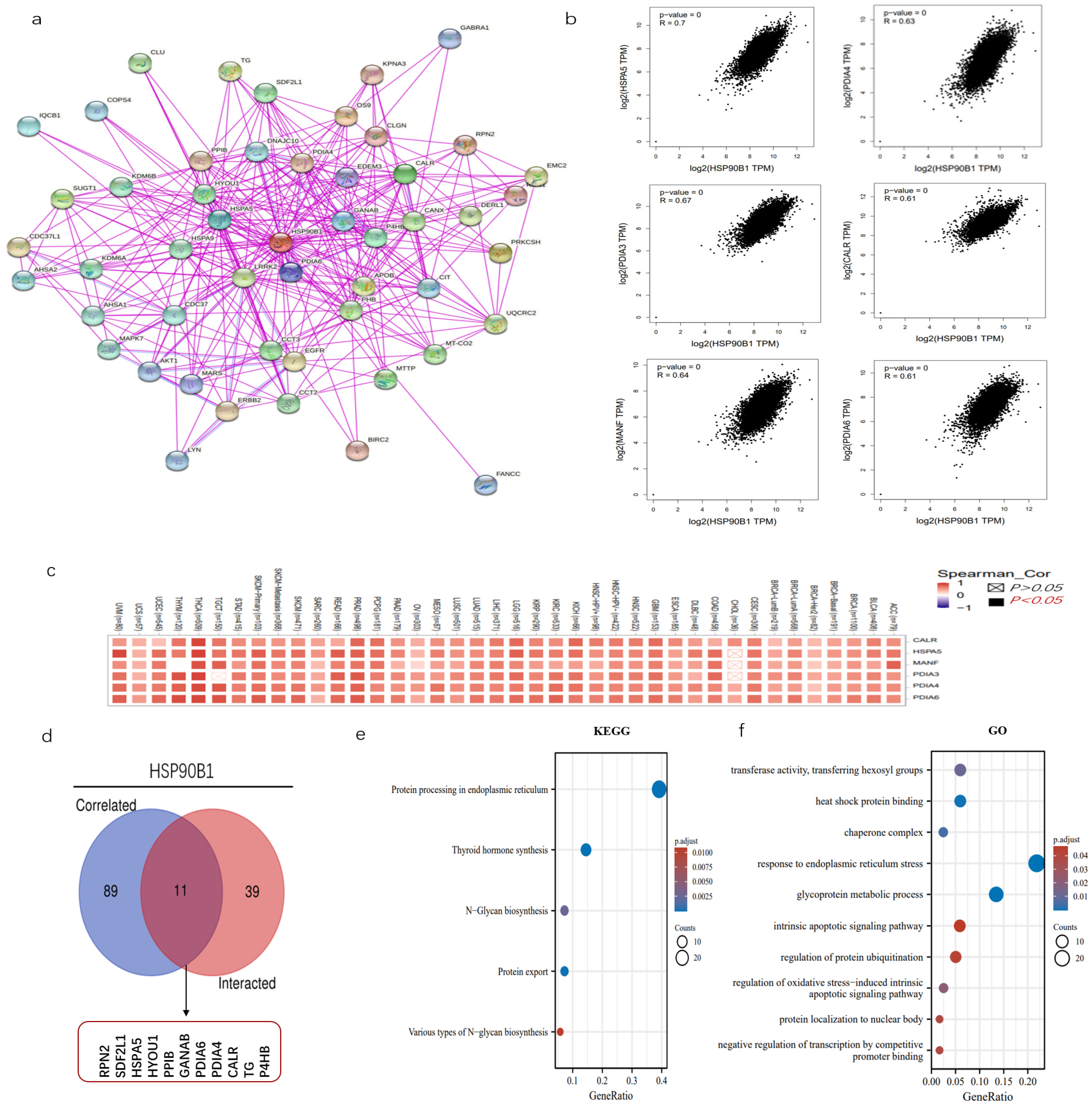
The examination of gene enrichment is an essential part of our research into the molecular mechanisms behind genes that are involved in cancer. STRING was used to perform an analysis on 50 experimentally validated proteins having the potential to interact with HSP90B1, and the resulting interaction network diagram of these proteins is shown in Figure 6a. Then, using the GEPIA2 program, we screened 100 genes that had a high correlation with HSP90B1, revealing six genes that had the greatest connection (Figure 6b). The corresponding heat map further proved that HSP90B1 was positively correlated with the above six genes (HSPA5, PDIA3, MANF, PDIA4, CALR, and PDIA6) in various tumours (Figure 6c). HSPA5, like HSP90B1, also belongs to the heat-shock protein family. Recent studies report that HSPA5 could influence the growth of tumours by controlling ferroptosis [21]. PDIA4 is a potential therapeutic target for the treatment of glioblastoma because it controls the growth of glioblastoma cells by activating the PI3K/AKT/m-TOR pathway and suppressing apoptosis [22]. The combination of CALR and PDIA3 has also been suggested as a possible prognostic biomarker for non-small-cell lung cancer [23], with PDIA6 modulating apoptosis and autophagy of non-small-cell lung cancer cells via the MAP4K1/JNK signalling pathway [24]. Meanwhile, MANF is overexpressed in hepatocellular carcinoma and can be used as a potential diagnostic and prognostic indicator for hepatocellular carcinoma [25]. Subsequently, the genes that could interact with HSP90B1 and those that were highly correlated with HSP90B1 were cross-analysed, and the following 11 genes were obtained: RPN2, SDF2L1, HSPA5, HYOU1, PPIB, GANAB, PDIA6, PDIA4, CALR, TG, and P4HB (Figure 6d). Furthermore, KEGG data showed that “thyroid hormone synthesis” and “protein processing in endoplasmic reticulum” could be involved in the effect of HSP90B1 on tumour pathogenesis (Figure 6e). Then, GO enrichment analysis further indicates the mechanism of HSP90B1 regulating tumour progression (Figure 6f).
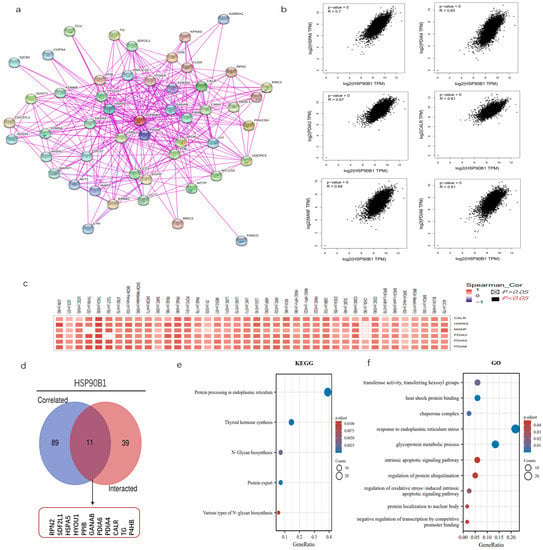
Figure 6.
Enrichment analysis of HSP90B1. (a) We first obtained 50 proteins that could interact with HSP90B1 using the STRING tool. (b) Additionally, 100 genes with a high correlation with HSP90B1 were obtained from the TCGA database, and the expression correlation between HSP90B1 and selected targeted genes (including HSPA5, PDIA3, MANF, PDIA4, CALR, and PDIA6) was analysed. (c) We analysed the relationship between these six genes and the incidence of each cancer. (d) We conducted a crossover analysis of 100 genes with a high correlation with HSP90B1 and 50 genes that could interact with HSP90B1. Enrichment analysis based on HSP90B1 with high correlation and interaction was performed using KEGG (e) and GO (f) pathway analyses.
2.7. Expression of HSP90B1 in Some Tumours
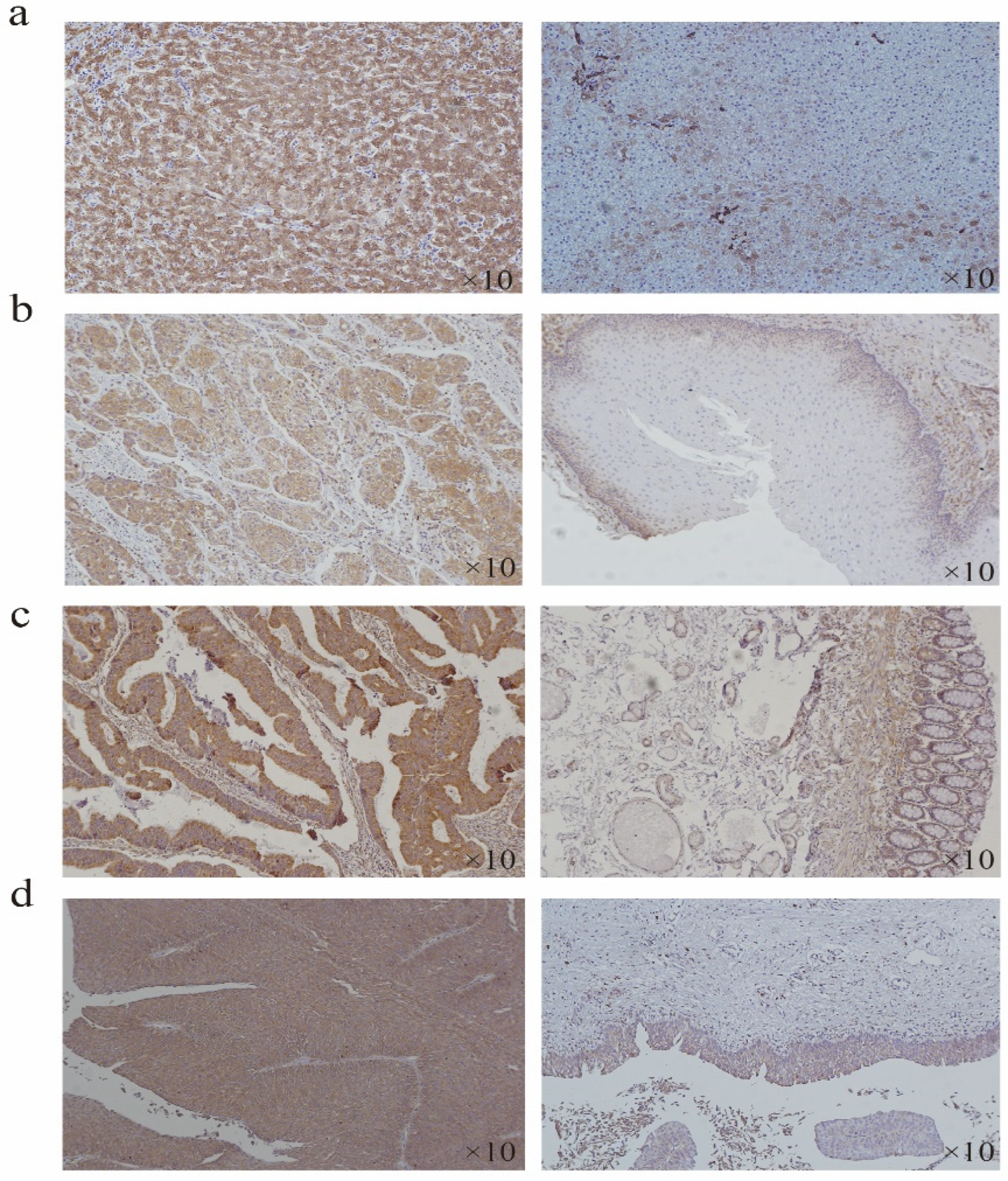
According to the expression difference of HSP90B1 RNA in tumours and corresponding normal tissues (Figure 1a), the tumour type with the most significant difference was selected for immunohistochemistry (IHC) analysis. The results showed that the staining intensity of HSP90B1 protein in liver cancer, oesophageal cancer, colorectal cancer, and bladder cancer was higher than that in the corresponding normal tissues. Moreover, HSP90B1 protein was mainly expressed in the cytoplasm and not in the nucleus (Figure 7a-d).
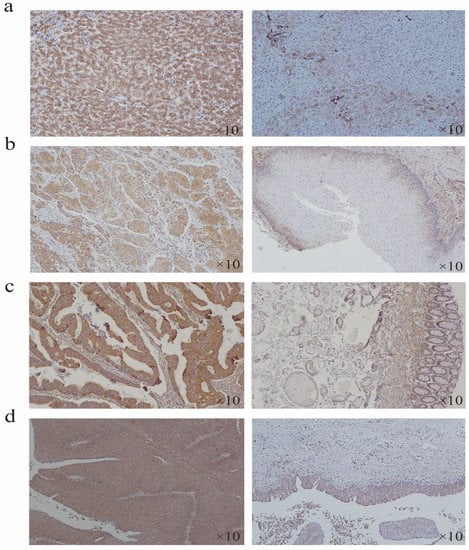
Figure 7.
Expression of HSP90B1 in liver cancer, oesophageal cancer, colorectal cancer, and bladder cancer. Immunohistochemistry was used to analyse the expression of HSP90B1 in liver cancer (a), oesophageal cancer (b), colorectal cancer (c), and bladder cancer (d).
3. Discussion
Cancer is a global public health issue and one of the primary causes of high morbidity and mortality [26]. Current approaches for treating cancer include surgical removal of the tumour, radiation therapy, chemotherapy, and targeted therapy. Despite advances in the treatments of cancer, there remain a significant proportion of people who do not benefit from them. This conundrum underlines the need to comprehensively understand the pathways that lead to the development of tumours [27]. Therefore, it is necessary to identify additional targets and biomarkers that could be used in the diagnosis and treatment of cancer [28]. In the field of precision medicine, discovering reliable cancer biomarkers and investigating their function in disease progression could aid in the development of cancer treatments.
HSPs are a category of stress proteins that are very well-conserved and found in a wide variety of species. In its role as a molecular chaperone, it plays a primary role in the assembly and sorting, transmembrane transport, and degradation of intracellular proteins. These proteins are induced in response to a variety of stresses, including traumatic injury, infectious diseases, hypoxia, and malignant tumours [29]. Ischemia and hypoxia are typically present in tumour tissues, consequently generating a quick stress response and increasing the expression level of HSPs. Previous studies report a connection between HSP90B1 and various illnesses, including cancer, pneumonia [30], polycystic ovary syndrome [31], ulcerative colitis [32], and others. Notably, an emphasis on the function of HSP90B1 in tumour development is currently observed. Furthermore, recent studies report that the aberrant expression of HSP90B1 in lung adenocarcinoma [13], bladder cancer [33], colorectal cancer [34], and tongue squamous cell carcinoma [35] is associated with a poor prognosis. Additionally, an internalized antibody targeting cell surface HSP90B1 was also shown to effectively inhibit tumour angiogenesis in colorectal cancer [36]. HSP90B1 can also affect the progression of thyroid cancer by regulating the expression and sub-localization of its client protein ITGA2 [37]. However, these studies are limited to some tumours, and thus, more studies are required to investigate their role in other tumours.
The overexpression of HSP90B1 on the cell membrane has been reported to boost cell proliferation and tumour development by increasing HER2 dimerization and the downstream signalling cascade. Moreover, HSP90B1 also interacts specifically with HER2 on the cytoplasmic membrane of human breast cancers [38]. Additionally, it has been shown that HSP90B1 induces apoptosis in prostate cancer cells, thereby preventing the cells from migrating [39]. Collectively, these studies suggest that cell surface HSP90B1 has potential as a therapeutic target for cancer treatment.
Despite the increasing number of research devoted to the investigation of the role of HSP90B1 in many disorders, including cancer, it remains unclear whether HSP90B1 plays a pro-oncogenic or anti-oncogenic role in tumour pathogenesis. Using the TCGA database, the current study’s all-encompassing method comprised an investigation of the HSP90B1 expression level in 33 different cancers. Additionally, we used the CPTAC and GEO databases to methodically gather and combine data on proteins and phosphor proteins in addition to other molecular characteristics and genetic changes.
On examining the levels of HSP90B1 expression in each tumour, it was observed to be highly expressed in the majority of cancers. The variations in HSP90B1 expression levels observed across the spectrum of tumour types are indicative of its diverse processes and functions. Furthermore, GEPIA2 revealed that poor overall survival in ACC, BLCA, GBM, KIRP, LGG, LUAD, and SARC was significantly associated with high HSP90B1 expression; however, in ACC, BLCA, CHOL, KIRC, KIRP, LUAD, LUSC, and UVM, poor disease-free survival was significantly correlated with high HSP90B1 expression. Based on these findings, HSP90B1 has potential as a biomarker for determining the outlook for patients with cancer. The investigation of the mutation driving mechanism revealed that the HSP90B1 gene alterations in UCEC were mostly missense mutations, while the sarcoma mutations were amplification mutations. Additionally, the mutation of HSP90B1 showed an important effect on the progression-free survival of patients with BLCA patients and the overall survival and relapse-free survival of patients with breast cancer.
An important step in the development of cancer is the phosphorylation–dephosphorylation cascade. By determining the amount of total protein and phosphorylated protein, the probable molecular pathways of HSP90B1 in UCEC, breast cancer, blear cell RCC, Colon cancer, LUAD, and ovarian cancer can be elucidated. Accordingly, the phosphorylation sites of HSP90B1 were significantly increased in breast cancer, LUAD, and clear cell RCC but significantly reduced in colon cancer. However, contradictory results were obtained at several locations for UCEC and ovarian cancer, suggesting that these cancers may have distinct causes. Nonetheless, studies on the correlation between HSP90B1 phosphorylation and tumorigenesis are insufficient and require further analyses. This study identified four novel phosphorylation sites, which could be one of the important pathways through which HSP90B1 functions in tumours. Subsequently, HSP90B1 expression and the immune invasion of cancer-associated fibroblasts in various tumours were analysed. In BRCA and PRAD, HSP90B1 expression was negatively correlated with tumour-associated fibroblast infiltration, while in HNSC and THYM, HSP90B1 expression was positively correlated with tumour-associated fibroblast infiltration.
Additionally, we also identified 11 genes that interacted with HSP90B1 and were highly correlated with HSP90B1: RPN2, SDF2L1, HSPA5, HYOU1, PPIB, GANAB, PDIA6, PDIA4, CALR, TG, and P4HB. RPN2 is speculated to be involved in the progression of malignant tumours by regulating various signalling pathways, such as STAT3, NF-κB, and PI3K-Akt [40,41]. It is also considered a promising prognostic biomarker for colorectal cancer, non-small-cell lung cancer, and gastric adenocarcinoma [42,43,44]. Furthermore, SDF2L1 mRNA is significantly upregulated by the unfolded protein response (UPR) and can interact with ER chaperone complexes, including HSPA5, to regulate chaperone activity. [45]. Thus, there exists a high possibility that SDF2L1 interacts with HSP90B1, which can be explored in our future studies. Chen et al. found that multiple stemness-related markers were downregulated on HSPA5 knockdown and demonstrated that HSPA5 is a chaperone protein associated with cancer stemness maintenance in head and neck cancer cells [46]. In addition to head and neck cancer, a potential function of GRP78 in cancer stemness has also been reported in breast cancer and glioma [47,48]. HYOU1 is a chaperone protein located in the ER and serves not only as a potential therapeutic target for cancer but also as an immunostimulatory adjuvant, owing to its anti-tumour immune response. It is also used as a molecular target for the treatment of many ER-related diseases [49]. PPIB induces chemoresistance in colorectal cancer by degrading wild-type p53 [50], and it can regulate hepatocellular carcinoma cell apoptosis and metastasis [51]. Currently, research on the GANAB gene in cancer is scarce. PDIA6 has been shown to promote pancreatic cancer progression and immune evasion through β-catenin and PD-L1 deubiquitination [52]. It can also affect the progression of bladder cancer, oral squamous cell carcinoma, and non-small-cell lung cancer [24,53,54]. PDIA4 is a novel ER stress chaperone that regulates adiponectin expression and inflammation in adipose tissue [55], and it is also involved in tumour progression by affecting apoptosis [56]. Furthermore, PDIA4 and PDIA6 regulate cisplatin-induced lung adenocarcinoma cell death resistance [57]. CALR can not only serve as a potential prognostic biomarker for lung cancer [58] but is also an important target for tumour immunotherapy [59]. The TG gene mainly plays a role in thyroid diseases, with very few studies being reported on this topic. P4HB is also a chaperone protein that has been associated with temozolomide resistance via UPR in gliomas [60]. Therefore, these 11 genes can regulate tumour progression in various ways; however, further experimental analysis is required to prove this conclusion. Furthermore, GO and KEGG analyses suggest that HSP90B1 could regulate the occurrence and development of tumours via various mechanisms, such as participating in ER stress response or regulating apoptosis.
4. Conclusions
Our comprehensive pan-cancer analysis of HSP90B1 revealed that the high expression of HSP90B1 was associated with the poor clinical prognosis of various human cancers, indicating that HSP90B1 can be used as an effective biomarker in cancer. Additionally, a statistical correlation was observed between the expression of HSP90B1 and cancer-related fibroblast infiltration, tumour mutation burden, and protein phosphorylation, which validated the role of HSP90B1 in tumorigenesis from various perspectives. However, this study has some limitations. Most of our research is based on data from online databases. Thus, more in vitro and in vivo experiments are required to verify our analyses.
5. Materials and methods
5.1. Gene Expression Analysis
We compared HSP90B1 mRNA expression between tumour and normal tissues, which were obtained from the TCGA database, using TIMER2 (http://timer.cistrome.org/ (accessed on 21 July 2022)-> Gene DE). Moreover, data from the GTEx and TCGA databases were used to examine the expression of HSP90B1 in different types of cancer and normal tissues. Furthermore, the difference in HSP90B1 protein expression between different tumour types and corresponding adjacent tissues was analysed using UALCAN [21] (http://ualcan.path.uab.edu/analysis -prot.html (accessed on 25 July 2022)->CPTAC analysis) and the CPTAC (Clinical Proteomic Tumour Analysis Consortium) dataset. HSP90B1 expression in different cancers, which were obtained from the Human Protein Atlas database, was analysed using immunohistochemical labelling. Moreover, the “Stage Plot” module of GEPIA2 (http://gepia2.cancer-pku.cn/ (accessed on 28 July 2022)) was used to examine the expression levels of HSP90B1 in distinct cancer types at different clinical stages.
5.2. Survival Prognosis Analysis
The overall survival (OS) and disease-free survival (DFS) were obtained for HSP90B1 in all TCGA tumours using the “survival map” module of GEPIA2.
5.3. Genetic Alteration Analysis
cBioPortal for Cancer Genomics (http://www.cbioportal.org (accessed on 21 July 2022)) was used to evaluate the genetic alterations of HSP90B1 in pan-cancer, including somatic mutations, structural variants, amplifications, profound deletions, and multiple alterations. Moreover, in TCGA patients with and without HSP90B1 gene mutation, the data on OS, DFS, PFS, and RFS were obtained.
5.4. Immuno-Infiltration Analysis
The association of HSP90B1 expression to immune invasion in all TCGA cancers was determined using the “Immune genes” module of TIMER2, which also allowed the selection of CD8+ T cells and cancer-associated fibroblasts from the immune system. Estimates of immune infiltration were made using the EPIC, MCPCOUNTER, and XCELL algorithms. p-values and partial correlation values were calculated using the purity-corrected Spearman rank correlation test.
5.5. Functional Enrichment Analysis
A search for “HSP90B1” and “Homo sapiens” in the STRING database was the first step in this analysis, and then, the following parameters were set: Network edges (“evidence”), the maximum number of participants to display (“no more than 50 participants”), and active interactions (“experiments”) were included in the “Low confidence (0.150)” minimum score requirement. Subsequently, 50 proteins were obtained that could bind to HSP90B1. GEPIA2 was used to analyse the 100 genes with the highest correlation with HSP90B1. Finally, HSP90B1-binding and HSP90B1-interacting genes were compared using the interactive Venn diagram viewer Jvenn. Additionally, the two sets of data were combined to perform KEGG and GO pathway analysis.
5.6. Immunohistochemistry
Antibodies for HSP90B1 (ab238126) were purchased from Abcam company. Samples were embedded in paraffin at a thickness of 4 μm. Deparaffinization and rehydration were performed on each slide. To eliminate aldehyde linkages from antigens, they were reextracted using a pressure cooker and 0.01 M citrate buffer (pH 6). The slides were incubated with HSP90B1 antibody (1: 2000) overnight. After incubating the HRP-labelled secondary antibody for 1 h, immunodetection was performed the following day using diaminobenzidine, according to the manufacturer’s instructions [61].
5.7. Statistical Analysis
t-Tests were used to evaluate differences in HSPA5 expression between cancer and normal tissues. Univariate Cox regression analysis was used to obtain the HR and p-value for the survival analysis. Patients were divided into two groups, with high and low HSPA5 expression, respectively, and Kaplan–Meier analysis was utilized to compare their survival rates. All statistical tests were conducted at the p < 0.05 significance level. * p < 0.05, ** p < 0.01, *** p < 0.001.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biom12101377/s1, Figure S1: Expression of HSP90B1 protein in various tumour tissues; Figure S2: The methylation expression level of HSP90B1 in various cancers and corresponding normal tissues.
Author Contributions
Y.W., project development, data analysis and collection, and manuscript writing; X.W., data analysis and collection. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Scientific research project of Jiangsu Provincial Health Commission (M2021005).
Institutional Review Board Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Nantong Tumour Hospital. The patients/participants provided their written informed consent to participate in this study.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets obtained from TCGA database (https://portal.gdc.cancer.gov/ accessed on 25 July 2022) and UALCAN database(http://ualcan.path.uab.edu/analysis.html accessed on 25 July 2022), partial analysis by Cbioportal for Cancer Genomics website (http://www.cbioportal.org accessed on 25 July 2022) and TIMER2.0 database (http://timer.cistrome.org/ accessed on 25 July 2022).
Acknowledgments
We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript. In addition, we would also like to thank Zhigang Chen for his help with the revision of the article.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ACC | Adrenocortical carcinoma |
| BLCA | Bladder urothelial carcinoma |
| BRCA | Breast invasive carcinoma |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma |
| CHOL | Cholangiocarcinoma |
| COAD | Colon adenocarcinoma |
| DLBC | Lymphoid neoplasm diffuse large B-cell lymphoma |
| ESCA | Oesophageal carcinoma |
| GBM | Glioblastoma multiforme |
| HNSC | Head and neck squamous cell carcinoma |
| KICH | Kidney chromophobe |
| KIRC | Kidney renal clear cell carcinoma |
| KIRP | Kidney renal papillary cell carcinoma |
| LAML | Acute myeloid leukaemia |
| LGG | Brain lower-grade glioma |
| LIHC | Liver hepatocellular carcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| MESO | Mesothelioma |
| OV | Ovarian serous cystadenocarcinoma |
| PAAD | Pancreatic adenocarcinoma |
| PCPG | Pheochromocytoma and paraganglioma |
| PRAD | Prostate adenocarcinoma |
| READ | Rectum adenocarcinoma |
| SARC | Sarcoma |
| SKCM | Skin cutaneous melanoma |
| STAD | Stomach adenocarcinoma |
| STES | Stomach and oesophageal carcinoma |
| TGCT | Testicular germ cell tumours |
| THCA | Thyroid carcinoma |
| THYM | Thymoma |
| UCEC | Uterine corpus endometrial carcinoma |
| UCS | Uterine carcinosarcoma |
| UVM | Uveal melanoma |
References
- Tomczak, K.; Czerwińska, P.; Wiznerowicz, M. Review the Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Chin, L.; Andersen, J.; Futreal, P.A. Cancer genomics: From discovery science to personalized medicine. Nat. Med. 2011, 17, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Huo, G.; Wang, Y.; Chen, J.; Song, Y.; Zhang, C.; Guo, H.; Zuo, R.; Zhu, F.; Cui, J.; Chen, W.; et al. A Pan-Cancer Analysis of the Oncogenic Role of Twinfilin Actin Binding Protein 1 in Human Tumors. Front. Oncol. 2021, 11, 692136. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Piel, W.H.; Gui, L.; Bruford, E.; Monteiro, A. The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics 2005, 86, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Cho, Y.; Lee, S. Cell Surface GRP94 as a Novel Emerging Therapeutic Target for Monoclonal Antibody Cancer Therapy. Cells 2021, 10, 670. [Google Scholar] [CrossRef]
- Graustein, A.D.; Misch, E.A.; Musvosvi, M.; Shey, M.; Shah, J.A.; Seshadri, C.; Aguoju, A.; Bowman, K.; Mulenga, H.; Veldsman, A.; et al. Toll-like receptor chaperone HSP90B1 and the immune response to Mycobacteria. PLoS ONE 2018, 13, e0208940. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, Z. Endoplasmic reticulum HSP90b1 (gp96, grp94) optimizes B-cell function via chaperoning integrin and TLR but not immunoglobulin. Blood 2008, 112, 1223–1230. [Google Scholar] [CrossRef]
- Liu, B.; Staron, M.; Hong, F.; Wu, B.X.; Sun, S.; Morales, C.; Crosson, C.E.; Tomlinson, S.; Kim, I.; Wu, D.; et al. Essential roles of grp94 in gut homeostasis via chaperoning canonical Wnt pathway. Proc. Natl. Acad. Sci. USA 2013, 110, 6877–6882. [Google Scholar] [CrossRef]
- Dejeans, N.; Glorieux, C.; Guenin, S.; Beck, R.; Sid, B.; Rousseau, R.; Bisig, B.; Delvenne, P.; Calderon, P.B.; Verrax, J. Overexpression of GRP94 in breast cancer cells resistant to oxidative stress promotes high levels of cancer cell proliferation and migration: Implications for tumor recurrence. Free Radic. Biol. Med. 2012, 52, 993–1002. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Liu, C.-G.; Mikhail, S.; Yang, C.S. Overexpression of glucose-regulated protein 94 (Grp94) in esophageal adenocarcinomas of a rat surgical model and humans. Carcinogenesis 2002, 23, 123–130. [Google Scholar] [CrossRef]
- Hu, T.; Xie, N.; Qin, C.; Wang, J.; You, Y. Glucose-regulated protein 94 is a novel glioma biomarker and promotes the aggressiveness of glioma via Wnt/β-catenin signaling pathway. Tumor Biol. 2015, 36, 9357–9364. [Google Scholar] [CrossRef] [PubMed]
- Rachidi, S.; Sun, S.; Wu, B.X.; Jones, E.; Drake, R.R.; Ogretmen, B.; Cowart, L.A.; Clarke, C.J.; Hannun, Y.A.; Chiosis, G.; et al. Endoplasmic reticulum heat shock protein gp96 maintains liver homeostasis and promotes hepatocellular carcinogenesis. J. Hepatol. 2014, 62, 879–888. [Google Scholar] [CrossRef]
- Duan, X.-F.; Xin, Y.-W. Overexpression of molecule GRP94 favors tumor progression in lung adenocarcinoma by interaction with regulatory T cells. Thorac. Cancer 2020, 11, 704–712. [Google Scholar] [CrossRef]
- Kim, K.; Lee, H.W.; Lee, E.H.; Park, M.I.; Lee, J.S.; Kim, M.S.; Kim, K.; Roh, M.S.; Pak, M.G.; Oh, J.E.; et al. Differential expression of HSP90 isoforms and their correlations with clinicopathologic factors in patients with colorectal cancer. Int. J. Clin. Exp. Pathol. 2019, 12, 978–986. [Google Scholar]
- Nomura, H.; Uzawa, K.; Yamano, Y.; Fushimi, K.; Ishigami, T.; Kato, Y.; Saito, K.; Nakashima, D.; Higo, M.; Kouzu, Y.; et al. Network-based analysis of calcium-binding protein genes identifies Grp94 as a target in human oral carcinogenesis. Br. J. Cancer 2007, 97, 792–801. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Lee, C.-H.; Tu, C.-C.; Wu, C.-H.; Huang, M.-T.; Wei, P.-L.; Chang, Y.-J. Glucose-regulated protein 94 mediates progression and metastasis of esophageal squamous cell carcinoma via mitochondrial function and the NF-kB/COX-2/VEGF axis. Oncotarget 2018, 9, 9425–9441. [Google Scholar] [CrossRef]
- Zheng, H.-C.; Takahashi, H.; Li, X.-H.; Hara, T.; Masuda, S.; Guan, Y.-F.; Takano, Y. Overexpression of GRP78 and GRP94 are markers for aggressive behavior and poor prognosis in gastric carcinomas. Hum. Pathol. 2008, 39, 1042–1049. [Google Scholar] [CrossRef]
- Yun, S.; Lee, S.; Lee, H.-Y.; Oh, H.; Kwak, Y.; Lee, H. Clinicopathologic and Prognostic Association of GRP94 Expression in Colorectal Cancer with Synchronous and Metachronous Metastases. Int. J. Mol. Sci. 2021, 22, 7042. [Google Scholar] [CrossRef]
- Pakravan, N.; Langroudi, L.; Hajimoradi, M.; Hassan, Z.M. Co-administration of GP96 and Her2/neu DNA vaccine in a Her2 breast cancer model. Cell Stress Chaperones 2010, 15, 977–984. [Google Scholar] [CrossRef]
- McShane, R.; Arya, S.; Stewart, A.J.; Caie, P.D.; Bates, M. Prognostic features of the tumour microenvironment in oesophageal ad-enocarcinoma. Biochim. Biophys. Acta. Rev. Cancer 2021, 1876, 188598. [Google Scholar] [CrossRef]
- Lv, M.; Cai, Y.; Hou, W.; Peng, K.; Xu, K.; Lu, C.; Yu, W.; Zhang, W.; Liu, L. The RNA-binding protein SND1 promotes the degradation of GPX4 by destabilizing the HSPA5 mRNA and suppressing HSPA5 expression, promoting ferroptosis in osteoarthritis chondrocytes. Agents Actions 2022, 71, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, W.; Liu, Y.; Ma, Z.; Xiang, W.; Wen, Y.; Zhang, D.; Li, Y.; Li, Y.; Li, T.; et al. PDIA4 promotes glioblastoma progression via the PI3K/AKT/m-TOR pathway. Biochem. Biophys. Res. Commun. 2022, 597, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, H.; Chen, R.; Zhang, Y.; Sun, X.-X.; Huang, W.; Bian, H.; Chen, Z.-N. Combination of CALR and PDIA3 is a potential prognostic biomarker for non-small cell lung cancer. Oncotarget 2017, 8, 96945–96957. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Liu, X.; Qi, X.; Liu, X.; Peng, F.; Li, H.; Fu, H.; Pei, S.; Chen, L.; Chi, X.; et al. PDIA6 modulates apoptosis and autophagy of non-small cell lung cancer cells via the MAP4K1/JNK signaling pathway. eBioMedicine 2019, 42, 311–325. [Google Scholar] [CrossRef]
- He, J.; Li, G.; Liu, X.; Ma, L.; Zhang, P.; Zhang, J.; Zheng, S.; Wang, J.; Liu, J. Diagnostic and Prognostic Values of MANF Expression in Hepatocellular Carcinoma. BioMed Res. Int. 2020, 2020, 1936385. [Google Scholar] [CrossRef]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef]
- Graf, R.P.; Fisher, V.; Mateo, J.; Gjoerup, O.V.; Madison, R.W.; Raskina, K.; Tukachinsky, H.; Creeden, J.; Cunningham, R.; Huang, R.S.; et al. Predictive Genomic Biomarkers of Hormonal Therapy Versus Chemotherapy Benefit in Metastatic Castration-resistant Prostate Cancer. Eur. Urol. 2021, 81, 37–47. [Google Scholar] [CrossRef]
- Gu, C.; Chang, W.; Wu, J.; Yao, Y.; Liu, G.; Yuan, Y.; Quan, W.; Sun, Z.; Shang, A.; Li, D. NCOA4: An Immunomodulation-Related Prognostic Biomarker in Colon Adenocarcinoma and Pan-Cancer. J. Oncol. 2022, 2022, 5242437. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Hageman, J.; Vos, M.J.; Kubota, H.; Tanguay, R.M.; Bruford, E.A.; Cheetham, M.E.; Chen, B.; Hightower, L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 2009, 14, 105–111. [Google Scholar] [CrossRef]
- Anas, A.A.; De Vos, A.F.; Hoogendijk, A.J.; Van Lieshout, M.H.P.; Van Heijst, J.W.J.; Florquin, S.; Li, Z.; Veer, C.V.; Van Der Poll, T. Endoplasmic reticulum chaperone gp96 in macrophages is essential for protective immunity during Gram-negative pneumonia. J. Pathol. 2015, 238, 74–84. [Google Scholar] [CrossRef]
- Li, L.; Mo, H.; Zhang, J.; Zhou, Y.; Peng, X.; Luo, X. The Role of Heat Shock Protein 90B1 in Patients with Polycystic Ovary Syndrome. PLoS ONE 2016, 11, e0152837. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Guo, A.-P.; Xu, J.-C.; You, Q.-D.; Xu, X.-L. Discovery of a Potent Grp94 Selective Inhibitor with Anti-Inflammatory Efficacy in a Mouse Model of Ulcerative Colitis. J. Med. Chem. 2018, 61, 9513–9533. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Xu, L.; He, W.; Dai, J.; Sun, F. Long noncoding RNA DLX6-AS1 promotes cell growth and invasiveness in bladder cancer via modulating the miR-223-HSP90B1 axis. Cell Cycle 2019, 18, 3288–3299. [Google Scholar] [CrossRef]
- Lee, J.S.; Roh, M.S.; Lee, H.W.; Lee, E.H.; Pak, M.G.; Kim, K.; Nam, H.-Y.; Kim, K.M.; Jung, S.B. Prognostic significance of glucose-related protein 94 in colorectal cancer. Pathol.-Res. Pr. 2020, 216, 153013. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feng, Y.; Chen, S. HSP90B1 overexpression is associated with poor prognosis in tongue squamous cell carcinoma. J. Stomatol. Oral Maxillofac. Surg. 2022. [Google Scholar] [CrossRef]
- Bin Cho, Y.; Kim, J.W.; Heo, K.; Kim, H.J.; Yun, S.; Lee, H.S.; Shin, H.G.; Shim, H.; Yu, H.; Kim, Y.-H.; et al. An internalizing antibody targeting of cell surface GRP94 effectively suppresses tumor angiogenesis of colorectal cancer. Biomed. Pharmacother. 2022, 150, 113051. [Google Scholar] [CrossRef]
- Gao, Y.; Pan, Y.; Wang, T.; Yao, Y.; Yuan, W.; Zhu, X.; Wang, K. MicroRNA-99a-3p/GRP94 axis affects metastatic progression of human papillary thyroid carcinoma by regulating ITGA2 expression and localization. Acta Biochim. Biophys. Sin. 2021, 53, 1650–1661. [Google Scholar] [CrossRef]
- Li, X.; Sun, L.; Hou, J.; Gui, M.; Ying, J.; Zhao, H.; Lv, N.; Meng, S. Cell membrane gp96 facilitates HER2 dimerization and serves as a novel target in breast cancer. Int. J. Cancer 2014, 137, 512–524. [Google Scholar] [CrossRef]
- Lu, T.; Wang, Y.; Xu, K.; Zhou, Z.; Gong, J.; Zhang, Y.; Gong, H.; Dai, Q.; Yang, J.; Xiong, B.; et al. Co-downregulation of GRP78 and GRP94 induces apoptosis and inhibits migration in prostate cancer cells. Open Life Sci. 2019, 14, 384–391. [Google Scholar] [CrossRef]
- Huang, L.; Jian, Z.; Gao, Y.; Zhou, P.; Zhang, G.; Jiang, B.; Lv, Y. RPN2 promotes metastasis of hepatocellular carcinoma cell and inhibits autophagy via STAT3 and NF-κB pathways. Aging 2019, 11, 6674–6690. [Google Scholar] [CrossRef]
- Han, C.; Chen, S.; Ma, H.; Wen, X.; Wang, Z.; Xu, Y.; Jin, X.; Yu, X.; Wang, M. RPN2 Predicts Poor Prognosis and Promotes Bladder Cancer Growth and Metastasis via the PI3K-Akt Pathway. OncoTargets Ther. 2021, 14, 1643–1657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yan, B.; Späth, S.S.; Qun, H.; Cornelius, S.; Guan, D.; Shao, J.; Hagiwara, K.; Van Waes, C.; Chen, Z.; et al. Integrated transcriptional profiling and genomic analyses reveal RPN2 and HMGB1 as promising biomarkers in colorectal cancer. Cell Biosci. 2015, 5, 53. [Google Scholar] [CrossRef]
- Fujita, Y.; Yagishita, S.; Takeshita, F.; Yamamoto, Y.; Kuwano, K.; Ochiya, T. Prognostic and therapeutic impact of RPN2-mediated tumor malignancy in non-small-cell lung cancer. Oncotarget 2015, 6, 3335–3345. [Google Scholar] [CrossRef]
- Fujimoto, D.; Goi, T.; Kurebayashi, H.; Kato, S.; Morikawa, M.; Koneri, K.; Murakami, M.; Hirono, Y. 252P Expression of Ribophorine II (RPN2) is a new prognostic factor in human gastric adenocarcinoma. Ann. Oncol. 2016, 27. [Google Scholar] [CrossRef]
- Hanafusa, K.; Wada, I.; Hosokawa, N. SDF2-like protein 1 (SDF2L1) regulates the endoplasmic reticulum localization and chaperone activity of ERdj3 protein. J. Biol. Chem. 2019, 294, 19335–19348. [Google Scholar] [CrossRef]
- Chen, H.-Y.; Chang, J.T.-C.; Chien, K.-Y.; Lee, Y.-S.; You, G.-R.; Cheng, A.-J. The Endogenous GRP78 Interactome in Human Head and Neck Cancers: A Deterministic Role of Cell Surface GRP78 in Cancer Stemness. Sci. Rep. 2018, 8, 536. [Google Scholar] [CrossRef] [PubMed]
- Conner, C.; Lager, T.W.; Guldner, I.H.; Wu, M.-Z.; Hishida, Y.; Hishida, T.; Ruiz, S.; Yamasaki, A.E.; Gilson, R.C.; Belmonte, J.C.I.; et al. Cell surface GRP78 promotes stemness in normal and neoplastic cells. Sci. Rep. 2020, 10, 3474. [Google Scholar] [CrossRef]
- Yang, W.; Xiu, Z.; He, Y.; Huang, W.; Li, Y.; Sun, T. Bip inhibition in glioma stem cells promotes radiation-induced immunogenic cell death. Cell Death Dis. 2020, 11, 786. [Google Scholar] [CrossRef]
- Rao, S.; Oyang, L.; Liang, J.; Yi, P.; Han, Y.; Luo, X.; Xia, L.; Lin, J.; Tan, S.; Hu, J.; et al. Biological Function of HYOU1 in Tumors and Other Diseases. OncoTargets Ther. 2021, 14, 1727–1735. [Google Scholar] [CrossRef]
- Choi, T.G.; Nguyen, M.N.; Kim, J.; Jo, Y.H.; Jang, M.; Nguyen, N.N.Y.; Yun, H.R.; Choe, W.; Kang, I.; Ha, J.; et al. Cyclophilin B induces chemoresistance by degrading wild-type p53 via interaction with MDM2 in colorectal cancer. J. Pathol. 2018, 246, 115–126. [Google Scholar] [CrossRef]
- Wu, H.; Xie, D.; Yang, Y.; Yang, Q.; Shi, X.; Yang, R. Ultrasound-Targeted Microbubble Destruction-Mediated miR-206 Overexpression Promotes Apoptosis and Inhibits Metastasis of Hepatocellular Carcinoma Cells Via Targeting PPIB. Technol. Cancer Res. Treat. 2020, 19, 1533033820959355. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xia, P.; Wang, Z.; Xu, J.; Zhang, L.; Jiang, Y. PDIA6 promotes pancreatic cancer progression and immune escape through CSN5-mediated deubiquitination of β-catenin and PD-L1. Neoplasia 2021, 23, 912–928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, L.; Luo, G. Long non-coding RNA PCAT6 regulates bladder cancer progression via the microRNA-143-3p/PDIA6 axis. Exp. Ther. Med. 2021, 22, 947. [Google Scholar] [CrossRef]
- Mao, L.; Wu, X.; Gong, Z.; Yu, M.; Huang, Z. PDIA6 contributes to aerobic glycolysis and cancer progression in oral squamous cell carcinoma. World J. Surg. Oncol. 2021, 19, 88 . [Google Scholar] [CrossRef]
- Su, S.; Chien, C.; Chen, Y.; Chiang, C.; Lin, F.; Kuo, F.; Huang, C.; Li, P.; Liu, J.; Lu, C.; et al. PDIA4, a novel ER stress chaperone, modulates adiponectin expression and inflammation in adipose tissue. BioFactors 2022. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, H.; Cheng, Q. PDIA4: The basic characteristics, functions and its potential connection with cancer. Biomed. Pharmacother. 2019, 122, 109688. [Google Scholar] [CrossRef]
- Tufo, G.; E Jones, A.W.; Wang, Z.; Hamelin, J.; Tajeddine, N.; Esposti, D.D.; Martel, C.; Boursier, C.; Gallerne, C.; Migdal, C.; et al. The protein disulfide isomerases PDIA4 and PDIA6 mediate resistance to cisplatin-induced cell death in lung adenocarcinoma. Cell Death Differ. 2014, 21, 685–695. [Google Scholar] [CrossRef]
- Gao, F.; Mu, X.; Wu, H.; Chen, L.; Liu, J.; Zhao, Y. Calreticulin (CALR)-induced activation of NF-ĸB signaling pathway boosts lung cancer cell proliferation. Bioengineered 2022, 13, 6856–6865. [Google Scholar] [CrossRef]
- Holmstrom, M.O.; Martinenaite, E.; Ahmad, S.M.; Met, O.; Friese, C.; Kjaer, L.; Riley, C.H.; Thor Straten, P.; Svane, I.M.; Hasselbalch, H.C.; et al. The calreticulin (CALR) exon 9 mutations are promising targets for cancer immune therapy. Leukemia 2018, 32, 429–437. [Google Scholar] [CrossRef]
- Sun, S.; Kiang, K.M.; Leung, G.K. Chaperone protein P4HB predicts temozolomide response and prognosis in malignant glioma. Oncol. Lett. 2022, 24, 1–10. [Google Scholar] [CrossRef]
- Yuan, Z.; Hu, H.; Zhu, Y.; Zhang, W.; Fang, Q.; Qiao, T.; Ma, T.; Wang, M.; Huang, R.; Tang, Q.; et al. Colorectal cancer cell intrinsic fibroblast activation protein alpha binds to Enolase1 and activates NF-κB pathway to promote metastasis. Cell Death Dis. 2021, 12, 543. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).