Changes to Urinary Proteome in High-Fat-Diet ApoE−/− Mice
Abstract
:1. Introduction
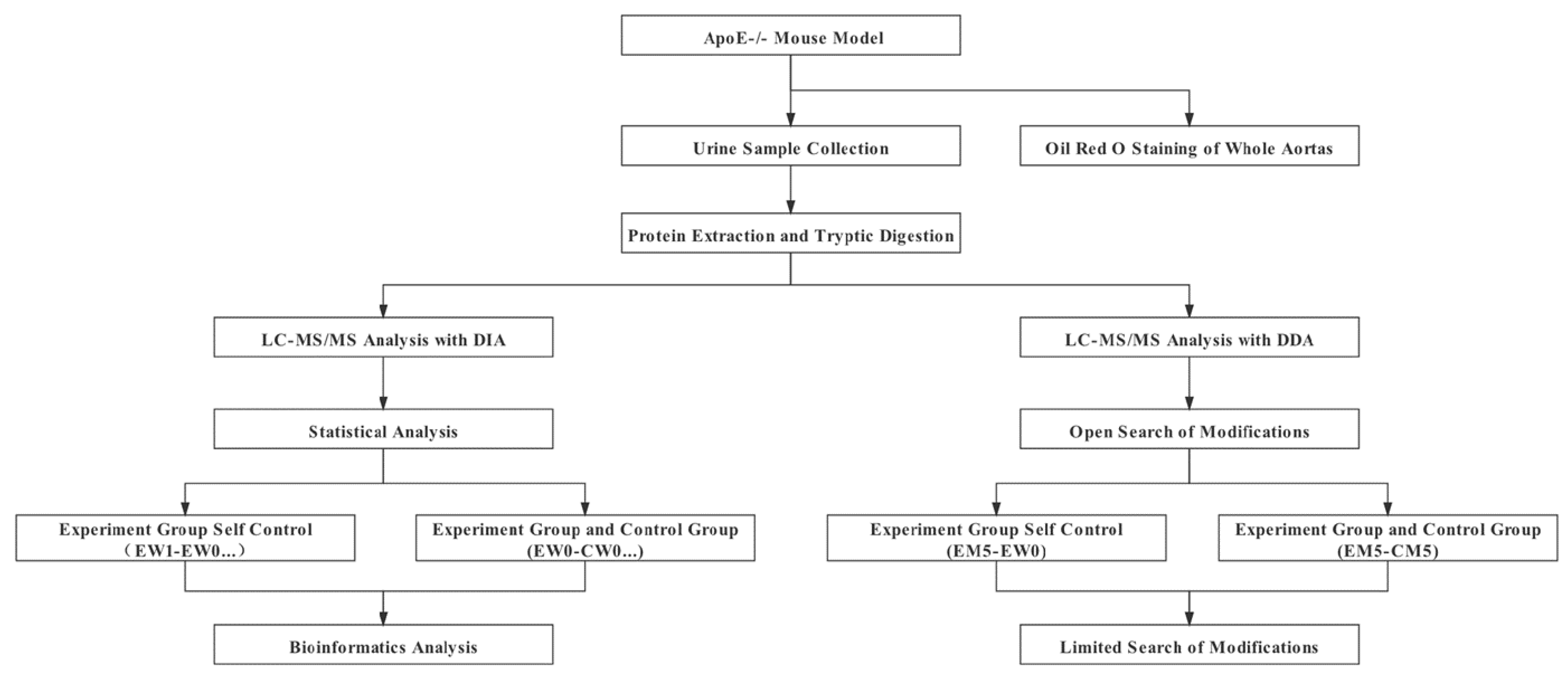
2. Materials and Methods
2.1. Experimental Animals
2.2. Histopathology
2.3. Urine Collection and Sample Preparation
2.4. Spin-Column Peptide Fractionation
2.5. LC-MS/MS Analysis
2.6. Label-free DIA Quantification
2.7. Protein Chemical Modifications Search
2.8. Statistical Analysis
3. Results
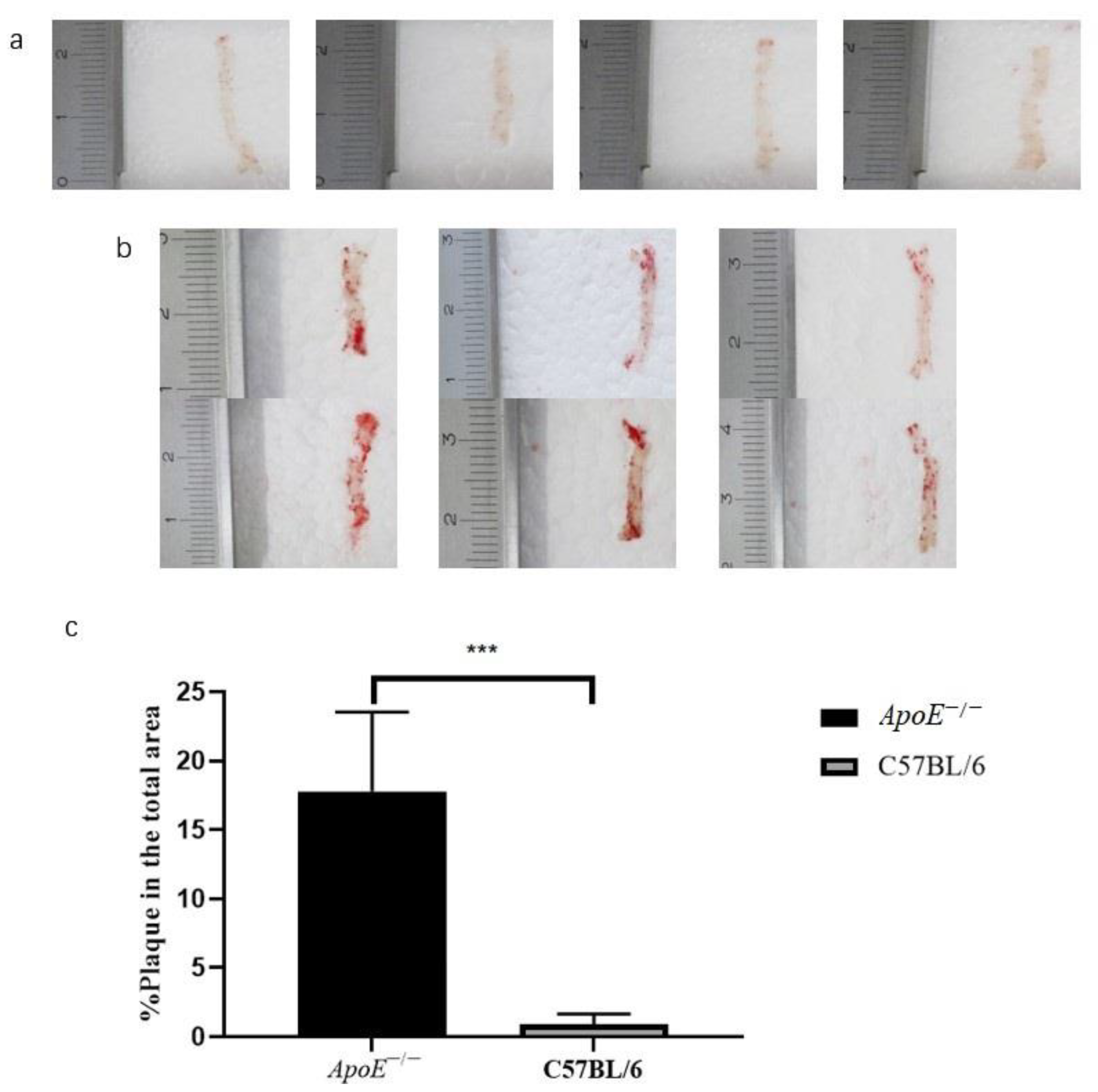
3.1. Histopathology
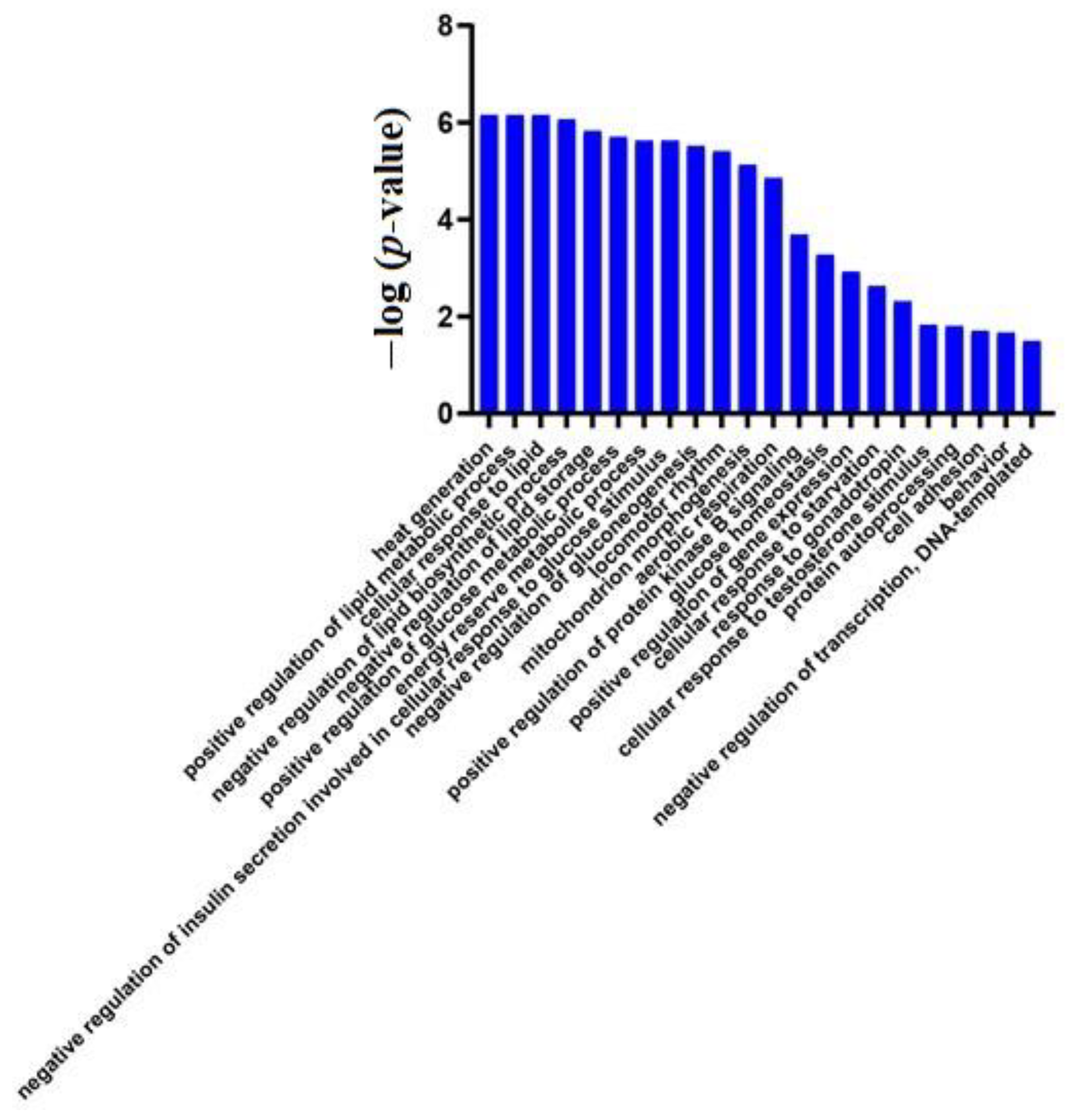
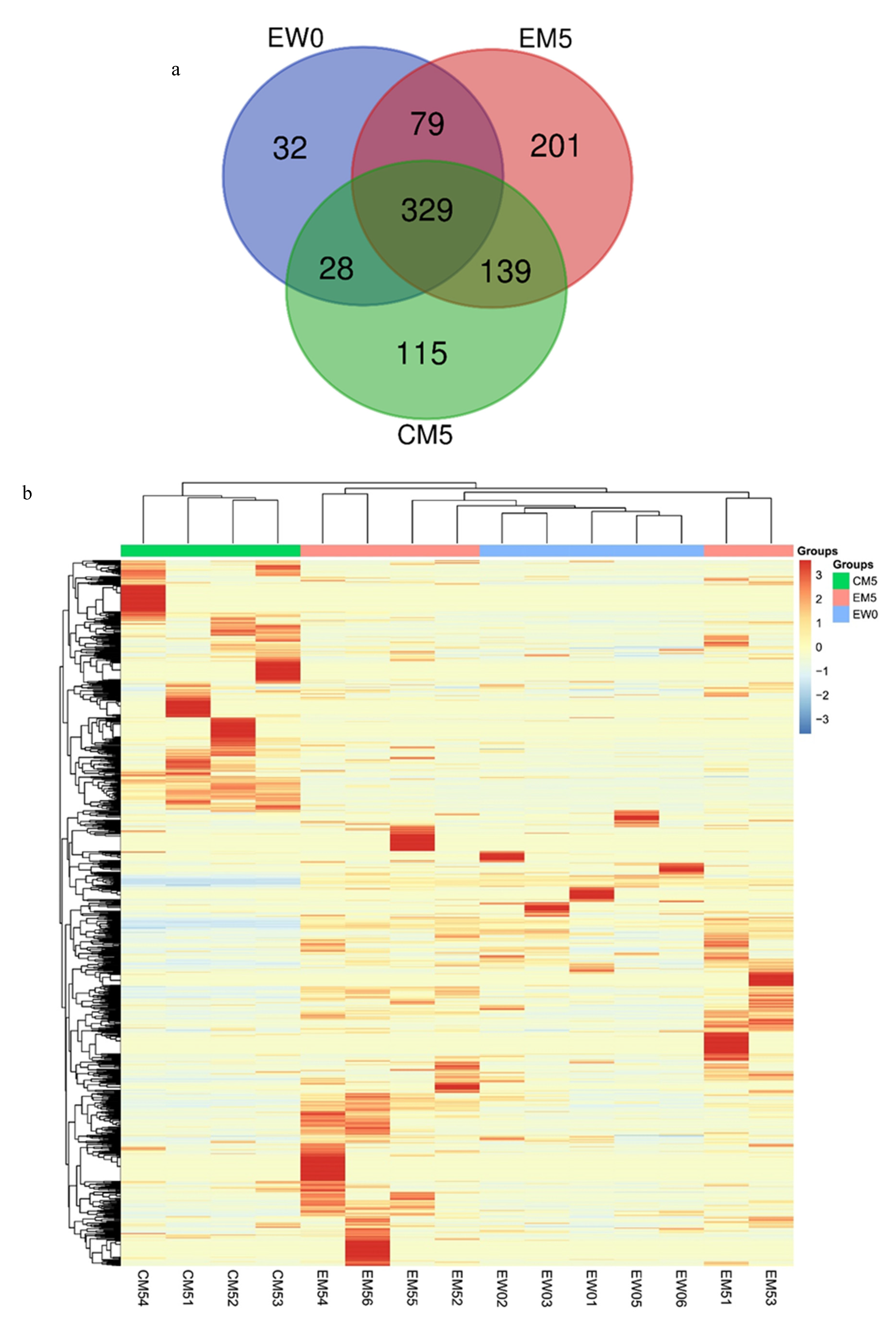
3.2. Differential Protein Screening and Functional Annotation
3.2.1. Comparison within the Experimental Group
Short-Term Effects of a High-Fat Diet
Urinary Proteome Changes in the Whole Process
3.2.2. Comparison between the Experimental Group and the Control Group
Effects of Genetic Factors
Urinary Proteome Changes during Whole Process
| UniProt | Human UniProt | Protein Name | p-Value | Fold Change | References |
|---|---|---|---|---|---|
| B5X0G2 | No | Major urinary protein 17 | 0.0008 | 7.92 | [20] |
| P11588 | No | Major urinary protein 1 | 0.0007 | 7.84 | [20] |
| A2BIM8 | No | Major urinary protein 18 | 0.0014 | 3.19 | [20] |
| Q9JI02 | No | Secretoglobin family 2B member 20 | 0.0488 | 2.75 | — |
| Q5FW60 | No | Major urinary protein 20 | 0.0121 | 2.65 | [20] |
| Q07797 | Q08380 | Galectin-3-binding protein | 0.0077 | 2.59 | [62,63] |
| Q61838 | No | Pregnancy zone protein | 0.0411 | 2.46 | [64] |
| P11591 | No | Major urinary protein 5 | 0.0136 | 2.40 | [20] |
| Q64695 | Q9UNN8 | Endothelial protein C receptor | 0.0150 | 2.23 | [65] |
| Q91WR8 | P59796 | Glutathione peroxidase 6 | 0.0469 | 1.99 | [66] |
| P06797 | P07711 | Cathepsin L1 | 0.0146 | 1.84 | [67] |
| P13597 | P05362 | Intercellular adhesion molecule 1 | 0.0432 | 1.66 | [68,69] |
| Q9JK39 | A8MVZ5 | Butyrophilin-like protein 10 | 0.0446 | 0.60 | — |
| P01898 | P01891 | H-2 class I histocompatibility antigen, Q10 alpha chain | 0.0429 | 0.59 | [70] |
| P55292 | Q02487 | Desmocollin-2 | 0.0160 | 0.57 | [71,72] |
| P23780 | P16278 | Beta-galactosidase | 0.0384 | 0.56 | — |
| Q60648 | P17900 | Ganglioside GM2 activator | 0.0382 | 0.56 | [73] |
| P00688 | P04746 | Pancreatic alpha-amylase | 0.0311 | 0.55 | [74] |
| P70269 | P14091 | Cathepsin E | 0.0299 | 0.54 | [75] |
| P11859 | P01019 | Angiotensinogen | 0.0338 | 0.51 | [22] |
| Q6UGQ3 | No | Secretoglobin family 2B member 2 | 0.0270 | 0.49 | — |
| O88322 | Q14112 | Nidogen-2 | 0.0004 | 0.43 | — |
| O88968 | P20062 | Transcobalamin-2 | 0.0155 | 0.39 | [25] |
| P11087 | P02452 | Collagen alpha-1(I) chain | 0.0080 | 0.34 | [23] |
| Q4KML4 | Q9P1F3 | Costars family protein ABRACL | 0.0181 | 0.28 | — |
| P35230 | Q06141 | Regenerating islet-derived protein 3-beta | 0.0012 | 0.24 | [26] |
| A2AEP0 | No | Odorant-binding protein 1b | 0.0213 | 0.20 | [76] |
| UniProt | Human UniProt | Protein Name | Fold Change | References | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EW0-CW0 | EW1-CW1 | EM1-CM1 | EM2-CM2 | EM3-CM3 | EM4-CM4 | EM5-CM5 | ||||
| P35230 | Q06141 | Regenerating islet-derived protein 3-beta | 5.22 | — | — | — | — | — | — | [26] |
| P13020 | P06396 | Gelsolin | 2.83 | — | — | — | — | — | — | [28] |
| P97426 | P12724 | Eosinophil cationic protein 1 | 2.57 | — | — | — | 1.77 | 7.57 | — | [77] |
| O88322 | Q14112 | Nidogen-2 | 2.32 | 1.96 | — | — | — | — | 0.39 | — |
| P29699 | P02765 | Alpha-2-HS-glycoprotein | 2.27 | — | — | — | — | — | — | [27] |
| P07758 | P01009 | Alpha-1-antitrypsin 1-1 | 2.11 | — | — | — | 0.58 | 0.25 | — | [23] |
| P07309 | P02766 | Transthyretin | 2.06 | — | — | — | — | — | — | [78] |
| P49183 | P24855 | Deoxyribonuclease-1 | 1.79 | — | 0.47 | — | — | — | — | [79] |
| P01864 | No | Ig gamma-2A chain C region secreted form | 1.55 | — | 0.30 | — | — | — | — | — |
| P19221 | P00734 | Prothrombin | 0.63 | — | — | — | — | — | 0.54 | [80] |
| P61110 | P61109 | Kidney androgen-regulated protein | 0.62 | — | — | — | — | 6.55 | — | [23] |
| Q9Z0M9 | O95998 | Interleukin-18-binding protein | 0.60 | — | — | — | 2.11 | — | — | [32] |
| P05533 | No | Lymphocyte antigen 6A-2/6E-1 | 0.60 | — | — | — | — | — | — | — |
| O09043 | O96009 | Napsin-A | 0.59 | — | — | — | 0.42 | — | — | — |
| P03953 | P00746 | Complement factor D | 0.59 | — | — | — | — | 2.60 | — | [81] |
| Q91VW3 | Q9H299 | SH3 domain-binding glutamic acid-rich-like protein 3 | 0.58 | — | — | 2.22 | — | — | — | — |
| P15379 | P16070 | CD44 antigen | 0.58 | — | 0.25 | — | — | 3.43 | — | [82] |
| P04441 | P04233 | H-2 class II histocompatibility antigen gamma chain | 0.54 | — | — | — | 4.59 | 13.02 | — | [31] |
| P25119 | P20333 | Tumour necrosis factor receptor superfamily member 1B | 0.54 | — | — | — | 1.74 | — | — | [83] |
| Q00993 | P30530 | Tyrosine-protein kinase receptor UFO | 0.53 | — | — | 0.51 | 2.16 | 3.62 | — | [84] |
| P07361 | P02763 | Alpha-1-acid glycoprotein 2 | 0.52 | — | — | — | — | 0.13 | — | [85] |
| P09470 | P12821 | Angiotensin-converting enzyme | 0.51 | — | — | — | — | — | — | [86] |
| Q91WR8 | P59796 | Glutathione peroxidase 6 | 0.47 | — | — | — | — | 3.34 | — | [66] |
| Q62395 | Q07654 | Trefoil factor 3 | 0.46 | — | — | — | — | — | — | [21] |
| Q9DAK9 | Q9NRX4 | 14 kDa phosphohistidine phosphatase | 0.46 | — | — | — | — | — | 2.02 | [87] |
| O88188 | O95711 | Lymphocyte antigen 86 | 0.45 | — | — | — | — | — | — | — |
| Q60932 | P21796 | Voltage-dependent anion-selective channel protein 1 | 0.44 | 0.45 | — | 3.68 | — | — | — | [88] |
| P11589 | No | Major urinary protein 2 | 0.44 | — | 3.82 | — | 1.98 | 6.19 | 3.16 | [20] |
| Q62266 | No | Cornifin-A | 0.43 | 0.49 | — | — | — | — | — | — |
| P17047 | P13473 | Lysosome-associated membrane glycoprotein 2 | 0.42 | — | — | — | 2.19 | 3.45 | 2.44 | [33] |
| P0CW03 | No | Lymphocyte antigen 6C2 | 0.41 | — | — | — | — | — | — | — |
| Q60590 | P02763 | Alpha-1-acid glycoprotein 1 | 0.41 | — | — | — | — | 0.08 | — | [85] |
| P01665 | No | Ig kappa chain V-III region PC 7043 | 0.41 | — | — | — | — | 7.91 | 2.38 | — |
| P04939 | No | Major urinary protein 3 | 0.39 | — | — | — | — | — | — | [20] |
| Q6SJQ5 | Q6UXZ3 | CMRF35-like molecule 3 | 0.35 | — | — | — | — | — | 0.56 | — |
| Q64695 | Q9UNN8 | Endothelial protein C receptor | 0.31 | — | — | — | — | — | — | [65] |
| E9Q557 | P15924 | Desmoplakin | 0.29 | — | — | — | 3.82 | — | — | — |
| P11591 | No | Major urinary protein 5 | 0.27 | — | — | — | — | 4.49 | 2.30 | [20] |
| P51437 | P49913 | Cathelicidin antimicrobial peptide | 0.24 | — | — | — | — | — | — | [89] |
| Q5FW60 | No | Major urinary protein 20 | 0.23 | — | — | — | — | — | 2.14 | [20] |
| A2BIM8 | No | Major urinary protein 18 | 0.20 | 0.50 | 6.10 | — | 2.10 | — | — | [20] |
| Q61646 | P00738 | Haptoglobin | 0.14 | — | — | — | — | 0.09 | — | [30] |
| P11588 | No | Major urinary protein 1 | 0.07 | 0.45 | — | — | 2.17 | — | — | [20] |
| B5X0G2 | No | Major urinary protein 17 | 0.06 | 0.34 | 28.79 | — | — | — | 4.06 | [20] |
| Q9JI02 | No | Secretoglobin family 2B member 20 | — | 2.83 | — | — | 0.23 | — | — | — |
| Q01279 | P00533 | Epidermal growth factor receptor | — | 1.52 | — | — | — | — | — | [90] |
| P10605 | P07858 | Cathepsin B | — | 0.63 | 0.44 | — | — | — | — | [91] |
| P50429 | P15848 | Arylsulfatase B | — | 0.56 | — | — | — | — | — | [92] |
| Q571E4 | P34059 | N-acetylgalactosamine-6-sulfatase | — | 0.52 | — | — | — | 3.57 | — | — |
| Q9JK39 | A8MVZ5 | Butyrophilin-like protein 10 | — | 0.50 | — | — | — | — | — | — |
| P23780 | P16278 | Beta-galactosidase | — | 0.48 | 0.15 | — | 0.28 | — | — | — |
| P70269 | P14091 | Cathepsin E | — | 0.42 | — | — | — | 2.14 | — | [75] |
| O35887 | O43852 | Calumenin | — | 0.40 | — | — | 2.88 | 4.38 | 1.88 | [93] |
| Q9EP95 | Q9BQ08 | Resistin-like alpha | — | 0.27 | — | — | — | — | 0.20 | [94] |
| P20152 | P08670 | Vimentin | — | — | 16.24 | — | — | — | — | [95] |
| Q8K0E8 | P02675 | Fibrinogen beta chain | — | — | 8.28 | — | — | — | — | [96] |
| P16858 | P04406 | Glyceraldehyde-3-phosphate dehydrogenase | — | — | 4.72 | — | — | — | — | [97] |
| Q9WTR5 | P55290 | Cadherin-13 | — | — | 2.78 | — | — | — | — | [34] |
| Q00897 | P01009 | Alpha-1-antitrypsin 1-4 | — | — | 2.57 | — | — | — | 3.27 | [23] |
| O09164 | P08294 | Extracellular superoxide dismutase [Cu–Zn] | — | — | 2.11 | 1.56 | — | 3.37 | — | [98] |
| P11276 | P02751 | Fibronectin | — | — | 0.58 | — | — | 2.15 | — | [35] |
| Q9Z0J0 | P61916 | NPC intracellular cholesterol transporter 2 | — | — | 0.57 | — | — | — | — | [99] |
| Q8BPB5 | Q12805 | EGF-containing fibulin-like extracellular matrix protein 1 | — | — | 0.53 | 0.46 | — | — | — | — |
| Q8BZT5 | Q9H756 | Leucine-rich repeat-containing protein 19 | — | — | 0.52 | 0.50 | 0.55 | — | — | [100] |
| P21614 | P02774 | Vitamin D-binding protein | — | — | 0.50 | — | — | — | 0.49 | [101] |
| P16675 | P10619 | Lysosomal protective protein | — | — | 0.49 | — | — | — | 2.51 | — |
| Q61147 | P00450 | Ceruloplasmin | — | — | 0.49 | — | — | 0.35 | — | [102] |
| P23953 | No | Carboxylesterase 1C | — | — | 0.48 | — | — | — | — | — |
| Q61398 | Q15113 | Procollagen C-endopeptidase enhancer 1 | — | — | 0.48 | — | — | — | — | [103] |
| O35664 | P48551 | Interferon alpha/beta receptor 2 | — | — | 0.47 | — | — | — | — | [104] |
| P11859 | P01019 | Angiotensinogen | — | — | 0.45 | — | — | 0.15 | — | [22] |
| P01898 | P01891 | H-2 class I histocompatibility antigen, Q10 alpha chain | — | — | 0.45 | — | — | — | 2.03 | [70] |
| C0HKG5 | No | Ribonuclease T2-A | — | — | 0.42 | 0.60 | — | — | — | — |
| Q61271 | P36896 | Activin receptor type-1B | — | — | 0.42 | — | — | 3.64 | — | — |
| O88968 | P20062 | Transcobalamin-2 | — | — | 0.40 | — | — | — | — | [25] |
| Q9Z0L8 | Q92820 | Gamma-glutamyl hydrolase | — | — | 0.39 | — | — | — | — | [105,106] |
| P35459 | Q14210 | Lymphocyte antigen 6D | — | — | 0.39 | 0.57 | — | — | — | — |
| Q61129 | P05156 | Complement factor I | — | — | 0.38 | — | — | — | — | — |
| P01878 | No | Ig alpha chain C region | — | — | 0.38 | — | 5.03 | — | — | — |
| P55292 | Q02487 | Desmocollin-2 | — | — | 0.38 | — | 3.72 | — | — | [71,72] |
| Q9JJS0 | Q9NQ36 | Signal peptide, CUB and EGF-like domain-containing protein 2 | — | — | 0.35 | — | — | — | — | [29] |
| Q9Z319 | Q9Y5Q5 | Atrial natriuretic peptide-converting enzyme | — | — | 0.35 | — | — | — | — | [107] |
| P09036 | P00995 | Serine protease inhibitor Kazal-type 1 | — | — | 0.34 | — | — | — | — | — |
| Q4KML4 | Q9P1F3 | Costars family protein ABRACL | — | — | 0.33 | — | 2.18 | — | — | — |
| Q925F2 | Q96AP7 | Endothelial cell-selective adhesion molecule | — | — | 0.32 | — | — | — | 0.48 | [108] |
| O89020 | P43652 | Afamin | — | — | 0.31 | 0.50 | — | — | — | [109] |
| Q9DAU7 | Q14508 | WAP four-disulfide core domain protein 2 | — | — | 0.31 | — | — | — | — | — |
| Q8BND5 | O00391 | Sulfhydryl oxidase 1 | — | — | 0.30 | 0.40 | — | — | 0.61 | — |
| P09803 | P12830 | Cadherin-1 | — | — | 0.29 | — | 2.78 | 2.34 | — | [30] |
| P02816 | P12273 | Prolactin-inducible protein homolog | — | — | 0.27 | — | 0.45 | — | — | [39] |
| Q91WR6 | Q9NU53 | Glycoprotein integral membrane protein 1 | — | — | 0.26 | 0.59 | — | — | 0.45 | — |
| Q3UDR8 | Q9GZM5 | Protein YIPF3 | — | — | 0.26 | — | — | — | — | — |
| Q6UGQ3 | No | Secretoglobin family 2B member 2 | — | — | 0.25 | — | — | — | 2.03 | — |
| Q9D3H2 | No | Odorant-binding protein 1a | — | — | 0.25 | — | 1.75 | — | — | [76] |
| P20060 | P07686 | Beta-hexosaminidase subunit beta | — | — | 0.23 | — | 0.35 | — | — | — |
| Q8K1H9 | Q9NY56 | Odorant-binding protein 2a | — | — | 0.21 | — | 0.31 | 0.21 | — | [76] |
| A2AEP0 | No | Odorant-binding protein 1b | — | — | 0.20 | — | — | — | — | [76] |
| Q8C6C9 | Q6P5S2 | Protein LEG1 homolog | — | — | 0.15 | — | 0.44 | — | — | — |
| P00688 | P04746 | Pancreatic alpha-amylase | — | — | 0.14 | — | — | 0.32 | — | [74] |
| P10287 | P22223 | Cadherin-3 | — | — | 0.13 | — | 3.19 | 5.62 | — | [30] |
| P56386 | P60022 | Beta-defensin 1 | — | — | — | 3.94 | — | 5.25 | 1.90 | [110] |
| P10639 | P10599 | Thioredoxin | — | — | — | 3.63 | — | — | — | [111] |
| O88844 | O75874 | Isocitrate dehydrogenase [NADP] cytoplasmic | — | — | — | 2.71 | — | — | — | [112] |
| Q00623 | P02647 | Apolipoprotein A-I | — | — | — | 2.07 | — | — | 0.36 | [113] |
| P0CG49 | P0CG47 | Polyubiquitin-B | — | — | — | 1.98 | — | — | — | [114] |
| Q03404 | Q03403 | Trefoil factor 2 | — | — | — | 1.69 | 2.11 | 8.94 | — | [21] |
| P15947 | P06870 | Kallikrein-1 | — | — | — | 0.66 | — | 1.74 | — | [115] |
| O55186 | P13987 | CD59A glycoprotein | — | — | — | 0.61 | — | 2.53 | — | [116] |
| Q921I1 | P02787 | Serotransferrin | — | — | — | 0.57 | 0.43 | 0.09 | 0.21 | [37] |
| Q60648 | P17900 | Ganglioside GM2 activator | — | — | — | 0.51 | — | — | 2.27 | [74] |
| O88792 | Q9Y624 | Junctional adhesion molecule A | — | — | — | 0.49 | — | — | — | [117] |
| P07724 | P02768 | Albumin | — | — | — | 0.38 | — | 0.38 | 0.43 | [118] |
| O70554 | No | Small proline-rich protein 2B | — | — | — | — | 7.98 | — | 3.26 | [119,120] |
| Q62267 | No | Cornifin-B | — | — | — | — | 5.69 | — | — | — |
| P35700 | Q06830 | Peroxiredoxin-1 | — | — | — | — | 3.75 | — | — | [121] |
| P18761 | P23280 | Carbonic anhydrase 6 | — | — | — | — | 3.37 | — | — | [122] |
| P01631 | No | Ig kappa chain V-II region 26-10 | — | — | — | — | 3.29 | 3.00 | — | — |
| P11087 | P02452 | Collagen alpha-1(I) chain | — | — | — | — | 3.21 | — | — | [23] |
| P01837 | P01834 | Immunoglobulin kappa constant | — | — | — | — | 3.19 | — | — | — |
| Q99N23 | No | Carbonic anhydrase 15 | — | — | — | — | 2.91 | — | — | [122] |
| P10126 | P68104 | Elongation factor 1-alpha 1 | — | — | — | — | 2.90 | — | — | [123] |
| P70663 | Q14515 | SPARC-like protein 1 | — | — | — | — | 2.74 | — | — | — |
| Q60847 | Q99715 | Collagen alpha-1(XII) chain | — | — | — | — | 2.68 | — | — | [23] |
| P29533 | P19320 | Vascular cell adhesion protein 1 | — | — | — | — | 2.62 | 2.12 | — | [124] |
| O55135 | P56537 | Eukaryotic translation initiation factor 6 | — | — | — | — | 2.55 | — | — | — |
| O54775 | O95388 | CCN family member 4 | — | — | — | — | 2.47 | — | 0.47 | [24] |
| P01843 | No | Ig lambda-1 chain C region | — | — | — | — | 2.45 | — | — | — |
| Q9DBV4 | Q9BRK3 | Matrix remodelling-associated protein 8 | — | — | — | — | 1.70 | — | 1.80 | — |
| O35608 | O15123 | Angiopoietin-2 | — | — | — | — | 1.61 | — | — | [125] |
| Q07797 | Q08380 | Galectin-3-binding protein | — | — | — | — | 1.59 | — | — | [62] |
| Q04519 | P17405 | Sphingomyelin phosphodiesterase | — | — | — | — | 0.57 | — | 1.89 | [126] |
| Q61838 | No | Pregnancy zone protein | — | — | — | — | 0.52 | — | — | [64] |
| Q08423 | P04155 | Trefoil factor 1 | — | — | — | — | — | 12.79 | — | [21] |
| O08997 | O00244 | Copper transport protein ATOX1 | — | — | — | — | — | 8.07 | — | [127] |
| P03977 | No | Ig kappa chain V-III region 50S10.1 | — | — | — | — | — | 4.79 | — | — |
| P42567 | P42566 | Epidermal growth factor receptor substrate 15 | — | — | — | — | — | 4.53 | — | [90] |
| Q91X17 | P07911 | Uromodulin | — | — | — | — | — | 4.22 | — | [128] |
| P57096 | O43653 | Prostate stem cell antigen | — | — | — | — | — | 3.53 | — | — |
| P70699 | P10253 | Lysosomal alpha-glucosidase | — | — | — | — | — | 3.37 | — | [129] |
| P01132 | P01133 | Pro-epidermal growth factor | — | — | — | — | — | 3.13 | — | — |
| P32507 | Q92692 | Nectin-2 | — | — | — | — | — | 3.03 | — | [130] |
| Q5SSE9 | Q86UQ4 | ATP-binding cassette sub-family A member 13 | — | — | — | — | — | 2.92 | — | — |
| P06797 | O60911 | Cathepsin L1 | — | — | — | — | — | 2.63 | — | [67,75] |
| Q8R242 | Q01459 | Di-N-acetylchitobiase | — | — | — | — | — | 2.47 | — | — |
| Q9Z0K8 | O95497 | Pantetheinase | — | — | — | — | — | 2.32 | 3.65 | — |
| O54782 | Q9Y2E5 | Epididymis-specific alpha-mannosidase | — | — | — | — | — | 1.99 | — | — |
| P22599 | P01009 | Alpha-1-antitrypsin 1-2 | — | — | — | — | — | 0.50 | — | [23] |
| P13634 | P00915 | Carbonic anhydrase 1 | — | — | — | — | — | 0.39 | 3.37 | [122] |
| P20918 | P00747 | Plasminogen | — | — | — | — | — | 0.32 | 0.63 | [131] |
| P07759 | P01011 | Serine protease inhibitor A3K | — | — | — | — | — | 0.26 | — | [38] |
| P02088 | P68871 | Hemoglobin subunit beta-1 | — | — | — | — | — | 0.24 | — | — |
| P01027 | P01024 | Complement C3 | — | — | — | — | — | 0.20 | — | [56] |
| Q00898 | P01009 | Alpha-1-antitrypsin 1-5 | — | — | — | — | — | 0.18 | — | [23] |
| P11672 | P80188 | Neutrophil gelatinase-associated lipocalin | — | — | — | — | — | 0.04 | — | [132] |
| P01887 | P61769 | Beta-2-microglobulin | — | — | — | — | — | — | 3.07 | [133] |
| O09114 | P41222 | Prostaglandin-H2 D-isomerase | — | — | — | — | — | — | 2.41 | [134] |
| Q9WUU7 | Q9UBR2 | Cathepsin Z | — | — | — | — | — | — | 2.32 | [135] |
| Q8BHC0 | Q9Y5Y7 | Lymphatic vessel endothelial hyaluronic acid receptor 1 | — | — | — | — | — | — | 0.61 | — |
| O70570 | P01833 | Polymeric immunoglobulin receptor | — | — | — | — | — | — | 0.55 | — |
| P26041 | P26038 | Moesin | — | — | — | — | — | — | 0.53 | [136] |
| Q7TMJ8 | Q96FE7 | Phosphoinositide-3-kinase-interacting protein 1 | — | — | — | — | — | — | 0.52 | [137] |
| P01660 | No | Ig kappa chain V-III region PC 3741/TEPC 111 | — | — | — | — | — | — | 0.43 | — |
| Q60928 | P19440 | Glutathione hydrolase 1 proenzyme | — | — | — | — | — | — | 0.39 | [106] |
| P61971 | P61970 | Nuclear transport factor 2 | — | — | — | — | — | — | 0.36 | — |
| P06330 | No | Ig heavy chain V region AC38 205.12 | — | — | — | — | — | — | 0.36 | — |
| Q921W8 | Q8WVN6 | Secreted and transmembrane protein 1A | — | — | — | — | — | — | 0.35 | — |
| Q8BX43 | Q969Z4 | Tumour necrosis factor receptor superfamily member 19L | — | — | — | — | — | — | 0.33 | [83] |
| Proteoglycan 4 | ||||||||||
| Q9JM99 | Q92954 | — | — | — | — | — | — | 0.26 | [137] | |
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rahman, M.S.; Woollard, K. Atherosclerosis. Adv. Exp. Med. Biol. 2017, 1003, 121–144. [Google Scholar] [CrossRef]
- Tabas, I.; Garcia-Cardena, G.; Owens, G.K. Recent insights into the cellular biology of atherosclerosis. J. Cell Biol. 2015, 209, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Cardiovascular Diseases (CVDs) Fact Sheet; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Barr, T.L.; VanGilder, R.L.; Seiberg, R.; Petrone, A.; Chantler, P.D.; Huang, C.C. Systemic Transcriptional Alterations of Innate and Adaptive Immune Signaling Pathways in Atherosclerosis, Ischemia Stroke, and Myocardial Infarction. J. Bioanal. Biomed. 2015, 7, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y. Urine-an untapped goldmine for biomarker discovery? Sci. China Life Sci. 2013, 56, 1145–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, Y.; Zhang, F.; An, M.; Yin, W.; Gao, Y. Early candidate biomarkers found from urine of glioblastoma multiforme rat before changes in MRI. Sci. China Life Sci. 2018, 61, 982–987. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Meng, W.; Gao, Y. Urine proteome changes in rats subcutaneously inoculated with approximately ten tumor cells. PeerJ 2019, 7, e7717. [Google Scholar] [CrossRef] [Green Version]
- Qin, W.; Wang, T.; Huang, H.; Gao, Y. Profiling of lysine-acetylated proteins in human urine. Sci. China Life Sci. 2019, 62, 1514–1520. [Google Scholar] [CrossRef]
- Zhao, M.; Li, M.; Li, X.; Shao, C.; Yin, J.; Gao, Y. Dynamic changes of urinary proteins in a focal segmental glomerulosclerosis rat model. Proteome Sci. 2014, 12, 42. [Google Scholar] [CrossRef] [Green Version]
- Hatters, D.M.; Peters-Libeu, C.A.; Weisgraber, K.H. Apolipoprotein E structure: Insights into function. Trends Biochem. Sci. 2006, 31, 445–454. [Google Scholar] [CrossRef]
- Rosenfeld, M.E.; Polinsky, P.; Virmani, R.; Kauser, K.; Rubanyi, G.; Schwartz, S.M. Advanced atherosclerotic lesions in the innominate artery of the ApoE knockout mouse. Arter. Thromb. Vasc. Biol. 2000, 20, 2587–2592. [Google Scholar] [CrossRef]
- Qu, G.; Wu, Q.; Zhao, B.; Miao, J.; Su, L. The promotion effect of novel magnetic nanoparticles on atherosclerotic plaque vulnerability in apolipoprotein E(-/-) mice. Toxicology 2019, 419, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Bruderer, R.; Bernhardt, O.M.; Gandhi, T.; Miladinovic, S.M.; Cheng, L.Y.; Messner, S.; Ehrenberger, T.; Zanotelli, V.; Butscheid, Y.; Escher, C.; et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteom. 2015, 14, 1400–1410. [Google Scholar] [CrossRef] [Green Version]
- Chi, H.; Liu, C.; Yang, H.; Zeng, W.F.; Wu, L.; Zhou, W.J.; Wang, R.M.; Niu, X.N.; Ding, Y.H.; Zhang, Y.; et al. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 2018, 36, 1059–1061. [Google Scholar] [CrossRef] [PubMed]
- Armitage, E.G.; Godzien, J.; Alonso-Herranz, V.; Lopez-Gonzalvez, A.; Barbas, C. Missing value imputation strategies for metabolomics data. Electrophoresis 2015, 36, 3050–3060. [Google Scholar] [CrossRef]
- Wang, S.; Li, W.; Hu, L.; Cheng, J.; Yang, H.; Liu, Y. NAguideR: Performing and prioritizing missing value imputations for consistent bottom-up proteomic analyses. Nucleic Acids Res. 2020, 48, e83. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An integrated proteome resource. Nucleic Acids Res. 2019, 47, D1211–D1217. [Google Scholar] [CrossRef] [Green Version]
- Kwak, J.; Strasser, E.; Luzynski, K.; Thoss, M.; Penn, D.J. Are MUPs a Toxic Waste Disposal System? PLoS ONE 2016, 11, e0151474. [Google Scholar] [CrossRef] [Green Version]
- De Giorgio, M.R.; Yoshioka, M.; Riedl, I.; Moreault, O.; Cherizol, R.G.; Shah, A.A.; Blin, N.; Richard, D.; St-Amand, J. Trefoil factor family member 2 (Tff2) KO mice are protected from high-fat diet-induced obesity. Obesity 2013, 21, 1389–1395. [Google Scholar] [CrossRef]
- Carroll, W.X.; Kalupahana, N.S.; Booker, S.L.; Siriwardhana, N.; Lemieux, M.; Saxton, A.M.; Moustaid-Moussa, N. Angiotensinogen gene silencing reduces markers of lipid accumulation and inflammation in cultured adipocytes. Front. Endocrinol. 2013, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von zur Muhlen, C.; Schiffer, E.; Sackmann, C.; Zurbig, P.; Neudorfer, I.; Zirlik, A.; Htun, N.; Iphofer, A.; Jansch, L.; Mischak, H.; et al. Urine proteome analysis reflects atherosclerotic disease in an ApoE-/- mouse model and allows the discovery of new candidate biomarkers in mouse and human atherosclerosis. Mol. Cell. Proteom. 2012, 11, M111.013847. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Dong, W.; Lin, Z.; Lu, J.; Wan, H.; Zhou, Z.; Liu, Z. CCN4 regulates vascular smooth muscle cell migration and proliferation. Mol. Cells 2013, 36, 112–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Celik, S.F.; Celik, E. Subclinical atherosclerosis and impaired cardiac autonomic control in pediatric patients with Vitamin B12 deficiency. Niger. J. Clin. Pract. 2018, 21, 1012–1016. [Google Scholar] [CrossRef] [PubMed]
- K, M.; Adole, P.S.; Vinod, K.V.; Balamurugan, N. Association of serum regenerating islet-derived protein 3-beta and oncostatin-M levels with the risk of acute coronary syndrome in patients with type 2 diabetes mellitus—A pilot study. Diabetes Metab. Syndr. 2020, 14, 1087–1092. [Google Scholar] [CrossRef]
- Muendlein, A.; Stark, N.; Rein, P.; Saely, C.H.; Geller-Rhomberg, S.; Geiger, K.; Vonbank, A.; Drexel, H. Are AHSG polymorphisms directly associated with coronary atherosclerosis? Clin. Chim. Acta 2012, 413, 287–290. [Google Scholar] [CrossRef]
- de la Cuesta, F.; Zubiri, I.; Maroto, A.S.; Posada, M.; Padial, L.R.; Vivanco, F.; Alvarez-Llamas, G.; Barderas, M.G. Deregulation of smooth muscle cell cytoskeleton within the human atherosclerotic coronary media layer. J. Proteom. 2013, 82, 155–165. [Google Scholar] [CrossRef]
- Ali, H.; Emoto, N.; Yagi, K.; Vignon-Zellweger, N.; Nakayama, K.; Hatakeyama, K.; Asada, Y.; Rikitake, Y.; Hirata, K. Localization and characterization of a novel secreted protein, SCUBE2, in the development and progression of atherosclerosis. Kobe J. Med. Sci. 2013, 59, E122–E131. [Google Scholar]
- Langlois, M.R.; Delanghe, J.R. Biological and clinical significance of haptoglobin polymorphism in humans. Clin. Chem. 1996, 42, 1589–1600. [Google Scholar] [CrossRef] [Green Version]
- Wigren, M.; Rattik, S.; Yao Mattisson, I.; Tomas, L.; Gronberg, C.; Soderberg, I.; Alm, R.; Sundius, L.; Ljungcrantz, I.; Bjorkbacka, H.; et al. Lack of Ability to Present Antigens on Major Histocompatibility Complex Class II Molecules Aggravates Atherosclerosis in ApoE(-/-) Mice. Circulation 2019, 139, 2554–2566. [Google Scholar] [CrossRef]
- Dinarello, C.A. Interleukin-18 and the pathogenesis of inflammatory diseases. Semin. Nephrol. 2007, 27, 98–114. [Google Scholar] [CrossRef]
- Wu, G.; Huang, J.; Wei, G.; Liu, L.; Pang, S.; Yan, B. LAMP-2 gene expression in peripheral leukocytes is increased in patients with coronary artery disease. Clin. Cardiol. 2011, 34, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Fujishima, Y.; Maeda, N.; Matsuda, K.; Masuda, S.; Mori, T.; Fukuda, S.; Sekimoto, R.; Yamaoka, M.; Obata, Y.; Kita, S.; et al. Adiponectin association with T-cadherin protects against neointima proliferation and atherosclerosis. FASEB J. 2017, 31, 1571–1583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenman, S.; von Smitten, K.; Vaheri, A. Fibronectin and atherosclerosis. Acta Med. Scand. Suppl. 1980, 642, 165–170. [Google Scholar] [CrossRef]
- Ichiki, T. Thyroid hormone and atherosclerosis. Vasc. Pharmacol. 2010, 52, 151–156. [Google Scholar] [CrossRef]
- Kibel, A.; Belovari, T.; Drenjancevic-Peric, I. The role of transferrin in atherosclerosis. Med. Hypotheses 2008, 70, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Wagsater, D.; Johansson, D.; Fontaine, V.; Vorkapic, E.; Backlund, A.; Razuvaev, A.; Mayranpaa, M.I.; Hjerpe, C.; Caidahl, K.; Hamsten, A.; et al. Serine protease inhibitor A3 in atherosclerosis and aneurysm disease. Int. J. Mol. Med. 2012, 30, 288–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauro, M.D.; Zorn, N.E. Prolactin induces proliferation of vascular smooth muscle cells through a protein kinase C-dependent mechanism. J. Cell Physiol. 1991, 148, 133–138. [Google Scholar] [CrossRef]
- Hartman, J.; Frishman, W.H. Inflammation and atherosclerosis: A review of the role of interleukin-6 in the development of atherosclerosis and the potential for targeted drug therapy. Cardiol. Rev. 2014, 22, 147–151. [Google Scholar] [CrossRef]
- Tinajero, M.G.; Gotlieb, A.I. Recent Developments in Vascular Adventitial Pathobiology: The Dynamic Adventitia as a Complex Regulator of Vascular Disease. Am. J. Pathol. 2020, 190, 520–534. [Google Scholar] [CrossRef] [Green Version]
- Natarelli, L.; Schober, A. MicroRNAs and the response to injury in atherosclerosis. Hamostaseologie 2015, 35, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Wight, T.N. The extracellular matrix and atherosclerosis. Curr. Opin. Lipidol. 1995, 6, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Murga, M.L.; Vinue, A.; Caeiro, J.R.; Guede, D.; Tarin, J.J.; Andres, V.; Cano, A. Impact of estrogens on atherosclerosis and bone in the apolipoprotein E-deficient mouse model. Menopause 2015, 22, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Isenovic, E.R.; Soskic, S.; Trpkovic, A.; Dobutovic, B.; Popovic, M.; Gluvic, Z.; Putnikovic, B.; Marche, P. Insulin, thrombine, ERK1/2 kinase and vascular smooth muscle cells proliferation. Curr. Pharm. Des. 2010, 16, 3895–3902. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Melendez, A.J. Role of cell adhesion molecules and immune-cell migration in the initiation, onset and development of atherosclerosis. Cell Adhes. Migr. 2007, 1, 171–175. [Google Scholar] [CrossRef] [Green Version]
- Whicher, J.; Biasucci, L.; Rifai, N. Inflammation, the acute phase response and atherosclerosis. Clin. Chem. Lab. Med. 1999, 37, 495–503. [Google Scholar] [CrossRef]
- van Dijk, R.A.; Rijs, K.; Wezel, A.; Hamming, J.F.; Kolodgie, F.D.; Virmani, R.; Schaapherder, A.F.; Lindeman, J.H. Systematic Evaluation of the Cellular Innate Immune Response During the Process of Human Atherosclerosis. J. Am. Heart Assoc. 2016, 5, e002860. [Google Scholar] [CrossRef] [Green Version]
- Tedgui, A.; Mallat, Z. Cytokines in atherosclerosis: Pathogenic and regulatory pathways. Physiol. Rev. 2006, 86, 515–581. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Touchard, A.; Henry, T.D.; Sangiorgi, G.; Spagnoli, L.G.; Mauriello, A.; Conover, C.; Schwartz, R.S. Extracellular proteases in atherosclerosis and restenosis. Arter. Thromb. Vasc. Biol. 2005, 25, 1119–1127. [Google Scholar] [CrossRef] [Green Version]
- Libby, P.; Buring, J.E.; Badimon, L.; Hansson, G.K.; Deanfield, J.; Bittencourt, M.S.; Tokgozoglu, L.; Lewis, E.F. Atherosclerosis. Nat. Rev. Dis. Prim. 2019, 5, 56. [Google Scholar] [CrossRef]
- Hansson, G.K.; Robertson, A.K.; Soderberg-Naucler, C. Inflammation and atherosclerosis. Annu. Rev. Pathol. 2006, 1, 297–329. [Google Scholar] [CrossRef]
- Sueishi, K.; Ichikawa, K.; Kato, K.; Nakagawa, K.; Chen, Y.X. Atherosclerosis: Coagulation and fibrinolysis. Semin. Thromb. Hemost. 1998, 24, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Linton, M.F.; Moslehi, J.J.; Babaev, V.R. Akt Signaling in Macrophage Polarization, Survival, and Atherosclerosis. Int. J. Mol. Sci. 2019, 20, 2703. [Google Scholar] [CrossRef] [Green Version]
- Ustundag, S.; Yilmaz, G.; Sevinc, C.; Akpinar, S.; Temizoz, O.; Sut, N.; Ustundag, A. Carotid intima media thickness is independently associated with urinary sodium excretion in patients with chronic kidney disease. Ren. Fail. 2015, 37, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Drapkina, O.M.; Gegenava, B.B.; Fomin, V.V. The role of the mLDL-induced activation of the complement system classical pathway and C3 expression stimulation in atherosclerosis. Ter. Arkhiv 2018, 90, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Jomova, K.; Valko, M. Advances in metal-induced oxidative stress and human disease. Toxicology 2011, 283, 65–87. [Google Scholar] [CrossRef]
- Nathan, L.; Chaudhuri, G. Estrogens and atherosclerosis. Annu. Rev. Pharmacol. Toxicol. 1997, 37, 477–515. [Google Scholar] [CrossRef]
- Wunderer, F.; Traeger, L.; Sigurslid, H.H.; Meybohm, P.; Bloch, D.B.; Malhotra, R. The role of hepcidin and iron homeostasis in atherosclerosis. Pharmacol. Res. 2020, 153, 104664. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.W.; Elvington, A.; Kessler, S.; Wohltmann, M.; Wu, G.F.; Randolph, G.J. B Cell-Mediated Antigen Presentation through MHC Class II Is Dispensable for Atherosclerosis Progression. Immunohorizons 2019, 3, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Qiao, L.; Wang, H.F.; Xiang, L.; Ma, J.; Zhu, Q.; Xu, D.; Zheng, H.; Peng, J.Q.; Zhang, S.; Lu, H.X.; et al. Deficient Chaperone-Mediated Autophagy Promotes Lipid Accumulation in Macrophage. J. Cardiovasc. Transl. Res. 2020, 14, 661–669. [Google Scholar] [CrossRef] [Green Version]
- Sano, H.; Hsu, D.K.; Yu, L.; Apgar, J.R.; Kuwabara, I.; Yamanaka, T.; Hirashima, M.; Liu, F.T. Human galectin-3 is a novel chemoattractant for monocytes and macrophages. J. Immunol. 2000, 165, 2156–2164. [Google Scholar] [CrossRef] [PubMed]
- Papaspyridonos, M.; McNeill, E.; de Bono, J.P.; Smith, A.; Burnand, K.G.; Channon, K.M.; Greaves, D.R. Galectin-3 is an amplifier of inflammation in atherosclerotic plaque progression through macrophage activation and monocyte chemoattraction. Arter. Thromb. Vasc. Biol. 2008, 28, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.C.; Chiabrando, G.A.; Vides, M.A. Pregnancy zone protein-tissue-type plasminogen activator complexes bind to low-density lipoprotein receptor-related protein (LRP). Arch. Biochem. Biophys. 2001, 389, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Navarro, S.; Bonet, E.; Estelles, A.; Montes, R.; Hermida, J.; Martos, L.; Espana, F.; Medina, P. The endothelial cell protein C receptor: Its role in thrombosis. Thromb. Res. 2011, 128, 410–416. [Google Scholar] [CrossRef]
- Lin, M.S.; Hsu, H.C.; Lin, L.C.; Li, H.Y.; Lee, B.C.; Lee, Y.T.; Chen, M.F. Higher glutathione peroxidase expression in thoracic aorta as a protective factor against oxidative stress and atherosclerosis in rabbits. Cardiology 2007, 108, 381–386. [Google Scholar] [CrossRef]
- Li, W.; Kornmark, L.; Jonasson, L.; Forssell, C.; Yuan, X.M. Cathepsin L is significantly associated with apoptosis and plaque destabilization in human atherosclerosis. Atherosclerosis 2009, 202, 92–102. [Google Scholar] [CrossRef]
- Ohta, T.; Saku, K.; Takata, K.; Adachi, N. Soluble vascular cell-adhesion molecule-1 and soluble intercellular adhesion molecule-1 correlate with lipid and apolipoprotein risk factors for coronary artery disease in children. Eur. J. Pediatr. 1999, 158, 592–598. [Google Scholar] [CrossRef]
- Rashad, N.M.; El-Shal, A.S.; Abomandour, H.G.; Aboelfath, A.; Rafeek, M.; Badr, M.S.; Ali, A.E.; Yousef, M.S.; Fathy, M.A.; Sharaf El Din, M. Intercellular adhesion molecule-1 expression and serum levels as markers of pre-clinical atherosclerosis in polycystic ovary syndrome. J. Ovarian Res. 2019, 12, 97. [Google Scholar] [CrossRef] [Green Version]
- Fortin, C.F.; McDonald, P.P.; Lesur, O.; Fulop, T., Jr. Aging and neutrophils: There is still much to do. Rejuvenation Res. 2008, 11, 873–882. [Google Scholar] [CrossRef]
- Li, Y.B.; Zhang, Q.H.; Chen, Z.; He, Z.J.; Yi, G.H. Oxidized low-density lipoprotein attenuated desmoglein 1 and desmocollin 2 expression via LOX-1/Ca(2+)/PKC-beta signal in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2015, 468, 380–386. [Google Scholar] [CrossRef]
- Jiang, K.; Rankin, C.R.; Nava, P.; Sumagin, R.; Kamekura, R.; Stowell, S.R.; Feng, M.; Parkos, C.A.; Nusrat, A. Galectin-3 regulates desmoglein-2 and intestinal epithelial intercellular adhesion. J. Biol. Chem. 2014, 289, 10510–10517. [Google Scholar] [CrossRef] [PubMed]
- Yanai, H.; Yoshida, H.; Tomono, Y.; Tada, N.; Chiba, H. The possible contribution of a general glycosphingolipid transporter, GM2 activator protein, to atherosclerosis. J. Atheroscler. Thromb. 2006, 13, 281–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozkok, A.; Elcioglu, O.C.; Cukadar, T.; Bakan, A.; Sasak, G.; Atilgan, K.G.; Alisir, S.; Kanbay, M.; Covic, A.; Odabas, A.R. Low serum pancreatic enzyme levels predict mortality and are associated with malnutrition-inflammation-atherosclerosis syndrome in patients with chronic kidney disease. Int. Urol. Nephrol. 2013, 45, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, T.; Kido, M.A.; Hatakeyama, J.; Okamoto, K.; Tsukuba, T.; Yamamoto, K. Defective adipose tissue development associated with hepatomegaly in cathepsin E-deficient mice fed a high-fat diet. Biochem. Biophys. Res. Commun. 2014, 446, 212–217. [Google Scholar] [CrossRef] [Green Version]
- Grolli, S.; Merli, E.; Conti, V.; Scaltriti, E.; Ramoni, R. Odorant binding protein has the biochemical properties of a scavenger for 4-hydroxy-2-nonenal in mammalian nasal mucosa. FEBS J. 2006, 273, 5131–5142. [Google Scholar] [CrossRef]
- Niccoli, G.; Ferrante, G.; Cosentino, N.; Conte, M.; Belloni, F.; Marino, M.; Baca, M.; Montone, R.A.; Sabato, V.; Schiavino, D.; et al. Eosinophil cationic protein: A new biomarker of coronary atherosclerosis. Atherosclerosis 2010, 211, 606–611. [Google Scholar] [CrossRef]
- Yang, C.S.; Wei, Y.S.; Tsai, H.L.; Cheong, I.S.; Chang, S.J.; Chou, H.C.; Lee, Y.R.; Chan, H.L. Proteomic analysis of prognostic plasma biomarkers in peripheral arterial occlusive disease. Mol. Biosyst. 2017, 13, 1297–1303. [Google Scholar] [CrossRef]
- Josefs, T.; Barrett, T.J.; Brown, E.J.; Quezada, A.; Wu, X.; Voisin, M.; Amengual, J.; Fisher, E.A. Neutrophil extracellular traps promote macrophage inflammation and impair atherosclerosis resolution in diabetic mice. JCI Insight 2020, 5, e134796. [Google Scholar] [CrossRef] [Green Version]
- Posma, J.J.; Posthuma, J.J.; Spronk, H.M. Coagulation and non-coagulation effects of thrombin. J. Thromb. Haemost. 2016, 14, 1908–1916. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.K.; Ha, T.Y.; McGregor, R.A.; Choi, M.S. Long-term curcumin administration protects against atherosclerosis via hepatic regulation of lipoprotein cholesterol metabolism. Mol. Nutr. Food Res. 2011, 55, 1829–1840. [Google Scholar] [CrossRef]
- Krolikoski, M.; Monslow, J.; Pure, E. The CD44-HA axis and inflammation in atherosclerosis: A temporal perspective. Matrix Biol. 2019, 78–79, 201–218. [Google Scholar] [CrossRef]
- Smeets, E.; Meiler, S.; Lutgens, E. Lymphocytic tumor necrosis factor receptor superfamily co-stimulatory molecules in the pathogenesis of atherosclerosis. Curr. Opin. Lipidol. 2013, 24, 518–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Meer, J.H.; van der Poll, T.; van ‘t Veer, C. TAM receptors, Gas6, and protein S: Roles in inflammation and hemostasis. Blood 2014, 123, 2460–2469. [Google Scholar] [CrossRef] [PubMed]
- Puerta, A.; Diez-Masa, J.C.; Martin-Alvarez, P.J.; Martin-Ventura, J.L.; Barbas, C.; Tunon, J.; Egido, J.; de Frutos, M. Study of the capillary electrophoresis profile of intact alpha-1-acid glycoprotein isoforms as a biomarker of atherothrombosis. Analyst 2011, 136, 816–822. [Google Scholar] [CrossRef]
- Jiang, F.; Yang, J.; Zhang, Y.; Dong, M.; Wang, S.; Zhang, Q.; Liu, F.F.; Zhang, K.; Zhang, C. Angiotensin-converting enzyme 2 and angiotensin 1-7: Novel therapeutic targets. Nat. Rev. Cardiol. 2014, 11, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Wieland, T.; Attwood, P.V. Alterations in reversible protein histidine phosphorylation as intracellular signals in cardiovascular disease. Front. Pharmacol. 2015, 6, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Gao, W.; Yang, Y.; Guo, S.; Wang, H.; Wang, W.; Zhang, S.; Zhou, Q.; Xu, H.; Yao, J.; et al. Inhibition of VDAC1 prevents Ca(2)(+)-mediated oxidative stress and apoptosis induced by 5-aminolevulinic acid mediated sonodynamic therapy in THP-1 macrophages. Apoptosis 2014, 19, 1712–1726. [Google Scholar] [CrossRef]
- Edfeldt, K.; Agerberth, B.; Rottenberg, M.E.; Gudmundsson, G.H.; Wang, X.B.; Mandal, K.; Xu, Q.; Yan, Z.Q. Involvement of the antimicrobial peptide LL-37 in human atherosclerosis. Arter. Thromb. Vasc. Biol. 2006, 26, 1551–1557. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Huang, Z.; Huang, W.; Chen, X.; Shan, P.; Zhong, P.; Khan, Z.; Wang, J.; Fang, Q.; Liang, G.; et al. Inhibition of epidermal growth factor receptor attenuates atherosclerosis via decreasing inflammation and oxidative stress. Sci. Rep. 2017, 8, 45917. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.F.; Herrington, D.M. The function of cathepsins B, D, and X in atherosclerosis. Am. J. Cardiovasc. Dis. 2016, 6, 163–170. [Google Scholar]
- Biros, E.; Moran, C.S.; Maguire, J.; Holliday, E.; Levi, C.; Golledge, J. Upregulation of arylsulfatase B in carotid atherosclerosis is associated with symptoms of cerebral embolization. Sci. Rep. 2017, 7, 4338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Romero, D.; Ruiz-Nodar, J.M.; Marin, F.; Tello-Montoliu, A.; Roldan, V.; Mainar, L.; Perez-Andreu, V.; Anton, A.I.; Bonaque, J.C.; Valdes, M.; et al. CALU A29809G polymorphism in coronary atherothrombosis: Implications for coronary calcification and prognosis. Ann. Med. 2010, 42, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Sakosda, H.; Fujishiro, M.; Anai, M.; Kushiyama, A.; Horike, N.; Kamata, H.; Ogihara, T.; Kurihara, H.; Uchijima, Y. Physiological significance of resistin and resistin-like molecules in the inflammatory process and insulin resistance. Curr. Diabetes Rev. 2006, 2, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Haversen, L.; Sundelin, J.P.; Mardinoglu, A.; Rutberg, M.; Stahlman, M.; Wilhelmsson, U.; Hulten, L.M.; Pekny, M.; Fogelstrand, P.; Bentzon, J.F.; et al. Vimentin deficiency in macrophages induces increased oxidative stress and vascular inflammation but attenuates atherosclerosis in mice. Sci. Rep. 2018, 8, 16973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oszajca, K.; Wronski, K.; Janiszewska, G.; Bienkiewicz, M.; Panek, M.; Bartkowiak, J.; Szemraj, J. Association analysis of genetic polymorphisms of factor V, factor VII and fibrinogen beta chain genes with human abdominal aortic aneurysm. Exp. Ther. Med. 2012, 4, 514–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.Q.; Ngo, A.; Hoffmann, P.; Ferrante, A.; Hii, C.S. Regulation of endothelial cell survival and death by the MAP kinase/ERK kinase kinase 3-glyceraldehyde-3-phosphate dehydrogenase signaling axis. Cell Signal. 2019, 58, 20–33. [Google Scholar] [CrossRef]
- Fukai, T.; Galis, Z.S.; Meng, X.P.; Parthasarathy, S.; Harrison, D.G. Vascular expression of extracellular superoxide dismutase in atherosclerosis. J. Clin. Investig. 1998, 101, 2101–2111. [Google Scholar] [CrossRef]
- Yu, X.H.; Jiang, N.; Yao, P.B.; Zheng, X.L.; Cayabyab, F.S.; Tang, C.K. NPC1, intracellular cholesterol trafficking and atherosclerosis. Clin. Chim. Acta 2014, 429, 69–75. [Google Scholar] [CrossRef]
- Sun, J.; Wang, Z.; Wang, X. Suppression of LRRC19 promotes cutaneous wound healing in pressure ulcers in mice. Organogenesis 2018, 14, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Gouni-Berthold, I.; Krone, W.; Berthold, H.K. Vitamin D and cardiovascular disease. Curr. Vasc. Pharmacol. 2009, 7, 414–422. [Google Scholar] [CrossRef]
- Arenas de Larriva, A.P.; Alonso, A.; Norby, F.L.; Roetker, N.S.; Folsom, A.R. Circulating ceruloplasmin, ceruloplasmin-associated genes and the incidence of venous thromboembolism in the Atherosclerosis Risk in Communities study. J. Thromb. Haemost. 2019, 17, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Pollard, R.D.; Blesso, C.N.; Zabalawi, M.; Fulp, B.; Gerelus, M.; Zhu, X.; Lyons, E.W.; Nuradin, N.; Francone, O.L.; Li, X.A.; et al. Procollagen C-endopeptidase Enhancer Protein 2 (PCPE2) Reduces Atherosclerosis in Mice by Enhancing Scavenger Receptor Class B1 (SR-BI)-mediated High-density Lipoprotein (HDL)-Cholesteryl Ester Uptake. J. Biol. Chem. 2015, 290, 15496–15511. [Google Scholar] [CrossRef] [PubMed]
- Teunissen, P.F.; Boshuizen, M.C.; Hollander, M.R.; Biesbroek, P.S.; van der Hoeven, N.W.; Mol, J.Q.; Gijbels, M.J.; van der Velden, S.; van der Pouw Kraan, T.C.; Horrevoets, A.J.; et al. MAb therapy against the IFN-alpha/beta receptor subunit 1 stimulates arteriogenesis in a murine hindlimb ischaemia model without enhancing atherosclerotic burden. Cardiovasc. Res. 2015, 107, 255–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachhawat, A.K.; Yadav, S. The glutathione cycle: Glutathione metabolism beyond the gamma-glutamyl cycle. IUBMB Life 2018, 70, 585–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Presnell, C.E.; Bhatti, G.; Numan, L.S.; Lerche, M.; Alkhateeb, S.K.; Ghalib, M.; Shammaa, M.; Kavdia, M. Computational insights into the role of glutathione in oxidative stress. Curr. Neurovasc. Res. 2013, 10, 185–194. [Google Scholar] [CrossRef]
- Ichiki, T.; Izumi, R.; Cataliotti, A.; Larsen, A.M.; Sandberg, S.M.; Burnett, J.C., Jr. Endothelial permeability in vitro and in vivo: Protective actions of ANP and omapatrilat in experimental atherosclerosis. Peptides 2013, 48, 21–26. [Google Scholar] [CrossRef] [Green Version]
- Inoue, M.; Ishida, T.; Yasuda, T.; Toh, R.; Hara, T.; Cangara, H.M.; Rikitake, Y.; Taira, K.; Sun, L.; Kundu, R.K.; et al. Endothelial cell-selective adhesion molecule modulates atherosclerosis through plaque angiogenesis and monocyte-endothelial interaction. Microvasc. Res. 2010, 80, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Franca, K.C.; Martinez, P.A.; Prado, M.L.; Lo, S.M.; Borges, B.E.; Zanata, S.M.; San Martin, A.; Nakao, L.S. Quiescin/sulfhydryl oxidase 1b (QSOX1b) induces migration and proliferation of vascular smooth muscle cells by distinct redox pathways. Arch. Biochem. Biophys. 2020, 679, 108220. [Google Scholar] [CrossRef]
- Bian, T.; Li, H.; Zhou, Q.; Ni, C.; Zhang, Y.; Yan, F. Human beta-Defensin 3 Reduces TNF-alpha-Induced Inflammation and Monocyte Adhesion in Human Umbilical Vein Endothelial Cells. Mediat. Inflamm. 2017, 2017, 8529542. [Google Scholar] [CrossRef] [Green Version]
- Couchie, D.; Vaisman, B.; Abderrazak, A.; Mahmood, D.F.D.; Hamza, M.M.; Canesi, F.; Diderot, V.; El Hadri, K.; Negre-Salvayre, A.; Le Page, A.; et al. Human Plasma Thioredoxin-80 Increases With Age and in ApoE(-/-) Mice Induces Inflammation, Angiogenesis, and Atherosclerosis. Circulation 2017, 136, 464–475. [Google Scholar] [CrossRef]
- Fu, X.; Huang, X.; Li, P.; Chen, W.; Xia, M. 7-Ketocholesterol inhibits isocitrate dehydrogenase 2 expression and impairs endothelial function via microRNA-144. Free Radic. Biol. Med. 2014, 71, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.D. Apolipoprotein A-I and its mimetics for the treatment of atherosclerosis. Curr. Opin. Investig. Drugs 2010, 11, 989–996. [Google Scholar] [PubMed]
- Hewing, B.; Ludwig, A.; Dan, C.; Potzsch, M.; Hannemann, C.; Petry, A.; Lauer, D.; Gorlach, A.; Kaschina, E.; Muller, D.N.; et al. Immunoproteasome subunit ss5i/LMP7-deficiency in atherosclerosis. Sci. Rep. 2017, 7, 13342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Huang, H.; Ma, Y.; Sun, Y.; Wang, G.; Tang, L. Association of the KLK1 rs5516 G allele and the ACE D allele with aortic aneurysm and atherosclerotic stenosis. Medicine 2016, 95, e5120. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.D.; Jackson, C.L.; Morgan, B.P.; Hughes, T.R. The membrane attack complex of complement drives the progression of atherosclerosis in apolipoprotein E knockout mice. Mol. Immunol. 2010, 47, 1098–1105. [Google Scholar] [CrossRef]
- Karshovska, E.; Zhao, Z.; Blanchet, X.; Schmitt, M.M.; Bidzhekov, K.; Soehnlein, O.; von Hundelshausen, P.; Mattheij, N.J.; Cosemans, J.M.; Megens, R.T.; et al. Hyperreactivity of junctional adhesion molecule A-deficient platelets accelerates atherosclerosis in hyperlipidemic mice. Circ. Res. 2015, 116, 587–599. [Google Scholar] [CrossRef] [Green Version]
- Allawi, A.A.D. Malnutrition, inflamation and atherosclerosis (MIA syndrome) in patients with end stage renal disease on maintenance hemodialysis (a single centre experience). Diabetes Metab. Syndr. 2018, 12, 91–97. [Google Scholar] [CrossRef]
- Segedy, A.K.; Pyle, A.L.; Li, B.; Zhang, Y.; Babaev, V.R.; Jat, P.; Fazio, S.; Atkinson, J.B.; Linton, M.F.; Young, P.P. Identification of small proline-rich repeat protein 3 as a novel atheroprotective factor that promotes adaptive Akt signaling in vascular smooth muscle cells. Arter. Thromb. Vasc. Biol. 2014, 34, 2527–2536. [Google Scholar] [CrossRef] [Green Version]
- Burke, R.M.; Lighthouse, J.K.; Quijada, P.; Dirkx, R.A., Jr.; Rosenberg, A.; Moravec, C.S.; Alexis, J.D.; Small, E.M. Small proline-rich protein 2B drives stress-dependent p53 degradation and fibroblast proliferation in heart failure. Proc. Natl. Acad. Sci. USA 2018, 115, E3436–E3445. [Google Scholar] [CrossRef] [Green Version]
- Madrigal-Matute, J.; Fernandez-Garcia, C.E.; Blanco-Colio, L.M.; Burillo, E.; Fortuno, A.; Martinez-Pinna, R.; Llamas-Granda, P.; Beloqui, O.; Egido, J.; Zalba, G.; et al. Thioredoxin-1/peroxiredoxin-1 as sensors of oxidative stress mediated by NADPH oxidase activity in atherosclerosis. Free Radic. Biol. Med. 2015, 86, 352–361. [Google Scholar] [CrossRef]
- Yuan, L.; Wang, M.; Liu, T.; Lei, Y.; Miao, Q.; Li, Q.; Wang, H.; Zhang, G.; Hou, Y.; Chang, X. Carbonic Anhydrase 1-Mediated Calcification Is Associated With Atherosclerosis, and Methazolamide Alleviates Its Pathogenesis. Front. Pharmacol. 2019, 10, 766. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Riazy, M.; Gold, M.; Tsai, S.H.; McNagny, K.; Proud, C.; Duronio, V. Impairing eukaryotic elongation factor 2 kinase activity decreases atherosclerotic plaque formation. Can. J. Cardiol. 2014, 30, 1684–1688. [Google Scholar] [CrossRef] [PubMed]
- Habas, K.; Shang, L. Alterations in intercellular adhesion molecule 1 (ICAM-1) and vascular cell adhesion molecule 1 (VCAM-1) in human endothelial cells. Tissue Cell 2018, 54, 139–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Moran, C.S.; Trollope, A.F.; Woodward, L.; Kinobe, R.; Rush, C.M.; Golledge, J. Angiopoietin-2 attenuates angiotensin II-induced aortic aneurysm and atherosclerosis in apolipoprotein E-deficient mice. Sci. Rep. 2016, 6, 35190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adada, M.; Luberto, C.; Canals, D. Inhibitors of the sphingomyelin cycle: Sphingomyelin synthases and sphingomyelinases. Chem. Phys. Lipids 2016, 197, 45–59. [Google Scholar] [CrossRef]
- Kohno, T.; Urao, N.; Ashino, T.; Sudhahar, V.; McKinney, R.D.; Hamakubo, T.; Iwanari, H.; Ushio-Fukai, M.; Fukai, T. Novel role of copper transport protein antioxidant-1 in neointimal formation after vascular injury. Arter. Thromb. Vasc. Biol. 2013, 33, 805–813. [Google Scholar] [CrossRef] [Green Version]
- Then, C.; Then, H.L.; Lechner, A.; Thorand, B.; Meisinger, C.; Heier, M.; Peters, A.; Koenig, W.; Rathmann, W.; Scherberich, J.; et al. Serum uromodulin and risk for cardiovascular morbidity and mortality in the community-based KORA F4 study. Atherosclerosis 2020, 297, 1–7. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, L.; Zhang, J.; Huang, J.; Wei, G.; Liu, L.; Zhang, J.; Yan, B. Altered expression of lysosomal hydrolase, acid alpha-glucosidase, gene in coronary artery disease. Coron. Artery Dis. 2016, 27, 104–108. [Google Scholar] [CrossRef]
- Tawa, H.; Rikitake, Y.; Takahashi, M.; Amano, H.; Miyata, M.; Satomi-Kobayashi, S.; Kinugasa, M.; Nagamatsu, Y.; Majima, T.; Ogita, H.; et al. Role of afadin in vascular endothelial growth factor- and sphingosine 1-phosphate-induced angiogenesis. Circ. Res. 2010, 106, 1731–1742. [Google Scholar] [CrossRef] [Green Version]
- Kremen, M.; Krishnan, R.; Emery, I.; Hu, J.H.; Slezicki, K.I.; Wu, A.; Qian, K.; Du, L.; Plawman, A.; Stempien-Otero, A.; et al. Plasminogen mediates the atherogenic effects of macrophage-expressed urokinase and accelerates atherosclerosis in apoE-knockout mice. Proc. Natl. Acad. Sci. USA 2008, 105, 17109–17114. [Google Scholar] [CrossRef] [Green Version]
- Hemdahl, A.L.; Gabrielsen, A.; Zhu, C.; Eriksson, P.; Hedin, U.; Kastrup, J.; Thoren, P.; Hansson, G.K. Expression of neutrophil gelatinase-associated lipocalin in atherosclerosis and myocardial infarction. Arter. Thromb. Vasc. Biol. 2006, 26, 136–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amighi, J.; Hoke, M.; Mlekusch, W.; Schlager, O.; Exner, M.; Haumer, M.; Pernicka, E.; Koppensteiner, R.; Minar, E.; Rumpold, H.; et al. Beta 2 microglobulin and the risk for cardiovascular events in patients with asymptomatic carotid atherosclerosis. Stroke 2011, 42, 1826–1833. [Google Scholar] [CrossRef] [PubMed]
- Ragolia, L.; Palaia, T.; Hall, C.E.; Maesaka, J.K.; Eguchi, N.; Urade, Y. Accelerated glucose intolerance, nephropathy, and atherosclerosis in prostaglandin D2 synthase knock-out mice. J. Biol. Chem. 2005, 280, 29946–29955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sena, B.F.; Figueiredo, J.L.; Aikawa, E. Cathepsin S As an Inhibitor of Cardiovascular Inflammation and Calcification in Chronic Kidney Disease. Front. Cardiovasc. Med. 2017, 4, 88. [Google Scholar] [CrossRef] [Green Version]
- Baeyens, N.; Latrache, I.; Yerna, X.; Noppe, G.; Horman, S.; Morel, N. Redundant control of migration and adhesion by ERM proteins in vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2013, 441, 579–585. [Google Scholar] [CrossRef]
- Nahon, J.E.; Hoekstra, M.; Van Eck, M. Total body proteoglycan 4 (Prg4) deficiency increases atherosclerosis susceptibility in apolipoprotein E knockout and low-density lipoprotein receptor knockout mice. Atherosclerosis 2018, 278, 315–316. [Google Scholar] [CrossRef]
- Jaisson, S.; Pietrement, C.; Gillery, P. Carbamylation-derived products: Bioactive compounds and potential biomarkers in chronic renal failure and atherosclerosis. Clin. Chem. 2011, 57, 1499–1505. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Liu, D.; Song, P.; Zou, M.H. Tryptophan-kynurenine pathway is dysregulated in inflammation, and immune activation. Front. Biosci. Landmark Ed. 2015, 20, 1116–1143. [Google Scholar] [CrossRef] [Green Version]
- Pamplona, R.; Dalfo, E.; Ayala, V.; Bellmunt, M.J.; Prat, J.; Ferrer, I.; Portero-Otin, M. Proteins in human brain cortex are modified by oxidation, glycoxidation, and lipoxidation. Effects of Alzheimer disease and identification of lipoxidation targets. J. Biol. Chem. 2005, 280, 21522–21530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nehler, M.R.; Taylor, L.M., Jr.; Porter, J.M. Homocysteinemia as a risk factor for atherosclerosis: A review. Cardiovasc. Surg. 1997, 5, 559–567. [Google Scholar] [CrossRef]
- Chen, S.S.; Tang, C.S.; Jin, H.F.; Du, J.B. Sulfur dioxide acts as a novel endogenous gaseous signaling molecule in the cardiovascular system. Chin. Med. J. 2011, 124, 1901–1905. [Google Scholar] [PubMed]
- Lee, S.E.; Park, Y.S. Korean Red Ginseng water extract inhibits COX-2 expression by suppressing p38 in acrolein-treated human endothelial cells. J. Ginseng Res. 2014, 38, 34–39. [Google Scholar] [CrossRef]
- McGrath, C.E.; Tallman, K.A.; Porter, N.A.; Marnett, L.J. Structure-activity analysis of diffusible lipid electrophiles associated with phospholipid peroxidation: 4-hydroxynonenal and 4-oxononenal analogues. Chem. Res. Toxicol. 2011, 24, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Nagai, R.; Unno, Y.; Hayashi, M.C.; Masuda, S.; Hayase, F.; Kinae, N.; Horiuchi, S. Peroxynitrite induces formation of N( epsilon )-(carboxymethyl) lysine by the cleavage of Amadori product and generation of glucosone and glyoxal from glucose: Novel pathways for protein modification by peroxynitrite. Diabetes 2002, 51, 2833–2839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuang, W.T.; Liu, Y.T.; Huang, C.S.; Lo, C.W.; Yao, H.T.; Chen, H.W.; Lii, C.K. Benzyl Isothiocyanate and Phenethyl Isothiocyanate Inhibit Adipogenesis and Hepatosteatosis in Mice with Obesity Induced by a High-Fat Diet. J. Agric. Food Chem. 2019, 67, 7136–7146. [Google Scholar] [CrossRef]
- Soares, E.S.A.K.; de Oliveira Cipriano Torres, D.; Santos Rocha, S.W.; dos Santos Gomes, F.O.; dos Santos Silva, B.; Donato, M.A.; Raposo, C.; Santos, A.C.; de Lima Mdo, C.; Galdino, S.L.; et al. Effect of new thiazolidine derivatives LPSF/GQ-02 and LPSF/GQ-16 on atherosclerotic lesions in LDL receptor-deficient mice (LDLR(-/-)). Cardiovasc. Pathol. 2013, 22, 81–90. [Google Scholar] [CrossRef]
- Liu, A.; Li, K.; Xu, L.; Si, M.; Teng, G.; Li, G.; Xue, J.; Liang, S.; Song, W. Metformin Delays the Development of Atherosclerosis in Type 1 Diabetes Mellitus via the Methylglyoxal Pathway. Diabetes Ther. 2020, 11, 633–642. [Google Scholar] [CrossRef] [Green Version]
- Becker, D.J.; Lowe, J.B. Fucose: Biosynthesis and biological function in mammals. Glycobiology 2003, 13, 41R–53R. [Google Scholar] [CrossRef]
- Yin, Z.; Zou, Y.; Wang, D.; Huang, X.; Xiong, S.; Cao, L.; Zhang, Y.; Sun, Y.; Zhang, N. Regulation of the Tec family of non-receptor tyrosine kinases in cardiovascular disease. Cell Death Discov. 2022, 8, 119. [Google Scholar] [CrossRef]
- Hatch, E.; Morrow, D.; Liu, W.; Cahill, P.A.; Redmond, E.M. Differential effects of alcohol and its metabolite acetaldehyde on vascular smooth muscle cell Notch signaling and growth. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H131–H137. [Google Scholar] [CrossRef]
- Heier, M.; Margeirsdottir, H.D.; Torjesen, P.A.; Seljeflot, I.; Stensaeth, K.H.; Gaarder, M.; Brunborg, C.; Hanssen, K.F.; Dahl-Jorgensen, K. The advanced glycation end product methylglyoxal-derived hydroimidazolone-1 and early signs of atherosclerosis in childhood diabetes. Diabetes Vasc. Dis. Res. 2015, 12, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Picard, S.; Parthasarathy, S.; Fruebis, J.; Witztum, J.L. Aminoguanidine inhibits oxidative modification of low density lipoprotein protein and the subsequent increase in uptake by macrophage scavenger receptors. Proc. Natl. Acad. Sci. USA 1992, 89, 6876–6880. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.F.; Chen, X.; Tang, X. Short-chain fatty acid, acylation and cardiovascular diseases. Clin. Sci. 2020, 134, 657–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Bao, Z.; Ding, Y.; Xu, S.; Du, R.; Yan, J.; Li, L.; Sun, Z.; Shao, C.; Gu, W. Nepsilon-carboxymethyl-lysine-induced PI3K/Akt signaling inhibition promotes foam cell apoptosis and atherosclerosis progression. Biomed. Pharmacother. 2019, 115, 108880. [Google Scholar] [CrossRef]
- Rodgers, K.J.; Dean, R.T. Metabolism of protein-bound DOPA in mammals. Int. J. Biochem. Cell Biol. 2000, 32, 945–955. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, Y.; Meng, W.; Wei, J.; Liu, Y.; Gao, Y. Changes to Urinary Proteome in High-Fat-Diet ApoE−/− Mice. Biomolecules 2022, 12, 1569. https://doi.org/10.3390/biom12111569
Hua Y, Meng W, Wei J, Liu Y, Gao Y. Changes to Urinary Proteome in High-Fat-Diet ApoE−/− Mice. Biomolecules. 2022; 12(11):1569. https://doi.org/10.3390/biom12111569
Chicago/Turabian StyleHua, Yuanrui, Wenshu Meng, Jing Wei, Yongtao Liu, and Youhe Gao. 2022. "Changes to Urinary Proteome in High-Fat-Diet ApoE−/− Mice" Biomolecules 12, no. 11: 1569. https://doi.org/10.3390/biom12111569






