A Machine Learning Approach for Recommending Herbal Formulae with Enhanced Interpretability and Applicability
Abstract
:1. Introduction
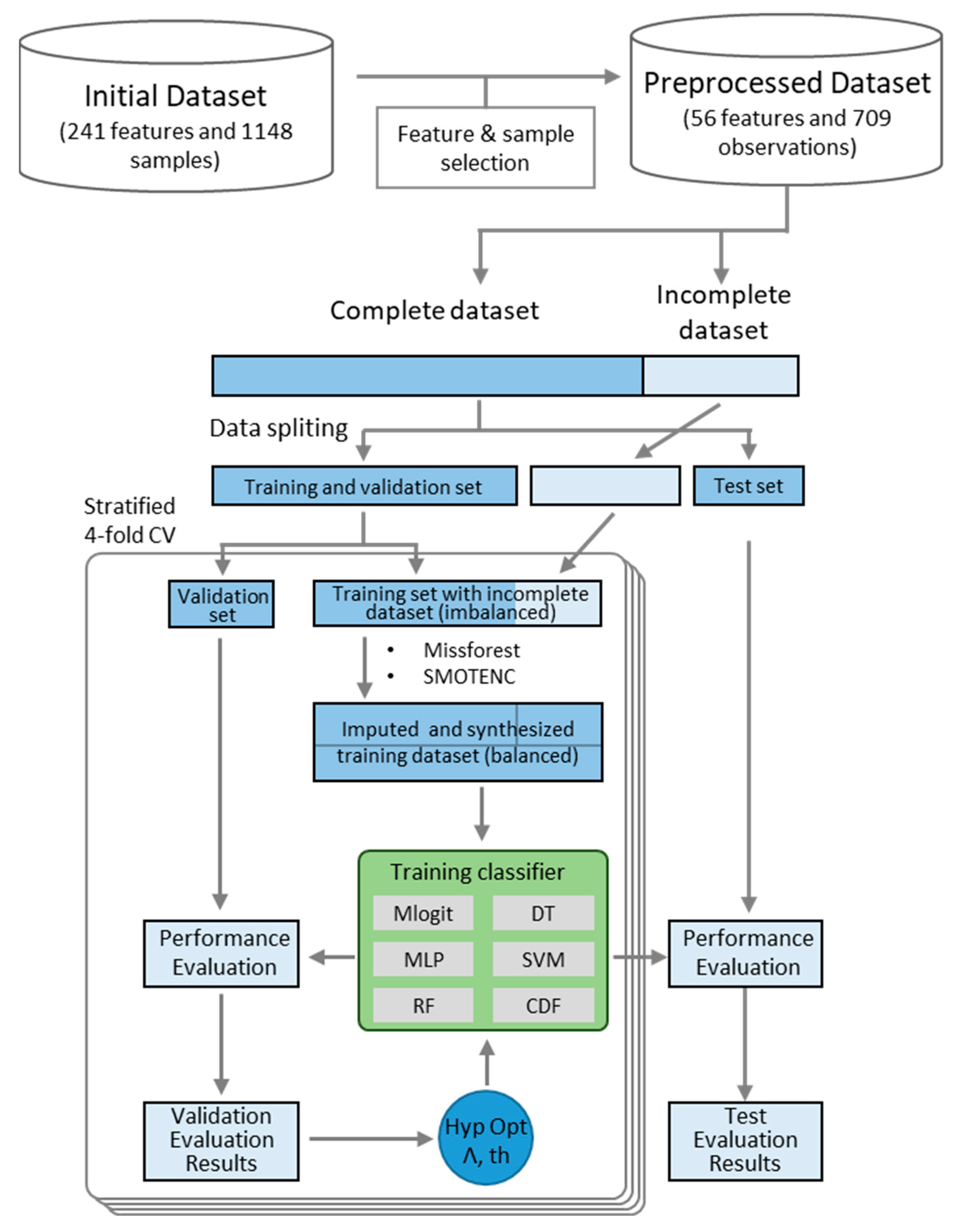
2. Materials and Methods
2.1. Data Collection and Selection Procedure
2.2. Classifier Models
2.3. Data Oversampling
2.4. Data Imputation
2.5. Case Study Selection and Data Processing
2.6. LIME
2.7. Performance Evaluation
3. Results
3.1. Dataset Construction and Description
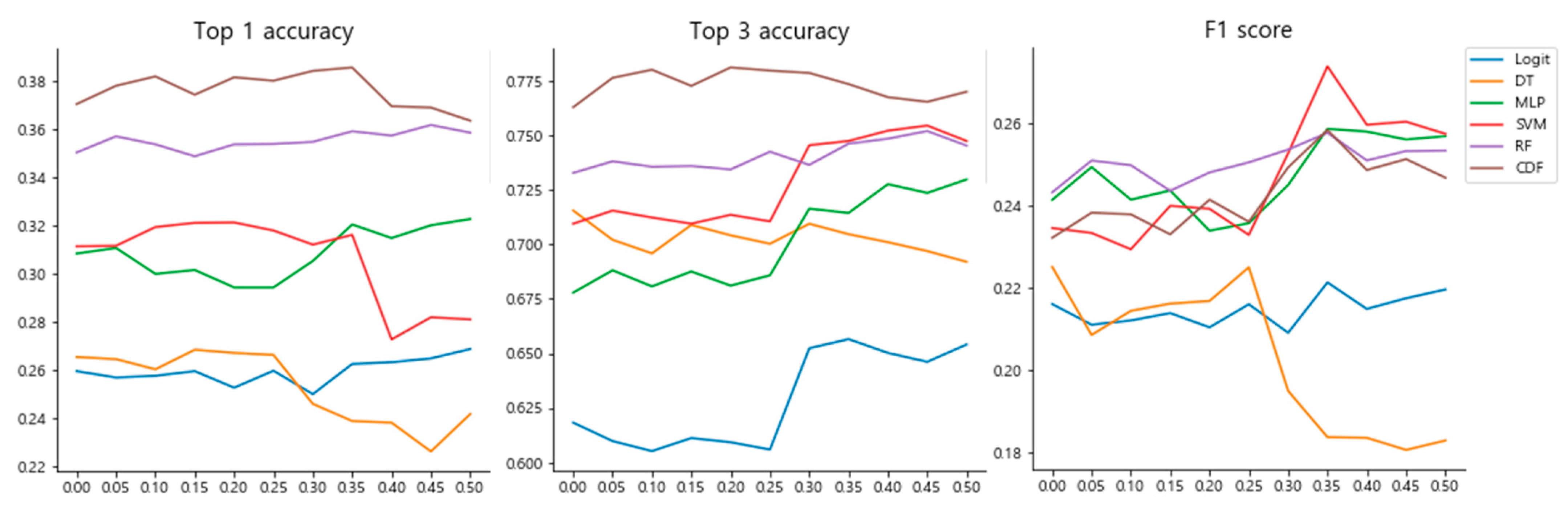
3.2. Hyperparameter Selection for the Classifiers
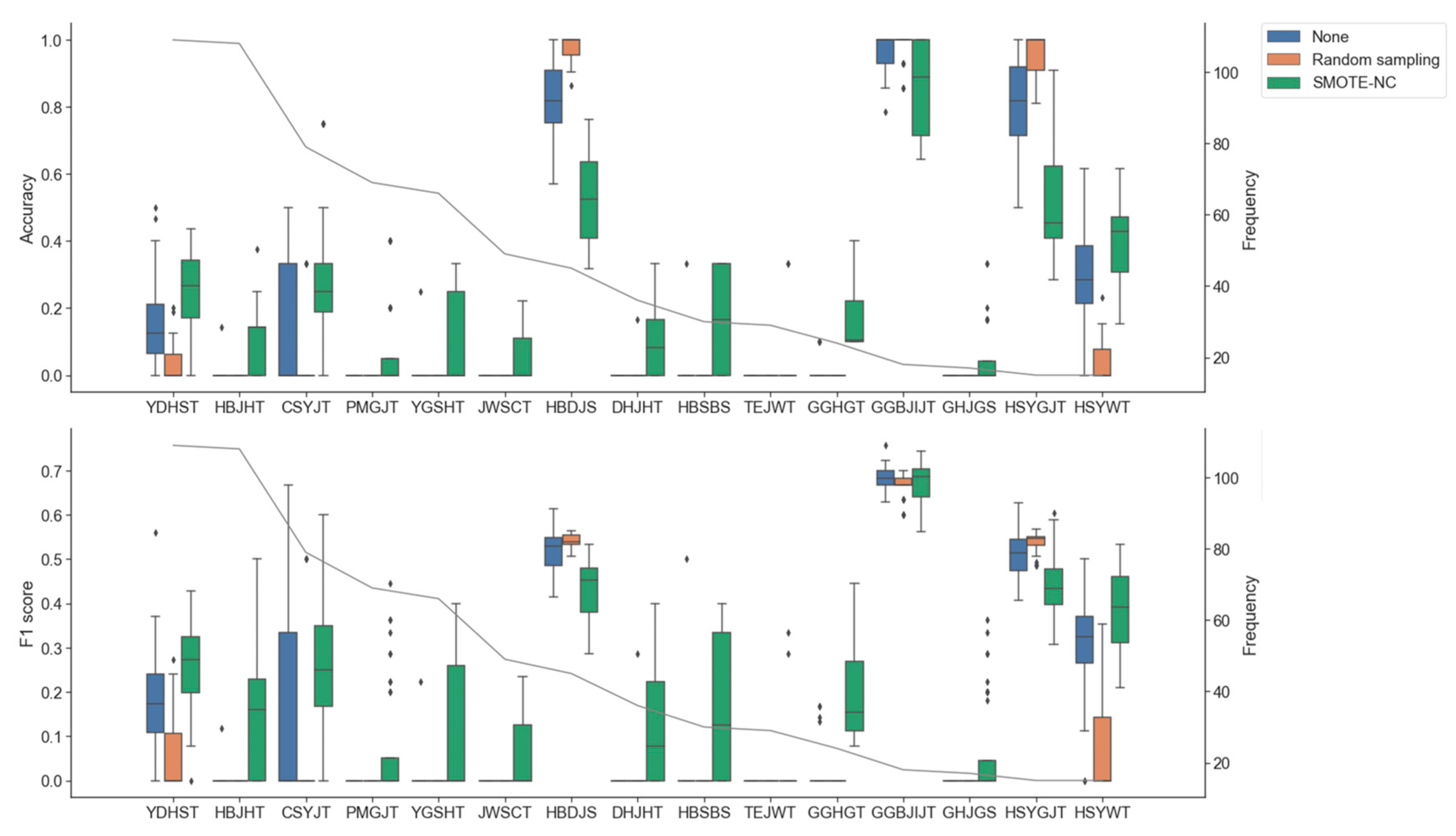
3.3. Impact of Oversampling and Data Imputation
3.4. Evaluating the Model Generalization Abilities on Unseen Datasets
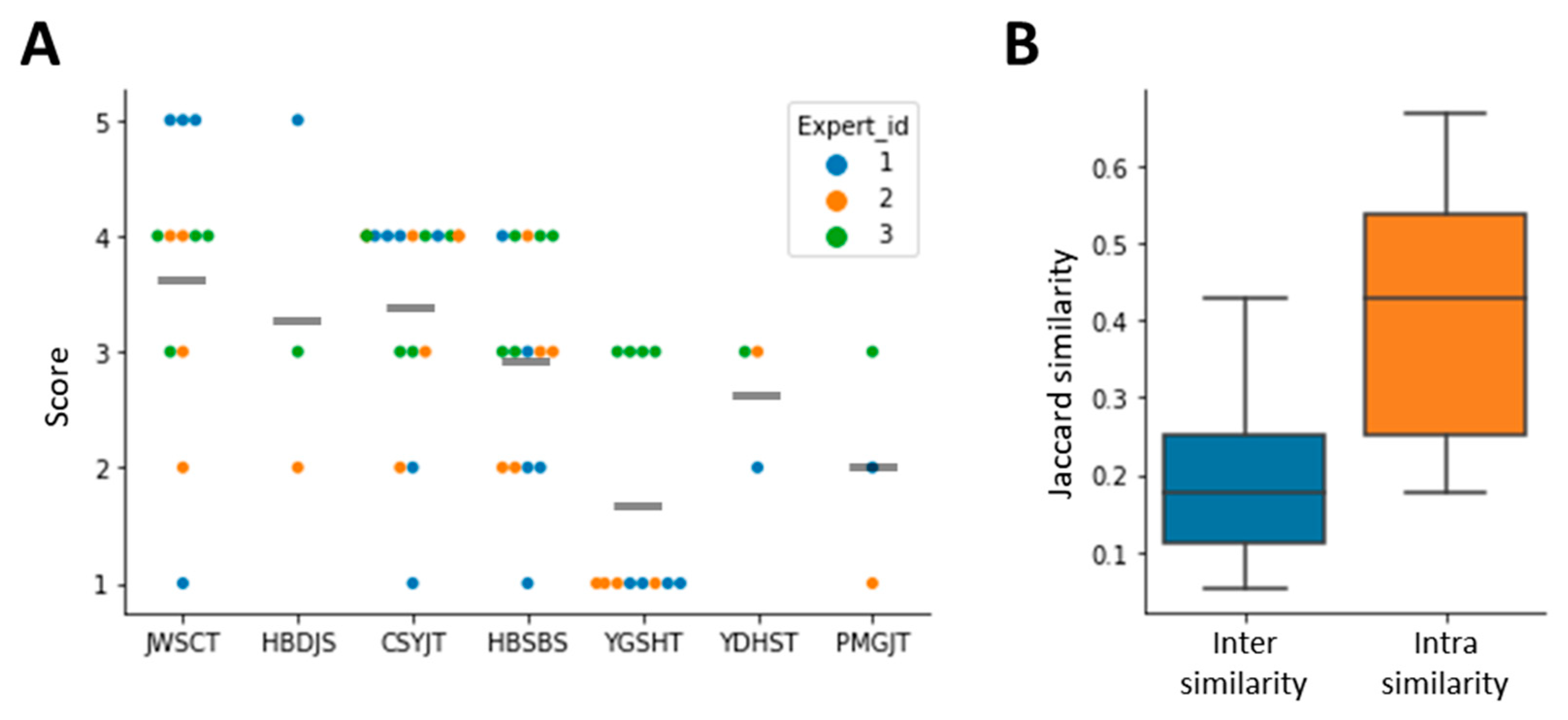
3.5. Model Interpretation Using Case Study Data
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, F.-S.; Weng, J.-K. Demystifying traditional herbal medicine with modern approach. Nat. Plants 2017, 3, 17109. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.-Y.; Kim, H.-G.; Lee, J.-S.; Cho, J.-H.; Ahn, Y.-C.; Lee, D.-S.; Son, C.-G. Anti-hepatofibrotic effects of CGX, a standardized herbal formula: A multicenter randomized clinical trial. Biomed. Pharmacother. 2020, 126, 110105. [Google Scholar] [CrossRef] [PubMed]
- Sul, J.-U.; Kim, M.K.; Leem, J.; Jo, H.-G.; Yoon, S.; Kim, J.; Lee, E.-J.; Yoo, J.-E.; Park, S.J.; Kim, Y. Il Efficacy and safety of gyejigachulbutang (Gui-Zhi-Jia-Shu-Fu-Tang, Keishikajutsubuto, TJ-18) for knee pain in patients with degenerative knee osteoarthritis: A randomized, placebo-controlled, patient and assessor blinded clinical trial. Trials 2019, 20, 140. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.; Liu, Z.; Li, N.; Li, Y.; Yang, F.; Pang, B.; Jin, X.; Zheng, W.; Zhang, J. Chinese medical drugs for coronavirus disease 2019: A systematic review and meta-analysis. Integr. Med. Res. 2020, 9, 100477. [Google Scholar] [CrossRef] [PubMed]
- Cheung, F. TCM: Made in China. Nature 2011, 480, S82–S83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, E.; Kim, Y.; Lee, E.J.; Yoo, H.R. Review on the development state and utilization of pattern identification questionnaire in Korean medicine by U code of Korean Classification of Disease. J. Physiol. Pathol. Korean Med. 2016, 30, 124–130. [Google Scholar] [CrossRef]
- Kang, B.-K.; Park, T.-Y.; Lee, J.A.; Moon, T.-W.; Ko, M.M.; Choi, J.; Lee, M.S. Reliability and validity of the Korean standard pattern identification for stroke (K-SPI-Stroke) questionnaire. BMC Complement. Altern. Med. 2012, 12, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, K.-T.; Kim, H.-T.; Hwang, E.-H.; Hwang, M.-S.; Heo, I.; Park, S.-Y.; Cho, J.-H.; Kim, K.-W.; Ha, I.-H.; Kim, M. Adaptation and dissemination of Korean medicine clinical practice guidelines for traffic injuries. Healthcare 2022, 10, 1166. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, Z. Exploration on generating traditional Chinese medicine prescriptions from symptoms with an end-to-end approach. In Proceedings of the CCF International Conference on Natural Language Processing and Chinese Computing, Dunhuang, China, 9–14 October 2019; pp. 486–498. [Google Scholar]
- Yao, L.; Zhang, Y.; Wei, B.; Zhang, W.; Jin, Z. A topic modeling approach for traditional Chinese medicine prescriptions. IEEE Trans. Knowl. Data Eng. 2018, 30, 1007–1021. [Google Scholar] [CrossRef]
- Yang, K.; Zhang, R.; He, L.; Li, Y.; Liu, W.; Yu, C.; Zhang, Y.; Li, X.; Liu, Y.; Xu, W. Multistage analysis method for detection of effective herb prescription from clinical data. Front. Med. 2018, 12, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yang, K.; Zeng, J.; Lai, X.; Wang, X.; Ji, C.; Li, Y.; Zhang, P.; Li, S. FordNet: Recommending traditional Chinese medicine formula via deep neural network integrating phenotype and molecule. Pharmacol. Res. 2021, 173, 105752. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Demonstrations, San Diego, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar] [CrossRef]
- Kim, J.Y.; Pham, D.D. Sasang constitutional medicine as a holistic tailored medicine. Evid.-Based Complement. Altern. Med. 2009, 6, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, H.J.; Baek, Y.; Kim, H.S.; Ryu, J.; Lee, S. Constitutional multicenter bank linked to Sasang constitutional phenotypic data. BMC Complement. Altern. Med. 2015, 15, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.H.; Feng, J. Deep forest. Natl. Sci. Rev. 2019, 6, 74–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Mukherjee, M.; Khushi, M. SMOTE-ENC: A novel SMOTE-based method to generate synthetic data for nominal and continuous features. Appl. Syst. Innov. 2021, 4, 18. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Bühlmann, P. Missforest-Non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bomin, K.; Hee-Geun, J. Effect of Modified Hyeongbangjiwhang-tang for essential tremor after total vaginal hysterectomy: Case report. J. Sasang Const. Med. 2018, 30, 59–66. [Google Scholar]
- Komal Kumar, N.; Vigneswari, D. A drug recommendation system for multi-disease in health care using machine learning. In Advances in Communication and Computational Technology; Springer: New York, NY, USA, 2021; pp. 1–12. [Google Scholar]
- Nagaraj, P.; Muneeswaran, V.; Deshik, G. Ensemble Machine Learning (Grid Search & Random Forest) based Enhanced Medical Expert Recommendation System for Diabetes Mellitus Prediction. In Proceedings of the 2022 3rd International Conference on Electronics and Sustainable Communication Systems (ICESC), Coimbatore, India, 17–19 August 2022; pp. 757–765. [Google Scholar]
- Han, H.; Li, W.; Wang, J.; Qin, G.; Qin, X. Enhance Explainability of Manifold Learning. Neurocomputing 2022, 500, 877–895. [Google Scholar] [CrossRef]
| Formula Name (Abbreviation) | Composition (per Serving) |
|---|---|
| Cheongsimyeonja-tang (CSYJT) | Nelumbinis Semen, Dioscoreae Rhizoma, Asparagi Tuber, Polygalae Radix, Acori Graminei Rhizoma, Zizyphi Semen, Longan Arillus, Thujae Semen, Scutellariae Radix, and Raphani Semen 7.5 g, Chrysanthemi Flos 1.125 g |
| Dokhwaljihwang-tang (DHJHT) | Rehmanniae Radix Preparata 15 g, Corni Fructus 7.5 g, Poria Sclerotium, and Alismatis Rhizoma 5.625 g, Moutan Cortex, Saposhnikoviae Radix, and Angelicae Continentalis Radix 3.75 g |
| Galgeunhaegi-tang (GGHGT) | Puerariae Radix 11.25 g, Cimicifugae Rhizoma 7.5 g, Scutellariae Radix, and Armeniacae Semen 5.625 g, Platycodonis Radix, Zizyphi Semen, Rhei Rhizoma, and Angelicae Dahuricae Radix 3.75 g |
| Gwakhyangjeonggi-san (GHJGS) | Agastachis Herba 5.625 g, Perillae Folium 3.75 g, Attactylodis Rhizoma, Attactylodis Rhizoma Alba, Pinelliae Tuber, Citri Unshius Pericarpium, Arecae Pericarpium, Citri Unshius Pericarpium Immaturus, Cinnamomi Cortex, Zingiberis Rhizoma, Alpiniae Oxyphyllae Fructus, and Glycyrrhizae Radix et Rhizoma 1.875 g |
| Gwangyebujaijung-tang (GGBJIJT) | Ginseng Radix 11.25 g, Attactylodis Rhizoma Alba, Zingiberis Rhizoma (stir-bake), and Cinnamomi Cortex 7.5 g, Paeoniae Radix Alba, Citri Unshius Pericarpium, and Glycyrrhizae Radix et Rhizoma (stir-bake) 3.75 g, Aconiti Lateralis Radix Preparata 3.75 or 7.5 g |
| Hyangbujapalmul-tang (HBJPMT) | Cyperi Rhizoma, Attractylodis Rhizoma Alba, Poria Sclerotium (white), Pinelliae Tuber, Citri Unshius Pericarpium, Magnoliae Cortex, and Amomi Fructus Rotundus 3.75 g, Ginseng Radix, Glycyrrhizae Radix et Rhizoma, Aucklandiae Radix, Amomi Fructus, and Alpiniae Oxyphyllae Fructus 1.875 g, Zingiberis Rhizoma Crudus 3 slices, Zizyphi Fructus 2 pulps |
| Hyangsayangwi-tang (HSYWT) | Ginseng Radix, Attactylodis Rhizoma Alba, Paeoniae Radix Alba, Glycyrrhizae Radix et Rhizoma (stir-bake), Cyperi Rhizoma, Citri Unshius Pericarpium, Zingiberis Rhizoma, Crataegi Fructus, Amoni Fructus, and Amomi Fructus Rotundus 3.75 g |
| Hyeongbangdojeok-san (HBDJS) | Rehmanniae Radix Crudus 11.25 g, Akebiae Caulis 7.5 g, Scrophulariae Radix, Trichosanthis Semen, Angelicae Decursivae Radix, Osterici Radix, Angelicae Continentalis Radix, Schizonepetae Spica, and Saposhnikoviae Radix 3.75 g |
| Hyeongbangjihwang-tang (HBJHT) | Rehmanniae Radix Preparata, Corni Fructus, Poria Sclerotium, and Alismatis Rhizoma 7.5 g, Plantaginis Semen, Osterici Radix, Angelicae Continetalis Radix, Schzonepetae Spca, and Saposhnikoviae Radix 3.75 g |
| Hyeongbangsabak-san (HBSBS) | Rehmanniae Radix Crudus 11.25 g, Poria Sclerotium 7.5 g, Gypsum Fibrosum, Anemarrhenae Rhizoma, Osterici Radix, Schizonepetae Spica, and Saposhnikoviae Radix 3.75 g |
| Jowiseungcheong-tang (JWSCT) | Coisis Semen, and Castaneae Semen 11.25 g, Raphani Semene 5.625 g, Ephedrae Herba, Platycodonis Radix, Liriopis Tuber, Schisandrae Fructure, Acori Graminei Rhizome, Polygalae Radix, Asparagi Tuber, Zizyphi semen, and Longan Arillus 3.75 g |
| Palmulgunja-tang (PMGJT) | Ginseng Radix 7.5 g, Astragali Radix, Attactylodis Thizoma Alba, Paeoniae Radix Alba, Angelicae Gigantis Radix, Cnidii Rhizoma, Citri Unshius Pericarpium, and Glycyrrhizae Radix et Rhizoma (stir-bake) 3.75 g, Zizyphi Fructurs 2 pulps |
| Taeeumjowi-tang (TEJWT) | Coisis Semen, and Castaneae Semen 11.25 g, Raphani Semen 7.5 g, Schisandrae Fructus, Liriopis Tuber, Acori Graminei Rhizoma, Platycodonis Radix, and Ephedrae Herba 3.75 g |
| Yanggyeoksanhwa-tang (YGSHT) | Rehmanniae Radix Crudus, Lonicerae Folium et Caulis, and Forsythiae Fructus 7.5 g, Gardeniae Fructus, Menthae Herba, Anemarrhenae Rhizoma, Gypsum Fibrosum, Saposhinikoviae Radix, and Schizonepetae Spica 3.75 g |
| Yeoldahanso-tang (YDHST) | Puerariae Radix 15 g, Scutellariae Radix, and Angelicae Tenuissimae Radix 7.5 g, Raphani Semen, Platycodonis Radix, Cimicifugae Rhizoma, and Angelicae Dahuricae Radix 3.75 g |
| Oversampling | Classifier Models | Top-1 Accuracy | Top-3 Accuracy | Macro-F1 | Macro-MCC # |
|---|---|---|---|---|---|
| None | Logit | 0.365 ± 0.003 | 0.674 ± 0.002 | 0.151 ± 0.002 | 0.116 ± 0.002 |
| DT | 0.347 ± 0.006 | 0.725 ± 0.004 | 0.222 ± 0.005 | 0.211 ± 0.007 | |
| MLP | 0.334 ± 0.005 | 0.691 ± 0.004 | 0.242 ± 0.006 | 0.198 ± 0.005 | |
| SVM | 0.389 ± 0.003 | 0.739 ± 0.002 | 0.237 ± 0.003 | 0.168 ± 0.006 | |
| RF | 0.388 ± 0.005 | 0.767 ± 0.003 | 0.193 ± 0.006 | 0.165 ± 0.006 | |
| CDF | 0.380 ± 0.004 | 0.786 ± 0.003 | 0.165 ± 0.003 | 0.140 ± 0.005 | |
| Random | Logit | 0.235 ± 0.002 | 0.589 ± 0.003 | 0.203 ± 0.003 | 0.161 ± 0.004 |
| Sampling | DT | 0.194 ± 0.006 | 0.693 ± 0.006 | 0.153 ± 0.005 | 0.120 ± 0.005 |
| MLP | 0.310 ± 0.005 | 0.668 ± 0.006 | 0.235 ± 0.004 | 0.191 ± 0.004 | |
| SVM | 0.304 ± 0.005 | 0.716 ± 0.003 | 0.208 ± 0.005 | 0.162 ± 0.005 | |
| RF | 0.357 ± 0.004 | 0.753 ± 0.005 | 0.221 ± 0.007 | 0.187 ± 0.008 | |
| CDF | 0.400 ± 0.002 | 0.754 ± 0.002 | 0.131 ± 0.002 | 0.125 ± 0.002 | |
| SMOTENC | Logit | 0.253 ± 0.003 | 0.617 ± 0.005 | 0.211 ± 0.004 | 0.167 ± 0.006 |
| DT | 0.265 ± 0.006 | 0.714 ± 0.005 | 0.220 ± 0.005 | 0.178 ± 0.008 | |
| MLP | 0.312 ± 0.005 | 0.683 ± 0.005 | 0.246 ± 0.005 | 0.195 ± 0.005 | |
| SVM | 0.310 ± 0.004 | 0.716 ± 0.004 | 0.237 ± 0.004 | 0.176 ± 0.006 | |
| RF | 0.349 ± 0.005 | 0.728 ± 0.006 | 0.238 ± 0.007 | 0.196 ± 0.010 | |
| CDF | 0.376 ± 0.006 | 0.773 ± 0.006 | 0.232 ± 0.008 | 0.189 ± 0.008 |
| Datasets | Classifier Models | Top-1 Accuracy | Top-3 Accuracy | Macro-F1 |
|---|---|---|---|---|
| Holdout | Logit | 0.247 ± 0.005 | 0.660 ± 0.005 | 0.227 ± 0.006 |
| test set | DT | 0.242 ± 0.013 | 0.698 ± 0.008 | 0.189 ± 0.012 |
| MLP | 0.326 ± 0.009 | 0.725 ± 0.012 | 0.270 ± 0.010 | |
| SVM | 0.263 ± 0.007 | 0.752 ± 0.008 | 0.249 ± 0.004 | |
| RF | 0.349 ± 0.008 | 0.766 ± 0.005 | 0.257 ± 0.007 | |
| CDF | 0.342 ± 0.005 | 0.785 ± 0.006 | 0.247 ± 0.008 | |
| Case study | Logit | 0.339 ± 0.023 | 0.835 ± 0.016 | 0.233 ± 0.027 |
| dataset | DT | 0.270 ± 0.037 | 0.765 ± 0.015 | 0.183 ± 0.028 |
| MLP | 0.291 ± 0.031 | 0.830 ± 0.019 | 0.278 ± 0.022 | |
| SVM | 0.287 ± 0.019 | 0.835 ± 0.017 | 0.202 ± 0.011 | |
| RF | 0.361 ± 0.022 | 0.865 ± 0.016 | 0.283 ± 0.017 | |
| CDF | 0.352 ± 0.031 | 0.896 ± 0.013 | 0.298 ± 0.025 |
| Feature Class | Weight | Feature Value | Feature Description |
|---|---|---|---|
| SC type | 0.39 | 3 (So-Yang type) | 1: Tae-yang type 2: So-eum type 3: So-yang type 4: Tae-yang type |
| Stool_frequency * | −0.02 | 2 | (frequency per day) |
| Stool_frequency † | −0.01 | 2 | (frequency per day) |
| Stool_form * | 0.01 | 4 (Mushy consistency with ragged edges) | 1: Separate hard lumps, 2: A sausage shape with cracks in the surface, 3: Soft blobs with clear-cut edges, 4: Mushy consistency with ragged edges, 5: Watery diarrhea |
| Stress † | −0.01 | 3 (Moderate) | 1: None, 2: Mild, 3: Moderate, 4: Severe, 5: Very severe |
| Fatigue * | 0.01 | 4 (Severe) | 1: None, 2: Mild, 3: Moderate, 4: Severe, 5: Very severe |
| Stool_tenesmus * | 0.01 | 3 (Moderate) | 1: None, 2: Mild, 3: Moderate, 4: Severe, 5: Very severe |
| Complexion_red † | 0.01 | 3 (Moderate) | 1: None, 2: Mild, 3: Moderate, 4: Severe, 5: Very severe |
| Digestion † | 0.01 | 4 (Poor) | 1: Very good, 2: Good, 3: Fair, 4: Poor, 5: Very poor |
| Stool_form† | 0.01 | 4 (Mushy consistency with ragged edges) | 1: Separate hard lumps, 2: A sausage shape with cracks in the surface, 3: Soft blobs with clear-cut edges, 4: Mushy consistency with ragged edges, 5: Watery diarrhea |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, W.-Y.; Lee, Y.; Lee, S.; Kim, Y.W.; Kim, J.-H. A Machine Learning Approach for Recommending Herbal Formulae with Enhanced Interpretability and Applicability. Biomolecules 2022, 12, 1604. https://doi.org/10.3390/biom12111604
Lee W-Y, Lee Y, Lee S, Kim YW, Kim J-H. A Machine Learning Approach for Recommending Herbal Formulae with Enhanced Interpretability and Applicability. Biomolecules. 2022; 12(11):1604. https://doi.org/10.3390/biom12111604
Chicago/Turabian StyleLee, Won-Yung, Youngseop Lee, Siwoo Lee, Young Woo Kim, and Ji-Hwan Kim. 2022. "A Machine Learning Approach for Recommending Herbal Formulae with Enhanced Interpretability and Applicability" Biomolecules 12, no. 11: 1604. https://doi.org/10.3390/biom12111604
APA StyleLee, W.-Y., Lee, Y., Lee, S., Kim, Y. W., & Kim, J.-H. (2022). A Machine Learning Approach for Recommending Herbal Formulae with Enhanced Interpretability and Applicability. Biomolecules, 12(11), 1604. https://doi.org/10.3390/biom12111604






