Clinical Significance and Immune Infiltration Analyses of the Cuproptosis-Related Human Copper Proteome in Gastric Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Cohort and Data Collection
2.2. Establishment of the CBP-Related Signature for Prognosis
2.3. Immunohistochemistry (IHC)
2.4. Immune Profile Analysis
2.5. Tumor Regression Grade (TRG) Assessment
2.6. Multiplex Immunofluorescence (mIF)
2.7. Statistical Analysis
3. Results
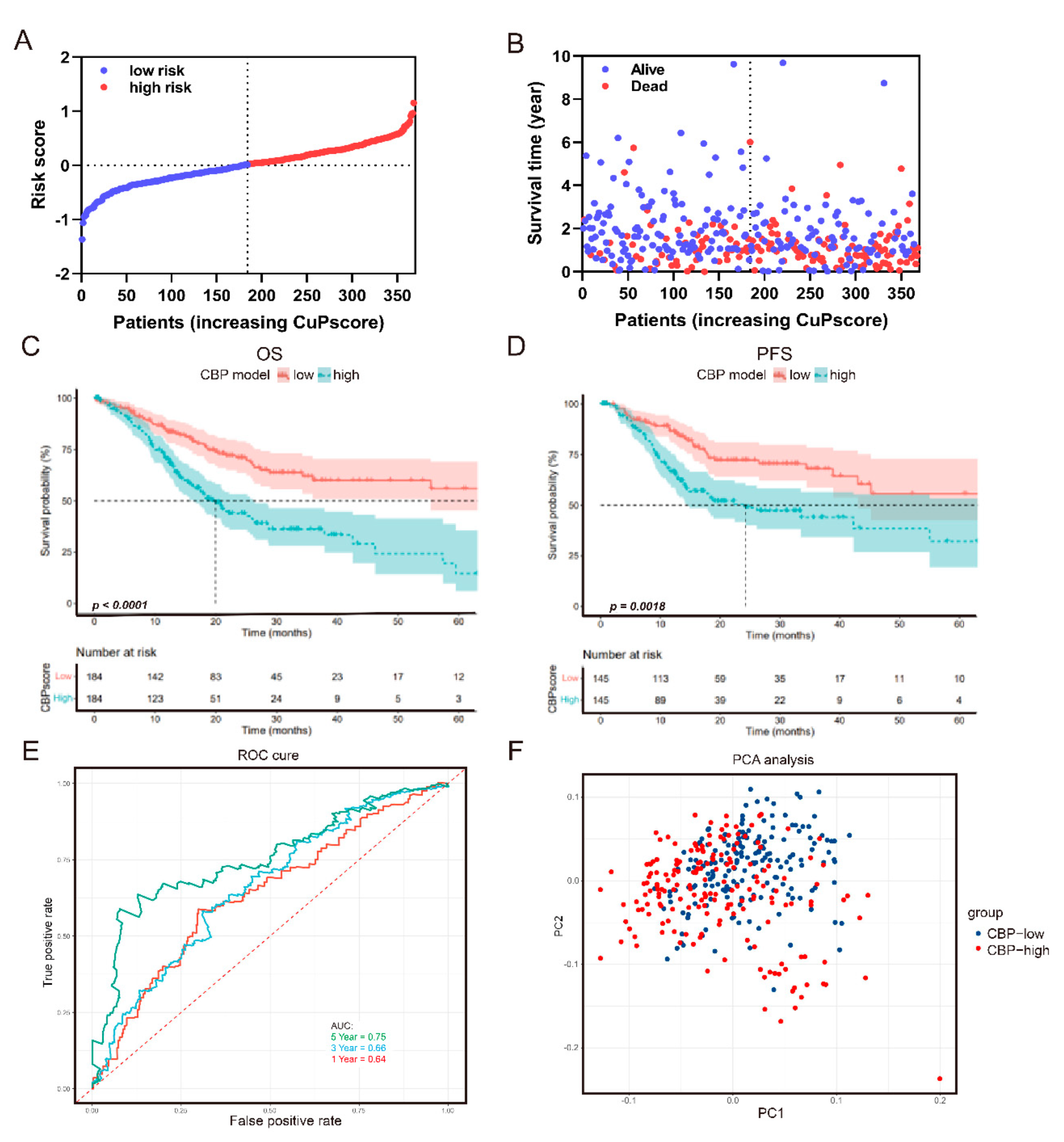
3.1. Construction of the CBP-Related Prognostic Signature in GC
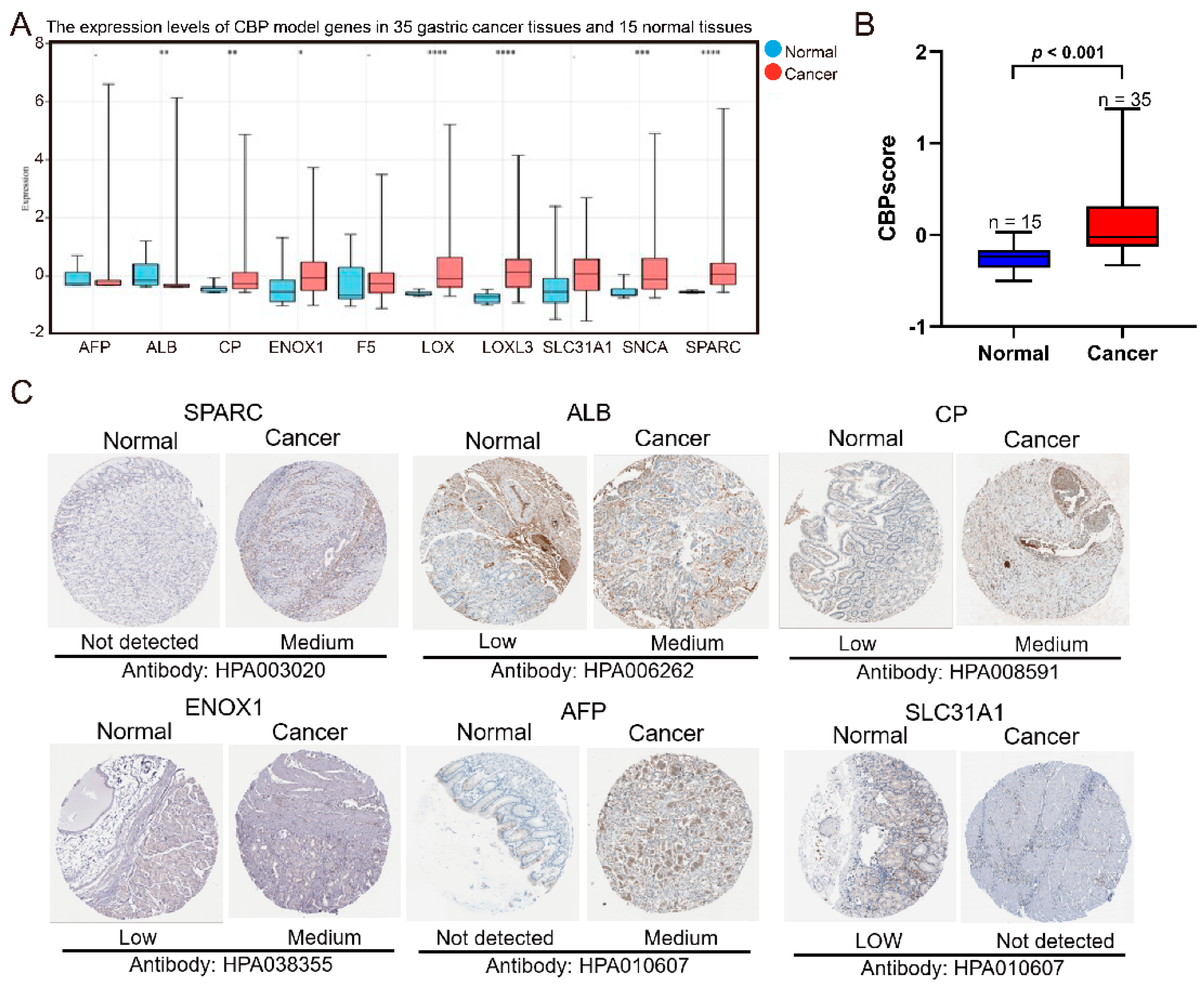
3.2. Verification of the Expression of Ten CBP Signature Genes between GC and Adjacent Normal Tissues
3.3. The Prognostic Value of the CBP Signature
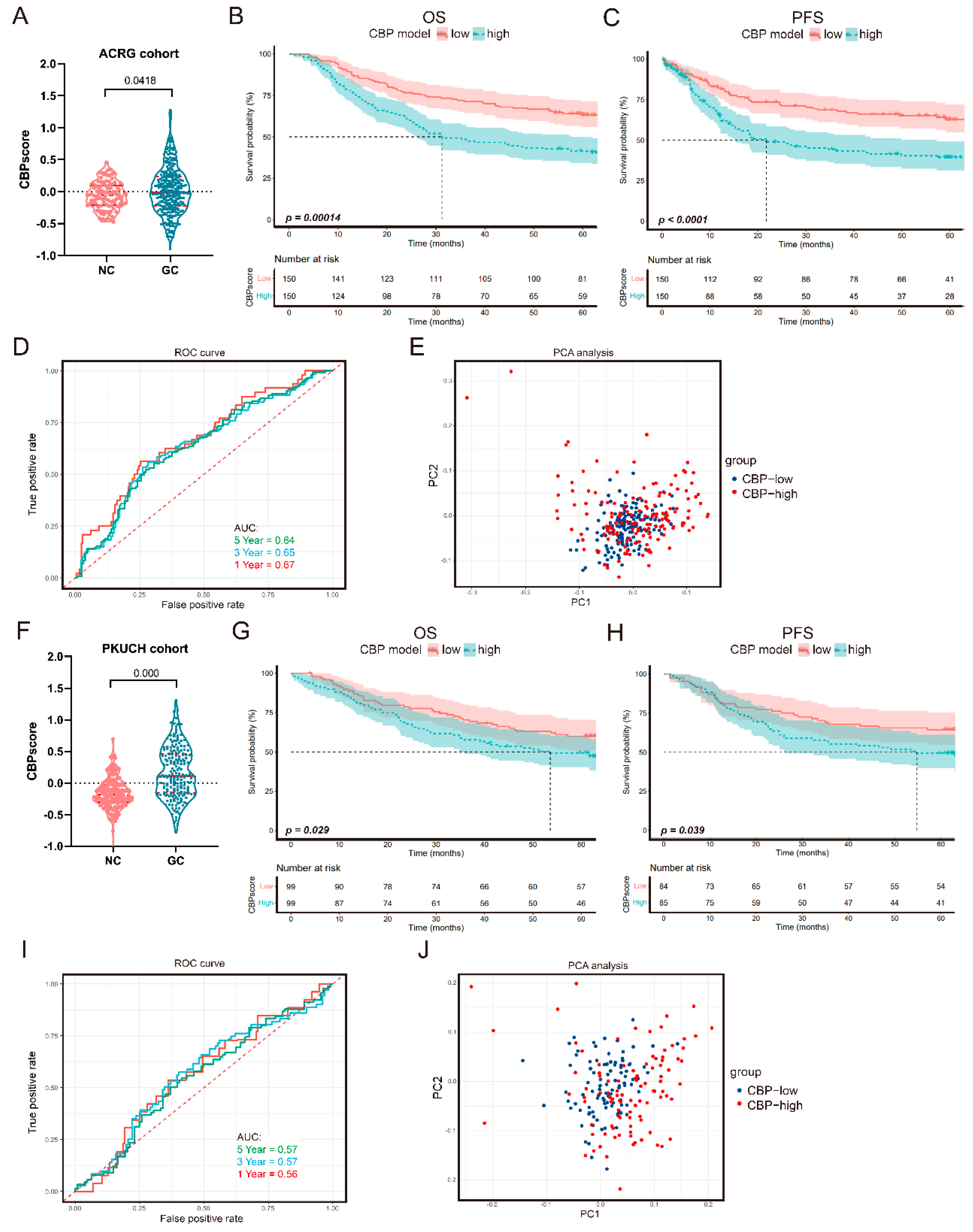
3.4. Verification of the CBP Prognostic Signature
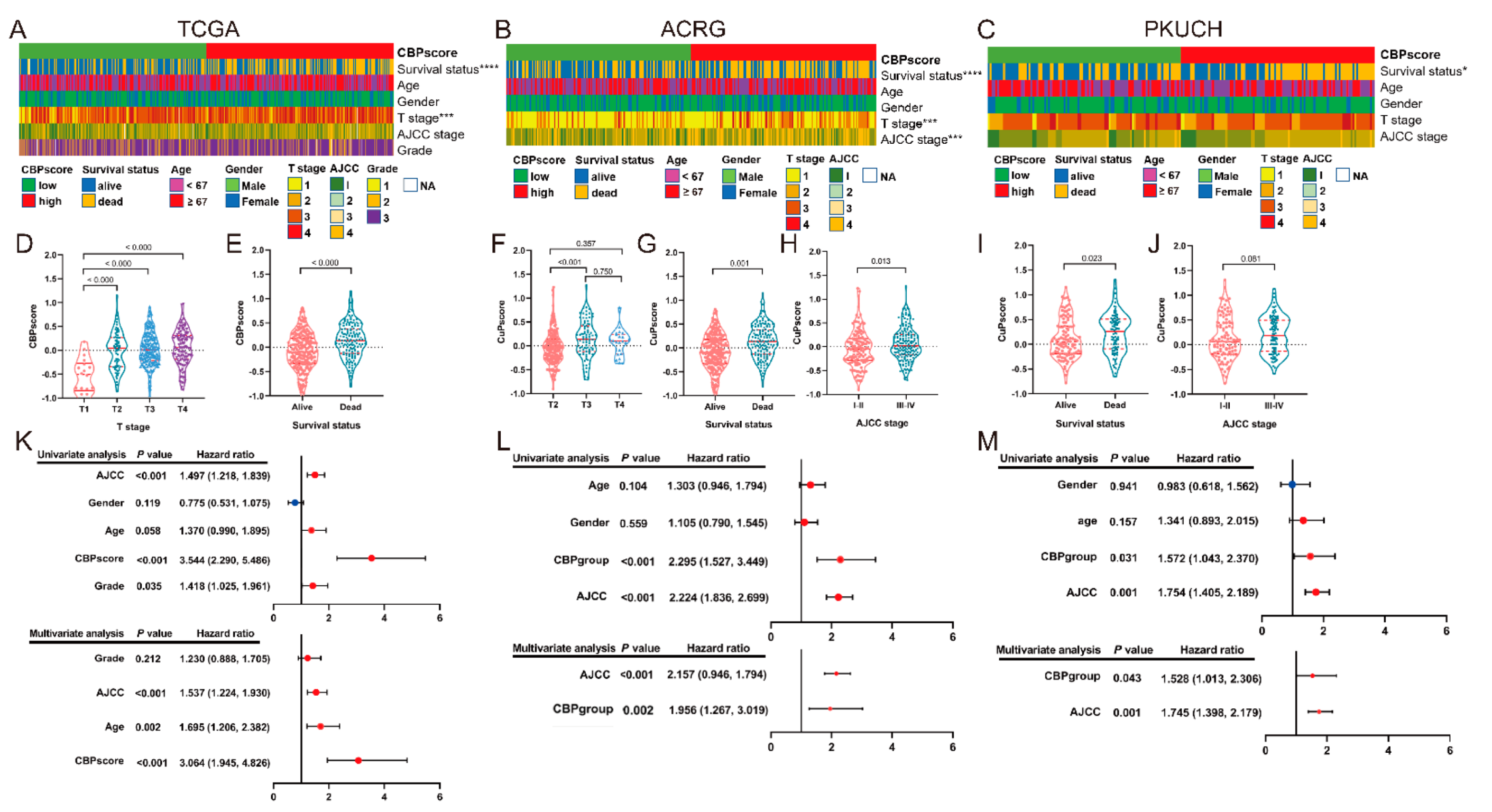
3.5. Correlation Analyses between the CBP Signature and Clinical Information
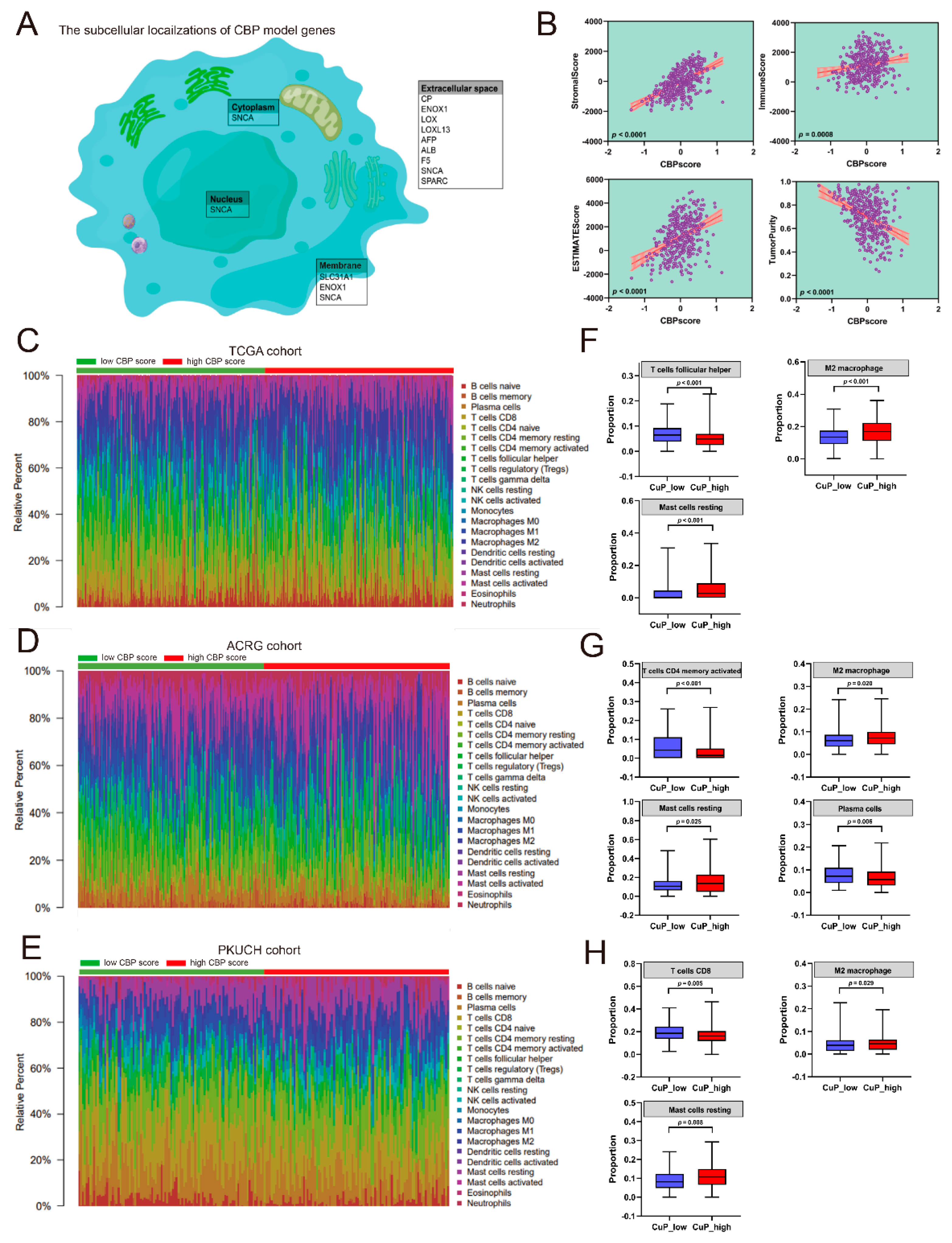
3.6. Correlation Analyses between the CBP Signature and Immune Infiltration
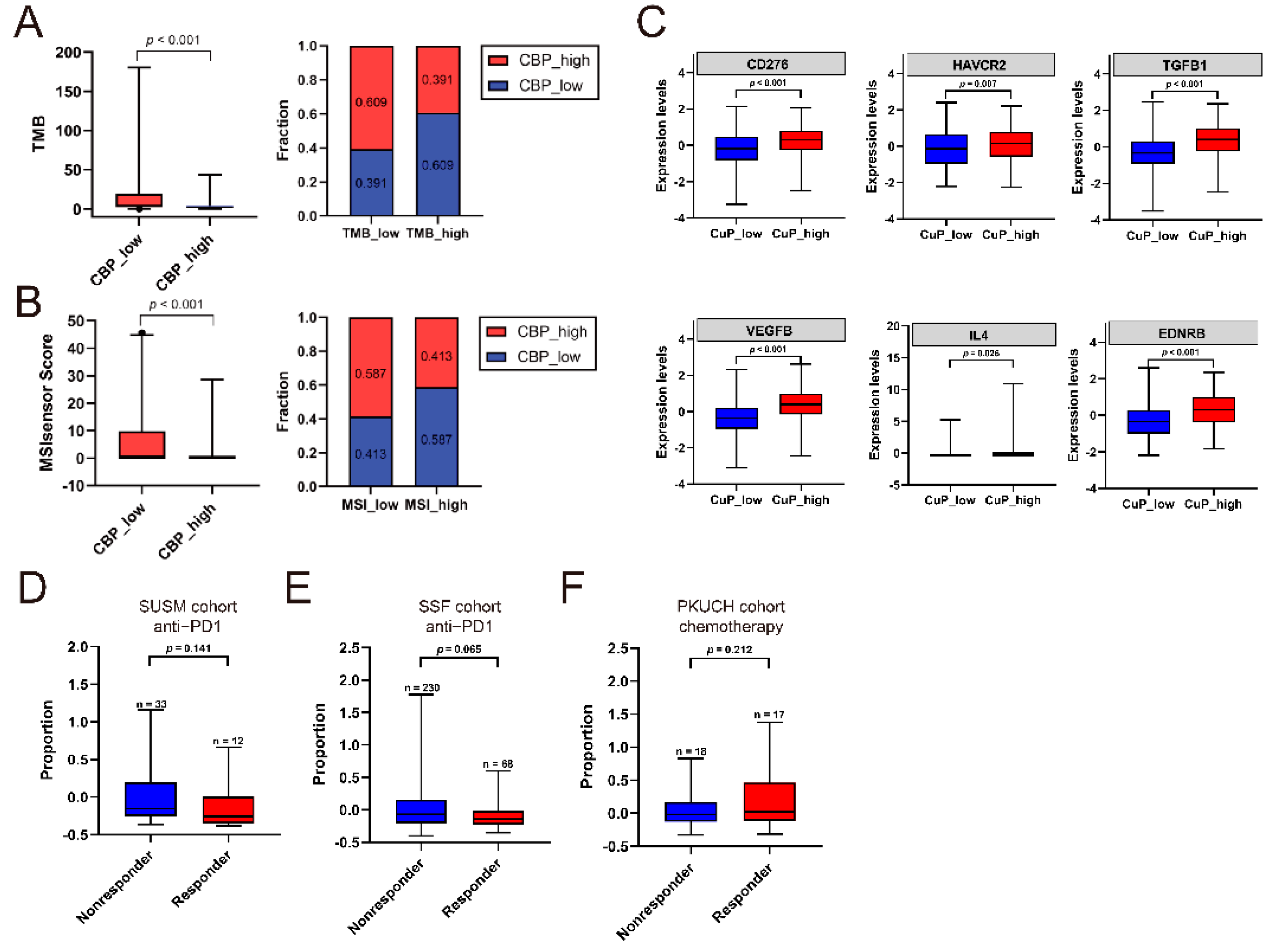
3.7. Association of CBP Signature, TMB/MSI and Response to Treatment
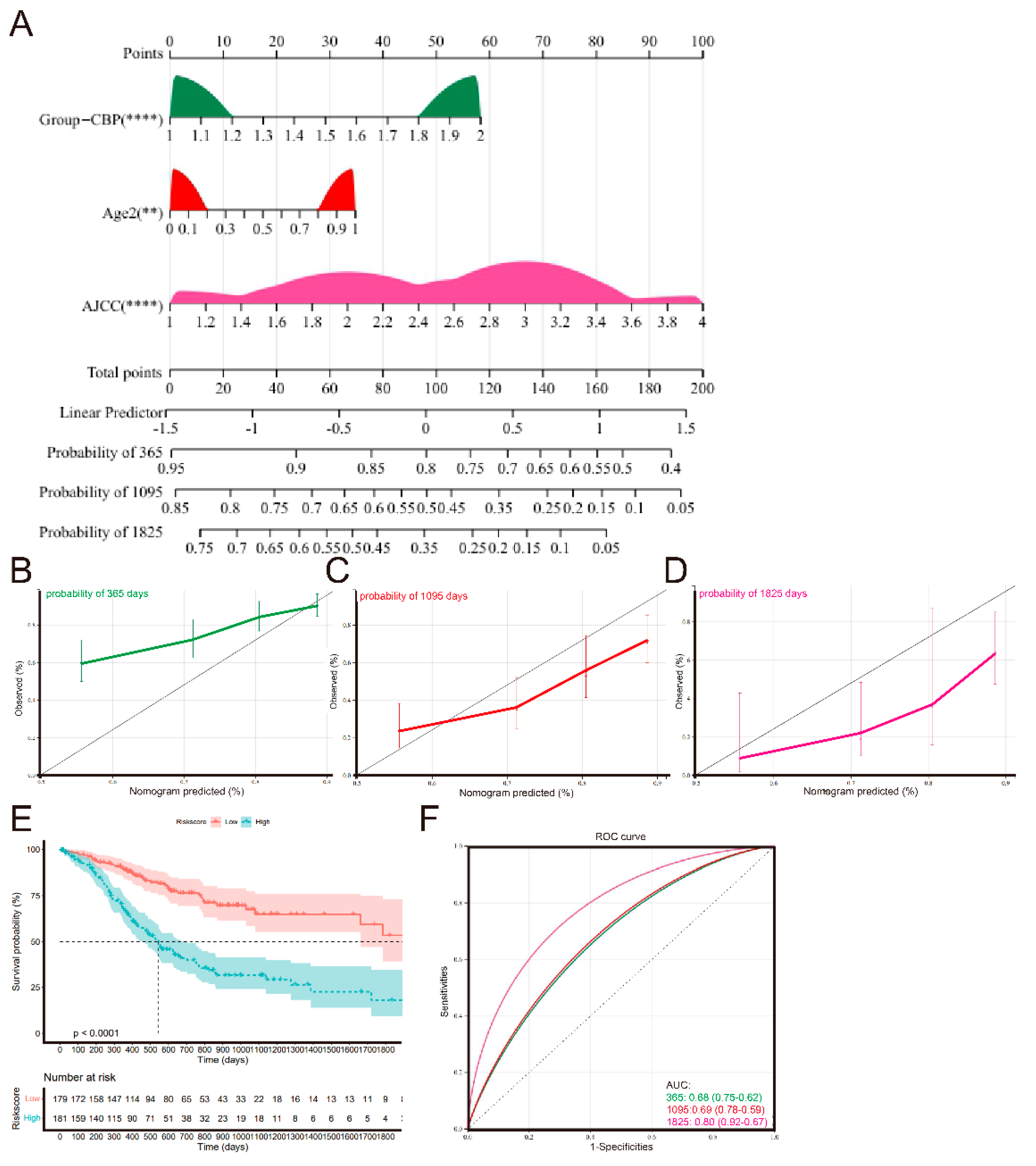
3.8. Establishment and Validation of a Nomogram Signature
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ying, X.; Liu, S.; Lyu, G.; Xu, Z.; Zhang, X.; Li, H.; Li, Q.; Wang, N.; Ji, J. Gastric cancer: Epidemiology, risk factors and prevention strategies. Chin. J. Cancer Res. 2020, 32, 695–704. [Google Scholar] [CrossRef]
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Gastroenterol. Rev. Przegląd Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Gastric Cancer Association; China Anti-Cancer Association. Chinese expert consensus on perioperative treatment of locally advanced gastric cancer (2021 version). Zhonghua Wei Chang Wai Ke Za Zhi 2021, 24, 741–748. [Google Scholar] [CrossRef]
- Janjigian, Y.Y.; Shitara, K.; Moehler, M.; Garrido, M.; Salman, P.; Shen, L.; Wyrwicz, L.; Yamaguchi, K.; Skoczylas, T.; Bragagnoli, A.C.; et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): A randomised, open-label, phase 3 trial. Lancet 2021, 398, 27–40. [Google Scholar] [CrossRef]
- Kang, Y.K.; Chen, L.T.; Ryu, M.H.; Oh, D.Y.; Oh, S.C.; Chung, H.C.; Lee, K.W.; Omori, T.; Shitara, K.; Sakuramoto, S.; et al. Nivolumab plus chemotherapy versus placebo plus chemotherapy in patients with HER2-negative, untreated, unresectable advanced or recurrent gastric or gastro-oesophageal junction cancer (ATTRACTION-4): A randomised, multicentre, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2022, 23, 234–247. [Google Scholar] [CrossRef]
- Strasser, A.; Vaux, D.L. Cell death in the origin and treatment of cancer. Mol. Cell 2020, 78, 1045–1054. [Google Scholar] [CrossRef]
- Badgley, M.A.; Kremer, D.M.; Maurer, H.C.; DelGiorno, K.E.; Lee, H.J.; Purohit, V.; Sagalovskiy, I.R.; Ma, A.; Kapilian, J.; Firl, C.E.M.; et al. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science 2020, 368, 85–89. [Google Scholar] [CrossRef]
- Fang, Y.; Tian, S.; Pan, Y.; Li, W.; Wang, Q.; Tang, Y.; Yu, T.; Wu, X.; Shi, Y.; Ma, P.; et al. Pyroptosis: A new frontier in cancer. Biomed. Pharmacother. 2020, 121, 109595. [Google Scholar] [CrossRef]
- Yu, J.; Li, S.; Qi, J.; Chen, Z.; Wu, Y.; Guo, J.; Wang, K.; Sun, X.; Zheng, J. Cleavage of GSDME by caspase-3 determines lobaplatin-induced pyroptosis in colon cancer cells. Cell Death Dis. 2019, 10, 193. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Cohen, L.J.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Xia, Y.; Liu, X.; Zhang, L.; Zhang, J.; Li, C.; Zhang, N.; Xu, H.; Li, Y. A new Schiff base coordinated copper(II) compound induces apoptosis and inhibits tumor growth in gastric cancer. Cancer Cell Int. 2019, 19, 81. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Guan, X.; Wang, M.; Wang, N.; Chen, Y.; Li, B.; Xu, Z.; Fu, F.; Zheng, Z.; Du, C. Disulfiram/Copper induces antitumor activity against gastric cancer via the ROS/MAPK and NPL4 pathways. Bioengineered 2022, 13, 6579–6589. [Google Scholar] [CrossRef]
- Blockhuys, S.; Celauro, E.; Hildesjo, C.; Feizi, A.; Stal, O.; Fierro-Gonzalez, J.C.; Wittung-Stafshede, P. Defining the human copper proteome and analysis of its expression variation in cancers. Metallomics 2017, 9, 112–123. [Google Scholar] [CrossRef] [Green Version]
- Koch, K.A.; Pena, M.M.; Thiele, D.J. Copper-binding motifs in catalysis, transport, detoxification and signaling. Chem. Biol. 1997, 4, 549–560. [Google Scholar] [CrossRef] [Green Version]
- Weaver, M.S.; Workman, G.; Sage, E.H. The copper binding domain of SPARC mediates cell survival in vitro via interaction with integrin beta1 and activation of integrin-linked kinase. J. Biol. Chem. 2008, 283, 22826–22837. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Xu, Z.; Hu, X.; Qian, L.; Li, Z.; Zhou, Y.; Dai, S.; Zeng, S.; Gong, Z. SNCA Is a Functionally low-expressed gene in lung adenocarcinoma. Genes 2018, 9, 16. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Zhou, R.; Zhao, Y.; Pan, Y.; Liang, H.; Zhang, J.S.; Tai, S.; Jin, L.; Teng, C.B. Blockage of SLC31A1-dependent copper absorption increases pancreatic cancer cell autophagy to resist cell death. Cell Prolif. 2019, 52, e12568. [Google Scholar] [CrossRef] [Green Version]
- Xing, X.; Shi, J.; Jia, Y.; Dou, Y.; Li, Z.; Dong, B.; Guo, T.; Cheng, X.; Li, X.; Du, H.; et al. Effect of neoadjuvant chemotherapy on the immune microenvironment in gastric cancer as determined by multiplex immunofluorescence and T cell receptor repertoire analysis. J. Immunother. Cancer 2022, 10, e003984. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martinez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, W.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Sano, T.; Coit, D.G.; Kim, H.H.; Roviello, F.; Kassab, P.; Wittekind, C.; Yamamoto, Y.; Ohashi, Y. Proposal of a new stage grouping of gastric cancer for TNM classification: International Gastric Cancer Association staging project. Gastric Cancer 2017, 20, 217–225. [Google Scholar] [CrossRef]
- Li, Z.; Gao, X.; Peng, X.; May Chen, M.J.; Li, Z.; Wei, B.; Wen, X.; Wei, B.; Dong, Y.; Bu, Z.; et al. Multi-omics characterization of molecular features of gastric cancer correlated with response to neoadjuvant chemotherapy. Sci. Adv. 2020, 6, eaay4211. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef]
- Andre, T.; Tougeron, D.; Piessen, G.; de la Fouchardiere, C.; Louvet, C.; Adenis, A.; Jary, M.; Tournigand, C.; Aparicio, T.; Desrame, J.; et al. Neoadjuvant nivolumab plus ipilimumab and adjuvant nivolumab in localized deficient mismatch repair/microsatellite instability-high gastric or esophagogastric junction adenocarcinoma: The GERCOR NEONIPIGA Phase II Study. J. Clin. Oncol. 2022, JCO2200686. [Google Scholar] [CrossRef]
- Li, Z.; Jia, Y.; Zhu, H.; Xing, X.; Pang, F.; Shan, F.; Li, S.; Wand, D.; Zhao, D.; Ma, T.; et al. Tumor mutation burden is correlated with response and prognosis in microsatellite-stable (MSS) gastric cancer patients undergoing neoadjuvant chemotherapy. Gastric Cancer 2021, 24, 1342–1354. [Google Scholar] [CrossRef]
- Glehen, O.; Gilly, F.N.; Arvieux, C.; Cotte, E.; Boutitie, F.; Mansvelt, B.; Bereder, J.M.; Lorimier, M.; Quenet, F.; Elias, D.; et al. Peritoneal carcinomatosis from gastric cancer: A multi-institutional study of 159 patients treated by cytoreductive surgery combined with perioperative intraperitoneal chemotherapy. Ann. Surg. Oncol. 2010, 17, 2370–2377. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Sun, J.; Zhang, X. A novel Cuproptosis-related LncRNA signature to predict prognosis in hepatocellular carcinoma. Sci. Rep. 2022, 12, 11325. [Google Scholar] [CrossRef]
- Bian, Z.; Fan, R.; Xie, L. A novel cuproptosis-related prognostic gene signature and validation of differential expression in clear cell renal cell carcinoma. Genes 2022, 13, 851. [Google Scholar] [CrossRef]
- Barresi, V.; Trovato-Salinaro, A.; Spampinato, G.; Musso, N.; Castorina, S.; Rizzarelli, E.; Condorelli, D.F. Transcriptome analysis of copper homeostasis genes reveals coordinated upregulation of SLC31A1,SCO1, and COX11 in colorectal cancer. FEBS Open Bio 2016, 6, 794–806. [Google Scholar] [CrossRef]
- Li, L.; Zhu, Z.; Zhao, Y.; Zhang, Q.; Wu, X.; Miao, B.; Gao, J.; Fei, S. FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics. Sci. Rep. 2019, 9, 7827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.K.; Shen, L.; Jiao, X.; Zhang, X.T. Predictive and prognostic value of serum AFP level and its dynamic changes in advanced gastric cancer patients with elevated serum AFP. World J. Gastroenterol. 2018, 24, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Kasashima, H.; Yashiro, M.; Okuno, T.; Miki, Y.; Kitayama, K.; Masuda, G.; Kinoshita, H.; Morisaki, T.; Fukuoka, T.; Hasegawa, T.; et al. Significance of the lysyl oxidase members lysyl oxidase like 1, 3, and 4 in gastric cancer. Digestion 2018, 98, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Nai, A.; Zeng, H.; Wu, Q.; He, Z.; Zeng, S.; Bashir, S.; Ma, F.; He, J.; Wan, W.; Xu, M. lncRNA/miR-29c-mediated high expression of LOX can influence the immune status and chemosensitivity and can forecast the poor prognosis of gastric cancer. Front. Cell Dev. Biol. 2021, 9, 760470. [Google Scholar] [CrossRef]
- Feng, F.; Tian, Y.; Xu, G.; Liu, Z.; Liu, S.; Zheng, G.; Guo, M.; Lian, X.; Fan, D.; Zhang, H. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer 2017, 17, 737. [Google Scholar] [CrossRef]
- Sun, C.; Mezzadra, R.; Schumacher, T.N. Regulation and function of the PD-L1 checkpoint. Immunity 2018, 48, 434–452. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Zhao, B. Efficacy of PD-1 or PD-L1 inhibitors and PD-L1 expression status in cancer: Meta-analysis. BMJ 2018, 362, k3529. [Google Scholar] [CrossRef] [Green Version]
- Hellmann, M.D.; Nathanson, T.; Rizvi, H.; Creelan, B.C.; Sanchez-Vega, F.; Ahuja, A.; Ni, A.; Novik, J.B.; Mangarin, L.M.B.; Abu-Akeel, M.; et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 2018, 33, 843–852.e4. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Wu, X.; Guo, T.; Jia, F.; Hu, Y.; Xing, X.; Gao, X.; Li, Z. Focal Adhesion-related signatures predict the treatment efficacy of chemotherapy and prognosis in patients with gastric cancer. Front. Oncol. 2022, 12, 808817. [Google Scholar] [CrossRef]
- Kim, J.H.; Seo, M.K.; Lee, J.A.; Yoo, S.Y.; Oh, H.J.; Kang, H.; Cho, N.Y.; Bae, J.M.; Kang, G.H.; Kim, S. Genomic and transcriptomic characterization of heterogeneous immune subgroups of microsatellite instability-high colorectal cancers. J. Immunother. Cancer 2021, 9, e003414. [Google Scholar] [CrossRef]
- Westcott, P.M.K.; Sacks, N.J.; Schenkel, J.M.; Ely, Z.A.; Smith, O.; Hauck, H.; Jaeger, A.M.; Zhang, D.; Backlund, C.M.; Beytagh, M.C.; et al. Low neoantigen expression and poor T-cell priming underlie early immune escape in colorectal cancer. Nat. Cancer 2021, 2, 1071–1085. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, X.; Guo, T.; Wu, X.; Gan, X.; Wang, Y.; Jia, F.; Zhang, Y.; Xing, X.; Gao, X.; Li, Z. Clinical Significance and Immune Infiltration Analyses of the Cuproptosis-Related Human Copper Proteome in Gastric Cancer. Biomolecules 2022, 12, 1459. https://doi.org/10.3390/biom12101459
Tang X, Guo T, Wu X, Gan X, Wang Y, Jia F, Zhang Y, Xing X, Gao X, Li Z. Clinical Significance and Immune Infiltration Analyses of the Cuproptosis-Related Human Copper Proteome in Gastric Cancer. Biomolecules. 2022; 12(10):1459. https://doi.org/10.3390/biom12101459
Chicago/Turabian StyleTang, Xiaohuan, Ting Guo, Xiaolong Wu, Xuejun Gan, Yiding Wang, Fangzhou Jia, Yan Zhang, Xiaofang Xing, Xiangyu Gao, and Ziyu Li. 2022. "Clinical Significance and Immune Infiltration Analyses of the Cuproptosis-Related Human Copper Proteome in Gastric Cancer" Biomolecules 12, no. 10: 1459. https://doi.org/10.3390/biom12101459





