Genome-Wide RNA Sequencing of Human Trabecular Meshwork Cells Treated with TGF-β1: Relevance to Pseudoexfoliation Glaucoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Trabecular Meshwork (HTM) Culture and Characterisation
2.2. TGF-β1 Treatment and RNA Extraction
2.3. Genome-Wide Expression Profiling with RNA Sequencing
2.4. RNA-Seq Data Analysis
2.5. Functional and Pathway Enrichment Analysis
2.6. LOXL1 Genotyping
2.7. RNA-Seq Data Validation
3. Results
3.1. Descriptive Features of RNA-Seq Data
3.2. LOXL1 Genotyping
3.3. Differential Gene Expression in Normal HTM Cells following TGF-β1 Treatment
3.4. Functional Enrichment and Pathway Analysis Reveals 53 Enriched Functional Clusters
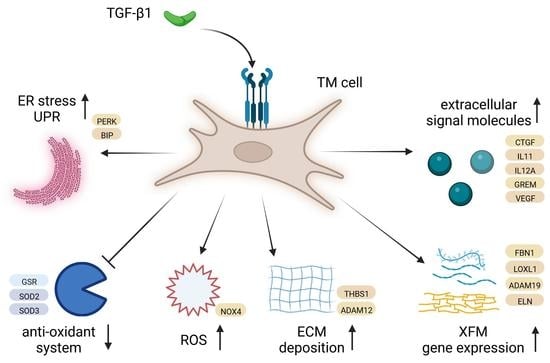
3.5. TGF-β1 Causes Differential Expression of Several Exfoliation Material Components
3.6. TGF-β1 Strongly Affects Remodelling of the Extracellular Matrix
3.7. TGF-β1 Modulates Expression of Extracellular Signal Molecules in HTM Cells
3.8. TGF-β1 Modulates Oxidative Stress and the Antioxidant System in HTM Cells
3.9. TGF-β1 Causes Upregulation of the Unfolded Protein Response in HTM Cells
3.10. TGF-β1 Increases Expression of Three XFS-Associated Genes from GWAS Studies
3.11. TGF-β1 May Modulate Retinoic Acid Signalling in HTM Cells
3.12. Validation of Differentially Expressed Genes by RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aboobakar, I.F.; Johnson, W.M.; Stamer, W.D.; Hauser, M.A.; Allingham, R.R. Major Review: Exfoliation Syndrome; Advances in Disease Genetics, Molecular Biology, and Epidemiology. Exp. Eye Res. 2017, 154, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Schlötzer-Schrehardt, U.; Khor, C.C. Pseudoexfoliation Syndrome and Glaucoma: From Genes to Disease Mechanisms. Curr. Opin. Ophthalmol. 2021, 32, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; Rao, A. Alternate Causes for Pathogenesis of Exfoliation Glaucoma, a Multifactorial Elastotic Disorder: A Literature Review. Curr. Issues Mol. Biol. 2022, 44, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Mullany, S.; Marshall, H.; Zhou, T.; Thomson, D.; Schmidt, J.M.; Qassim, A.; Knight, L.S.W.; Hollitt, G.; Berry, E.C.; Nguyen, T.; et al. RNA Sequencing of Lens Capsular Epithelium Implicates Novel Pathways in Pseudoexfoliation Syndrome. Investig. Ophthalmol. Vis. Sci. 2022, 63, 26. [Google Scholar] [CrossRef] [PubMed]
- Zenkel, M.; Schlötzer-Schrehardt, U. The Composition of Exfoliation Material and the Cells Involved in Its Production. J. Glaucoma 2014, 23, S12–S14. [Google Scholar] [CrossRef] [PubMed]
- Borrás, T. Growth Factors, Oxidative Damage, and Inflammation in Exfoliation Syndrome. J. Glaucoma 2018, 27, S54–S60. [Google Scholar] [CrossRef] [PubMed]
- Thorleifsson, G.; Magnusson, K.P.; Sulem, P.; Walters, G.B.; Gudbjartsson, D.F.; Stefansson, H.; Jonsson, T.; Jonasdottir, A.; Jonasdottir, A.; Stefansdottir, G.; et al. Common Sequence Variants in the LOXL1 Gene Confer Susceptibility to Exfoliation Glaucoma. Science (1979) 2007, 317, 1397–1400. [Google Scholar] [CrossRef]
- Pasquale, L.R.; Jiwani, A.Z.; Zehavi-Dorin, T.; Majd, A.; Rhee, D.J.; Chen, T.; Turalba, A.; Shen, L.; Brauner, S.; Grosskreutz, C.; et al. Solar Exposure and Residential Geographic History in Relation to Exfoliation Syndrome in the United States and Israel. JAMA Ophthalmol. 2014, 132, 1439–1445. [Google Scholar] [CrossRef] [Green Version]
- Tanito, M.; Kaidzu, S.; Takai, Y.; Ohira, A. Status of Systemic Oxidative Stresses in Patients with Primary Open-Angle Glaucoma and Pseudoexfoliation Syndrome. PLoS ONE 2012, 7, e49680. [Google Scholar] [CrossRef] [Green Version]
- Roedl, J.B.; Bleich, Ã.S.; Reulbach, U.; Rejdak, R.; Naumann, G.O.H.; Kruse, F.E.; Schlötzer-Schrehardt, U.; Kornhuber, J.; Jü nemann, A.G.M. Vitamin Deficiency and Hyperhomocysteinemia in Pseudoexfoliation Glaucoma. J. Neural Transm. 2007, 114, 571–575. [Google Scholar] [CrossRef]
- Kang, J.H.; Loomis, S.J.; Wiggs, J.L.; Willett, W.C.; Pasquale, L.R. A Prospective Study of Folate, Vitamin B₆, and Vitamin B₁₂ Intake in Relation to Exfoliation Glaucoma or Suspected Exfoliation Glaucoma. JAMA Ophthalmol. 2014, 132, 549–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zenkel, M.; Hoja, U.; Gießl, A.; Berner, D.; Hohberger, B.; Weller, J.M.; König, L.; Hübner, L.; Ostermann, T.A.; Gusek-Schneider, G.C.; et al. Dysregulated Retinoic Acid Signaling in the Pathogenesis of Pseudoexfoliation Syndrome. Int. J. Mol. Sci. 2022, 23, 5977. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, A.M.; Ritch, R.; Wolosin, J.M. Exfoliation Syndrome: A Disease of Autophagy and LOXL1 Proteopathy. J. Glaucoma 2018, 27, S44–S53. [Google Scholar] [CrossRef] [PubMed]
- Schlötzer-Schrehardt, U.; Zenkel, M.; Küchle, M.; Sakai, L.Y.; Naumann, G.O.H. Role of Transforming Growth Factor-Β1 and Its Latent Form Binding Protein in Pseudoexfoliation Syndrome. Exp. Eye Res. 2001, 73, 765–780. [Google Scholar] [CrossRef]
- Heldin, C.H.; Landström, M.; Moustakas, A. Mechanism of TGF-β Signaling to Growth Arrest, Apoptosis, and Epithelial-Mesenchymal Transition. Curr. Opin. Cell Biol. 2009, 21, 166–176. [Google Scholar] [CrossRef]
- Sharma, S.; Chataway, T.; Klebe, S.; Griggs, K.; Martin, S.; Chegeni, N.; Dave, A.; Zhou, T.; Ronci, M.; Voelcker, N.H.; et al. Novel Protein Constituents of Pathological Ocular Pseudoexfoliation Syndrome Deposits Identified with Mass Spectrometry. Mol. Vis. 2018, 24, 801. [Google Scholar]
- Zenkel, M.; Kruse, F.E.; Naumann, G.O.H.; Schlötzer-Schrehardt, U. Impaired Cytoprotective Mechanisms in Eyes with Pseudoexfoliation Syndrome/Glaucoma. Investig. Ophthalmol. Vis. Sci. 2007, 48, 5558–5566. [Google Scholar] [CrossRef]
- Zenkel, M.; Pöscbl, E.; von der Mark, K.; Hofmann-Rummelt, C.; Naumann, G.O.H.; Kruse, F.E.; Schlötzer-Schrebardt, U. Differential Gene Expression in Pseudoexfoliation Syndrome. Investig. Ophthalmol. Vis. Sci. 2005, 46, 3742–3752. [Google Scholar] [CrossRef] [Green Version]
- Want, A.; Gillespie, S.R.; Wang, Z.; Gordon, R.; Iomini, C.; Ritch, R.; Wolosin, J.M.; Bernstein, A.M. Autophagy and Mitochondrial Dysfunction in Tenon Fibroblasts from Exfoliation Glaucoma Patients. PLoS ONE 2016, 11, e0157404. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, M.; Sahay, P.; Rao, A. Primary Human Trabecular Meshwork Model for Pseudoexfoliation. Cells 2021, 10, 3448. [Google Scholar] [CrossRef]
- Tzavlaki, K.; Moustakas, A. TGF-β Signaling. Biomolecules 2020, 10, 487. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.M.; Gaston Pravia, K.A. Oxidative Stress and Glutathione in TGF-β-Mediated Fibrogenesis. Free Radic. Biol. Med. 2010, 48, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, J.C.; Bhattacharya, S.; Clark, A.F.; Zode, G.S. Increased Endoplasmic Reticulum Stress in Human Glaucomatous Trabecular Meshwork Cells and Tissues. Investig. Ophthalmol. Vis. Sci. 2015, 56, 3860–3868. [Google Scholar] [CrossRef] [Green Version]
- Keller, K.E.; Bhattacharya, S.K.; Borrás, T.; Brunner, T.M.; Chansangpetch, S.; Clark, A.F.; Dismuke, W.M.; Du, Y.; Elliott, M.H.; Ethier, C.R.; et al. Consensus Recommendations for Trabecular Meshwork Cell Isolation, Characterization and Culture. Exp. Eye Res. 2018, 171, 164. [Google Scholar] [CrossRef] [PubMed]
- Callaghan, B.; Lester, K.; Lane, B.; Fan, X.; Goljanek-Whysall, K.; Simpson, D.A.; Sheridan, C.; Willoughby, C.E. Genome-Wide Transcriptome Profiling of Human Trabecular Meshwork Cells Treated with TGF-Β2. Sci. Rep. 2022, 12, 9564. [Google Scholar] [CrossRef]
- Babraham Bioinformatics Fastqc. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 6 September 2022).
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Dobin, A.; Gingeras, T.R. Mapping RNA-Seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11–14. [Google Scholar] [CrossRef] [Green Version]
- GENCODE GRCh38.P13. Available online: https://www.gencodegenes.org/human/ (accessed on 6 September 2022).
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucera, M.; Isserlin, R.; Arkhangorodsky, A.; Bader, G.D. AutoAnnotate: A Cytoscape App for Summarizing Networks with Semantic Annotations. F1000Res 2016, 5, 1717. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor Package for Pathway-Based Data Integration and Visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aung, T.; Ozaki, M.; Lee, M.C.; Schlötzer-Schrehardt, U.; Thorleifsson, G.; Mizoguchi, T.; Igo, R.P.; Haripriya, A.; Williams, S.E.; Astakhov, Y.S.; et al. Genetic Association Study of Exfoliation Syndrome Identifies a Protective Rare Variant at LOXL1 and Five New Susceptibility Loci. Nat. Genet. 2017, 49, 993–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. DbSNP: The NCBI Database of Genetic Variation. Nucleic Acids Res. 2001, 29, 308. [Google Scholar] [CrossRef] [Green Version]
- Schlötzer-Schrehardt, U.; Lommatzsch, J.; Küchle, M.; Konstas, A.G.P.; Naumann, G.O.H. Matrix Metalloproteinases and Their Inhibitors in Aqueous Humor of Patients with Pseudoexfoliation Syndrome/Glaucoma and Primary Open-Angle Glaucoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 1117–1125. [Google Scholar] [CrossRef] [Green Version]
- Dan, J. Plasminogen Activator Inhibitor-1 in the Aqueous Humor of Patients With and Without Glaucoma. Arch. Ophthalmol. 2005, 123, 220. [Google Scholar] [CrossRef] [Green Version]
- Kroeger, H.; Chiang, W.C.; Felden, J.; Nguyen, A.; Lin, J.H. ER Stress and Unfolded Protein Response in Ocular Health and Disease. FEBS J. 2019, 286, 399. [Google Scholar] [CrossRef] [Green Version]
- Browne, J.G.; Ho, S.L.; Kane, R.; Oliver, N.; Clark, A.F.; O’Brien, C.J.; Crean, J.K. Connective Tissue Growth Factor Is Increased in Pseudoexfoliation Glaucoma. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3660–3666. [Google Scholar] [CrossRef] [Green Version]
- Borazan, M.; Karalezli, A.; Kucukerdonmez, C.; Bayraktar, N.; Kulaksizoglu, S.; Akman, A.; Akova, Y.A. Aqueous Humor and Plasma Levels of Vascular Endothelial Growth Factor and Nitric Oxide in Patients with Pseudoexfoliation Syndrome and Pseudoexfoliation Glaucoma. J. Glaucoma 2010, 19, 207–211. [Google Scholar] [CrossRef]
- Zenkel, M.; Schlötzer-Schrehardt, U. Expression and Regulation of LOXL1 and Elastin-Related Genes in Eyes with Exfoliation Syndrome. J Glaucoma 2014, 23, S48–S50. [Google Scholar] [CrossRef]
- Schlötzer-Schrehardt, U. Molecular Biology of Exfoliation Syndrome. J. Glaucoma 2018, 27, S32–S37. [Google Scholar] [CrossRef] [PubMed]
- Acott, T.S.; Kelley, M.J. Extracellular Matrix in the Trabecular Meshwork. Exp. Eye Res. 2008, 86, 543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prendes, M.A.; Harris, A.; Wirostko, B.M.; Gerber, A.L.; Siesky, B. The Role of Transforming Growth Factor β in Glaucoma and the Therapeutic Implications. Br. J. Ophthalmol. 2013, 97, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Ramsey, K.E.; Stephan, D.A.; Russell, P. Gene and Protein Expression Changes in Human Trabecular Meshwork Cells Treated with Transforming Growth Factor-β. Investig. Ophthalmol. Vis. Sci. 2004, 45, 4023–4034. [Google Scholar] [CrossRef] [PubMed]
- Smyth, A.; Irnaten, M.; Powell, S.K.; O’Brien, C.J. Elevated Expression of ADAM 12 and 19 in Human Glaucoma Lamina Cribrosa Cells. Investig. Ophthalmol. Vis. Sci. 2021, 62, 2770. [Google Scholar]
- De Groef, L.; van Hove, I.; Dekeyster, E.; Stalmans, I.; Moons, L. MMPs in the Trabecular Meshwork: Promising Targets for Future Glaucoma Therapies? Investig. Ophthalmol. Vis. Sci. 2013, 54, 7756–7763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeberg, M.A.T.; Farhat, Y.M.; Easa, A.; Kallenbach, J.G.; Malcolm, D.W.; Buckley, M.R.; Benoit, D.S.W.; Awad, H.A. Serpine1 Knockdown Enhances MMP Activity after Flexor Tendon Injury in Mice: Implications for Adhesions Therapy. Sci. Rep. 2018, 8, 5810. [Google Scholar] [CrossRef] [Green Version]
- Oh, D.J.; Kang, M.H.; Ooi, Y.H.; Choi, K.R.; Sage, E.H.; Rhee, D.J. Overexpression of SPARC in Human Trabecular Meshwork Increases Intraocular Pressure and Alters Extracellular Matrix. Investig. Ophthalmol. Vis. Sci. 2013, 54, 3309. [Google Scholar] [CrossRef] [Green Version]
- Wallace, D.M.; Murphy-Ullrich, J.E.; Downs, J.C.; O’Brien, C.J. The Role of Matricellular Proteins in Glaucoma. Matrix Biol. 2014, 37, 174–182. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A Single–Cell Type Transcriptomics Map of Human Tissues. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wu, W.; Zhang, W.; Lu, Y.W.; Tou, E.; Ye, J.; Gao, P.; Jourd’heuil, D.; Singer, H.A.; Wu, M.; et al. Selective Expression of TSPAN2 in Vascular Smooth Muscle Is Independently Regulated by TGF-Β1/SMAD and Myocardin/Serum Response Factor. FASEB J. 2017, 31, 2576–2591. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Yamagishi, R.; Honjo, M.; Igarashi, N.; Shimizu, S.; Aihara, M. Effects of Topical TGF-Β1, TGF-Β2, ATX, and LPA on IOP Elevation and Regulation of the Conventional Aqueous Humor Outflow Pathway. Mol. Vis. 2021, 27, 61. [Google Scholar]
- Ho, S.L.; Dogar, G.F.; Wang, J.; Crean, J.; Wu, Q.D.; Oliver, N.; Weitz, S.; Murray, A.; Cleary, P.E.; O’Brien, C. Elevated Aqueous Humour Tissue Inhibitor of Matrix Metalloproteinase-1 and Connective Tissue Growth Factor in Pseudoexfoliation Syndrome. Br. J. Ophthalmol. 2005, 89, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Lipson, K.E.; Wong, C.; Teng, Y.; Spong, S. CTGF Is a Central Mediator of Tissue Remodeling and Fibrosis and Its Inhibition Can Reverse the Process of Fibrosis. Fibrogenesis Tissue Repair. 2012, 5, S24. [Google Scholar] [CrossRef] [Green Version]
- Fuchshofer, R.; Stephan, D.A.; Russell, P.; Tamm, E.R. Gene Expression Profiling of TGFβ2- and/or BMP7-Treated Trabecular Meshwork Cells: Identification of Smad7 as a Critical Inhibitor of TGF-Β2 Signaling. Exp. Eye Res. 2009, 88, 1020. [Google Scholar] [CrossRef] [Green Version]
- Wordinger, R.J.; Lambert, W.; Agarwal, R.; Talati, M.; Clark, A.F. Human Trabecular Meshwork Cells Secrete Neurotrophins and Express Neurotrophin Receptors (Trk). Investig. Ophthalmol. Vis. Sci. 2000, 41, 3833–3841. [Google Scholar]
- Takano, S.; Uchida, K.; Itakura, M.; Iwase, D.; Aikawa, J.; Inoue, G.; Mukai, M.; Miyagi, M.; Murata, K.; Sekiguchi, H.; et al. Transforming Growth Factor-β Stimulates Nerve Growth Factor Production in Osteoarthritic Synovium. BMC Musculoskelet Disord. 2019, 20, 204. [Google Scholar] [CrossRef]
- Flügel-Koch, C.; Ohlmann, A.; Fuchshofer, R.; Welge-Lüssen, U.; Tamm, E.R. Thrombospondin-1 in the Trabecular Meshwork: Localization in Normal and Glaucomatous Eyes, and Induction by TGF-Β1 and Dexamethasone in Vitro. Exp. Eye Res. 2004, 79, 649–663. [Google Scholar] [CrossRef]
- Murphy-Ullrich, J.E.; Suto, M.J. Thrombospondin-1 Regulation of Latent TGF-β Activation: A Therapeutic Target for Fibrotic Disease. Matrix Biol. 2018, 68–69, 28. [Google Scholar] [CrossRef]
- Sethi, A.; Jain, A.; Zode, G.S.; Wordinger, R.J.; Clark, A.F. Role of TGFβ/Smad Signaling in Gremlin Induction of Human Trabecular Meshwork Extracellular Matrix Proteins. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wordinger, R.J.; Fleenor, D.L.; Hellberg, P.E.; Pang, I.H.; Tovar, T.O.; Zode, G.S.; Fuller, J.A.; Clark, A.F. Effects of TGF-Β2, BMP-4, and Gremlin in the Trabecular Meshwork: Implications for Glaucoma. Investig. Ophthalmol. Vis. Sci. 2007, 48, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Diez, R.; Lavoz, C.; Carvajal, G.; Rayego-Mateos, S.; Rodrigues Diez, R.R.; Ortiz, A.; Egido, J.; Mezzano, S.; Ruiz-Ortega, M. Gremlin Is a Downstream Profibrotic Mediator of Transforming Growth Factor-Beta in Cultured Renal Cells. Nephron. Exp. Nephrol. 2013, 122, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Gajda-Deryło, B.; Stahnke, T.; Struckmann, S.; Warsow, G.; Birke, K.; Birke, M.T.; Hohberger, B.; Rejdak, R.; Fuellen, G.; Jünemann, A.G. Comparison of Cytokine/Chemokine Levels in Aqueous Humor of Primary Open-Angle Glaucoma Patients with Positive or Negative Outcome Following Trabeculectomy. Biosci. Rep. 2019, 39, BSR20181894. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.; Dong, J.; D’Agostino, G.; Viswanathan, S.; Widjaja, A.A.; Lim, W.-W.; Ko, N.S.J.; Tan, J.; Chothani, S.P.; Huang, B.; et al. Interleukin-11 Is a Therapeutic Target in Idiopathic Pulmonary Fibrosis. Sci. Transl. Med. 2019, 11, eaaw1237. [Google Scholar] [CrossRef]
- Yashiro, R.; Nagasawa, T.; Kiji, M.; Hormdee, D.; Kobayashi, H.; Koshy, G.; Nitta, H.; Ishikawa, I. Transforming Growth Factor-β Stimulates Interleukin-11 Production by Human Periodontal Ligament and Gingival Fibroblasts. J. Clin. Periodontol. 2006, 33, 165–171. [Google Scholar] [CrossRef]
- Burgos-Blasco, B.; Vidal-Villegas, B.; Saenz-Frances, F.; Morales-Fernandez, L.; Perucho-Gonzalez, L.; Garcia-Feijoo, J.; Martinez-de-la-Casa, J.M. Tear and Aqueous Humour Cytokine Profile in Primary Open-Angle Glaucoma. Acta Ophthalmol. 2020, 98, e768–e772. [Google Scholar] [CrossRef]
- Vidal-Villegas, B.; Burgos-Blasco, B.; Santiago Alvarez, J.L.; Espino-Paisán, L.; Fernández-Vigo, J.; Andrés-Guerrero, V.; García-Feijoo, J.; Martínez-de-la-Casa, J.M. Proinflammatory Cytokine Profile Differences between Primary Open-Angle and Pseudoexfoliative Glaucoma. Ophthalmic. Res. 2021, 65, 111–120. [Google Scholar] [CrossRef]
- Mao, W.; Cameron Millar, J.; Wang, W.H.; Silverman, S.M.; Liu, Y.; Wordinger, R.J.; Rubin, J.S.; Pang, I.H.; Clark, A.F. Existence of the Canonical Wnt Signaling Pathway in the Human Trabecular Meshwork. Investig. Ophthalmol. Vis. Sci. 2012, 53, 7043. [Google Scholar] [CrossRef]
- Newman, D.R.; Sills, W.S.; Hanrahan, K.; Ziegler, A.; Tidd, K.M.G.; Cook, E.; Sannes, P.L. Expression of WNT5A in Idiopathic Pulmonary Fibrosis and Its Control by TGF-β and WNT7B in Human Lung Fibroblasts. J. Histochem. Cytochem. 2016, 64, 99. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yamagata, K.; Nishita, M.; Endo, M.; Arfian, N.; Rikitake, Y.; Emoto, N.; Hirata, K.I.; Tanaka, Y.; Minami, Y. Activation of Wnt5a-Ror2 Signaling Associated with Epithelial-to-Mesenchymal Transition of Tubular Epithelial Cells during Renal Fibrosis. Genes Cells 2013, 18, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Schlötzer-Schrehardt, U.; Ritch, R.; Call, M.; Chu, F.B.; Dong, F.; Rice, T.; Zhang, J.; Kao, W.W.-Y. Transient Expression of Wnt5a Elicits Ocular Features of Pseudoexfoliation Syndrome in Mice. PLoS ONE 2019, 14, e0212569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, I.H.; Millar, J.C.; Clark, A.F. Elevation of Intraocular Pressure in Rodents Using Viral Vectors Targeting The Trabecular Meshwork. Exp. Eye Res. 2015, 141, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Dursun, A.; Ozec, A.V.; Dogan, O.; Dursun, F.G.; Toker, M.I.; Topalkara, A.; Arici, M.K.; Erdogan, H. Evaluation of Choroidal Thickness in Patients with Pseudoexfoliation Syndrome and Pseudoexfoliation Glaucoma. J. Ophthalmol. 2016, 2016, 3545180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jobling, M.F.; Mott, J.D.; Finnegan, M.T.; Jurukovski, V.; Erickson, A.C.; Walian, P.J.; Taylor, S.E.; Ledbetter, S.; Lawrence, C.M.; Rifkin, D.B.; et al. Isoform-Specific Activation of Latent Transforming Growth Factor β (LTGF-β) by Reactive Oxygen Species. Radiat. Res. 2006, 166, 839–848. [Google Scholar] [CrossRef]
- Liu, R.M.; Desai, L.P. Reciprocal Regulation of TGF-β and Reactive Oxygen Species: A Perverse Cycle for Fibrosis. Redox Biol. 2015, 6, 565–577. [Google Scholar] [CrossRef] [Green Version]
- Michaeloudes, C.; Sukkar, M.B.; Khorasani, N.M.; Bhavsar, P.K.; Chung, K.F. TGF-β Regulates Nox4, MnSOD and Catalase Expression, and IL-6 Release in Airway Smooth Muscle Cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2011, 300, L295. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, S.M.; Lerner, S.F.; Brunzini, R.; Evelson, P.A.; Llesuy, S.F. Antioxidant Status in the Aqueous Humour of Patients with Glaucoma Associated with Exfoliation Syndrome. Eye 2008, 23, 1691–1697. [Google Scholar] [CrossRef] [Green Version]
- Takayanagi, Y.; Takai, Y.; Kaidzu, S.; Tanito, M. Evaluation of Redox Profiles of the Serum and Aqueous Humor in Patients with Primary Open-Angle Glaucoma and Exfoliation Glaucoma. Antioxidants 2020, 9, 1305. [Google Scholar] [CrossRef]
- Ryoo, I.-G.; Ha, H.; Kwak, M.-K. Inhibitory Role of the KEAP1-NRF2 Pathway in TGFβ1-Stimulated Renal Epithelial Transition to Fibroblastic Cells: A Modulatory Effect on SMAD Signaling. PLoS ONE 2014, 9, e93265. [Google Scholar] [CrossRef] [PubMed]
- Arsalane, K.; Dubois, C.M.; Muanza, T.; Bégin, R.; Boudreau, F.; Asselin, C.; Cantin, A.M. Transforming Growth Factor-Β1 Is a Potent Inhibitor of Glutathione Synthesis in the Lung Epithelial Cell Line A549: Transcriptional Effect on the GSH Rate-Limiting Enzyme γ-Glutamylcysteine Synthetase. Am. J. Respir. Cell Mol. Biol. 1997, 17, 599–607. [Google Scholar] [CrossRef]
- Péterfi, Z.; Donkó, Á.; Orient, A.; Sum, A.; Prókai, Á.; Molnár, B.; Veréb, Z.; Rajnavölgyi, É.; Kovács, K.J.; Müller, V.; et al. Peroxidasin Is Secreted and Incorporated into the Extracellular Matrix of Myofibroblasts and Fibrotic Kidney. Am. J. Pathol. 2009, 175, 725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanmer, K.L.; Mavri-Damelin, D. Peroxidasin Is a Novel Target of the Redox-Sensitive Transcription Factor Nrf2. Gene 2018, 674, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Micheal, S.; Siddiqui, S.N.; Zafar, S.N.; Iqbal, A.; Khan, M.I.; den Hollander, A.I. Identification of Novel Variants in LTBP2 and PXDN Using Whole-Exome Sequencing in Developmental and Congenital Glaucoma. PLoS ONE 2016, 11, e0159259. [Google Scholar] [CrossRef] [Green Version]
- Brown, K.D.; Shah, M.H.; Liu, G.S.; Chan, E.C.; Crowston, J.G.; Peshavariya, H.M. Transforming Growth Factor Β1–Induced NADPH Oxidase-4 Expression and Fibrotic Response in Conjunctival Fibroblasts. Investig. Ophthalmol. Vis. Sci. 2017, 58, 3011–3017. [Google Scholar] [CrossRef] [Green Version]
- Rao, V.R.; Stubbs, E.B. TGF-Β2 Promotes Oxidative Stress in Human Trabecular Meshwork Cells by Selectively Enhancing NADPH Oxidase 4 Expression. Investig. Ophthalmol. Vis. Sci. 2021, 62, 4. [Google Scholar] [CrossRef]
- Shah, M.H.; Chan, E.C.; van Bergen, N.J.; Pandav, S.S.; Ng, S.; Crowston, J.G.; Peshavariya, H.M. Nox4 Facilitates TGFβ1-Induced Fibrotic Response in Human Tenon’s Fibroblasts and Promotes Wound Collagen Accumulation in Murine Model of Glaucoma Filtration Surgery. Antioxidants 2020, 9, 1126. [Google Scholar] [CrossRef]
- Kropski, J.A.; Blackwell, T.S. Endoplasmic Reticulum Stress in the Pathogenesis of Fibrotic Disease. J. Clin. Investig. 2018, 128, 64–73. [Google Scholar] [CrossRef] [Green Version]
- Maddineni, P.; Kasetti, R.B.; Kodati, B.; Yacoub, S.; Zode, G.S. Sodium 4-Phenylbutyrate Reduces Ocular Hypertension by Degrading Extracellular Matrix Deposition via Activation of MMP9. Int. J. Mol. Sci. 2021, 22, 10095. [Google Scholar] [CrossRef]
- Zode, G.S.; Bugge, K.E.; Mohan, K.; Grozdanic, S.D.; Peters, J.C.; Koehn, D.R.; Anderson, M.G.; Kardon, R.H.; Stone, E.M.; Sheffield, V.C. Topical Ocular Sodium 4-Phenylbutyrate Rescues Glaucoma in a Myocilin Mouse Model of Primary Open-Angle Glaucoma. Investig. Ophthalmol. Vis. Sci. 2012, 53, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Osakue, D.; Yang, E.; Zhou, Y.; Gong, H.; Xia, X.; Du, Y. Endoplasmic Reticulum Stress Response of Trabecular Meshwork Stem Cells and Trabecular Meshwork Cells and Protective Effects of Activated PERK Pathway. Investig. Ophthalmol. Vis. Sci. 2019, 60, 265–273. [Google Scholar] [CrossRef] [Green Version]
- Zenkel, M.; Krysta, A.; Pasutto, F.; Juenemann, A.; Kruse, F.E.; Schlötzer-Schrehardt, U. Regulation of Lysyl Oxidase-like 1 (LOXL1) and Elastin-Related Genes by Pathogenic Factors Associated with Pseudoexfoliation Syndrome. Investig. Ophthalmol. Vis. Sci. 2011, 52, 8488–8495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greene, A.G.; Eivers, S.B.; McDonnell, F.; Dervan, E.W.J.; O’Brien, C.J.; Wallace, D.M. Differential Lysyl Oxidase like 1 Expression in Pseudoexfoliation Glaucoma Is Orchestrated via DNA Methylation. Exp. Eye Res. 2020, 201, 108349. [Google Scholar] [CrossRef] [PubMed]
- Schlötzer-Schrehardt, U.; Pasutto, F.; Sommer, P.; Hornstra, I.; Kruse, F.E.; Naumann, G.O.H.; Reis, A.; Zenkel, M. Genotype-Correlated Expression of Lysyl Oxidase-Like 1 in Ocular Tissues of Patients with Pseudoexfoliation Syndrome/Glaucoma and Normal Patients. Am. J. Pathol. 2008, 173, 1724. [Google Scholar] [CrossRef] [Green Version]
- Block, C.J.; Mitchell, A.V.; Wu, L.; Glassbrook, J.; Craig, D.; Chen, W.; Dyson, G.; DeGracia, D.; Polin, L.; Ratnam, M.; et al. RNA Binding Protein RBMS3 Is a Common EMT Effector That Modulates Triple-Negative Breast Cancer Progression via Stabilizing PRRX1 MRNA. Oncogene 2021, 40, 6430. [Google Scholar] [CrossRef]
- Fritz, D.; Stefanovic, B. RNA Binding Protein RBMS3 Is Expressed in Activated Hepatic Stellate Cells and Liver Fibrosis and Increases Expression of Transcription Factor Prx1. J. Mol. Biol. 2007, 371, 585. [Google Scholar] [CrossRef] [Green Version]
- Jayasena, C.S.; Bronner, M.E. Rbms3 Functions in Craniofacial Development by Posttranscriptionally Modulating TGF-β Signaling. J. Cell Biol. 2012, 199, 453–466. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Xi, P.-W.; Li, X.-X.; Sun, X.; Zhou, W.-B.; Xia, T.-S.; Shi, L.; Hu, Y.; Ding, Q.; Wei, J.-F. The RNA Binding Protein RBMS3 Inhibits the Metastasis of Breast Cancer by Regulating Twist1 Expression. J. Exp. Clin. Cancer Res. 2019, 38, 105. [Google Scholar] [CrossRef] [Green Version]
- Rosenthal, R.; Fromm, M. Endothelin Antagonism as an Active Principle for Glaucoma Therapy. Br. J. Pharmacol. 2011, 162, 806–816. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R.; Halder, G. The Two Faces of Hippo: Targeting the Hippo Pathway for Regenerative Medicine and Cancer Treatment. Nat. Rev. Drug Discov. 2014, 13, 63–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo Park, H.; Guan, K.-L. The Regulation of Hippo Pathway and Implications for Anticancer Drug Development The Hippo Pathway: A New Link from the Outside to Inside. Trends Pharmacol. Sci. 2013, 34, 581–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, R.; Irnaten, M.; Hopkins, A.; O’Callaghan, J.; Stamer, W.D.; Clark, A.F.; Wallace, D.; O’Brien, C.J. Matrix Mechanotransduction via Yes-Associated Protein in Human Lamina Cribrosa Cells in Glaucoma. Investig. Ophthalmol. Vis. Sci. 2022, 63, 16. [Google Scholar] [CrossRef] [PubMed]
- Hurley, D.J.; Normile, C.; Irnaten, M.; O’brien, C. The Intertwined Roles of Oxidative Stress and Endoplasmic Reticulum Stress in Glaucoma. Antioxidants 2022, 11, 886. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.-S.; Peterson, C.; Jun, A.; Foster, J. Targeting the Integrated Stress Response in Ophthalmology. Curr. Eye Res. 2021, 46, 1075–1088. [Google Scholar] [CrossRef]
- Kasetti, R.B.; Patel, P.D.; Maddineni, P.; Patil, S.; Kiehlbauch, C.; Millar, J.C.; Searby, C.C.; Raghunathan, V.K.; Sheffield, V.C.; Zode, G.S. ATF4 Leads to Glaucoma by Promoting Protein Synthesis and ER Client Protein Load. Nat. Commun. 2020, 11, 5594. [Google Scholar] [CrossRef]
- Kasetti, R.B.; Maddineni, P.; Kiehlbauch, C.; Patil, S.; Searby, C.C.; Levine, B.; Sheffield, V.C.; Zode, G.S. Autophagy Stimulation Reduces Ocular Hypertension in a Murine Glaucoma Model via Autophagic Degradation of Mutant Myocilin. JCI Insight 2021, 6, e143359. [Google Scholar] [CrossRef]
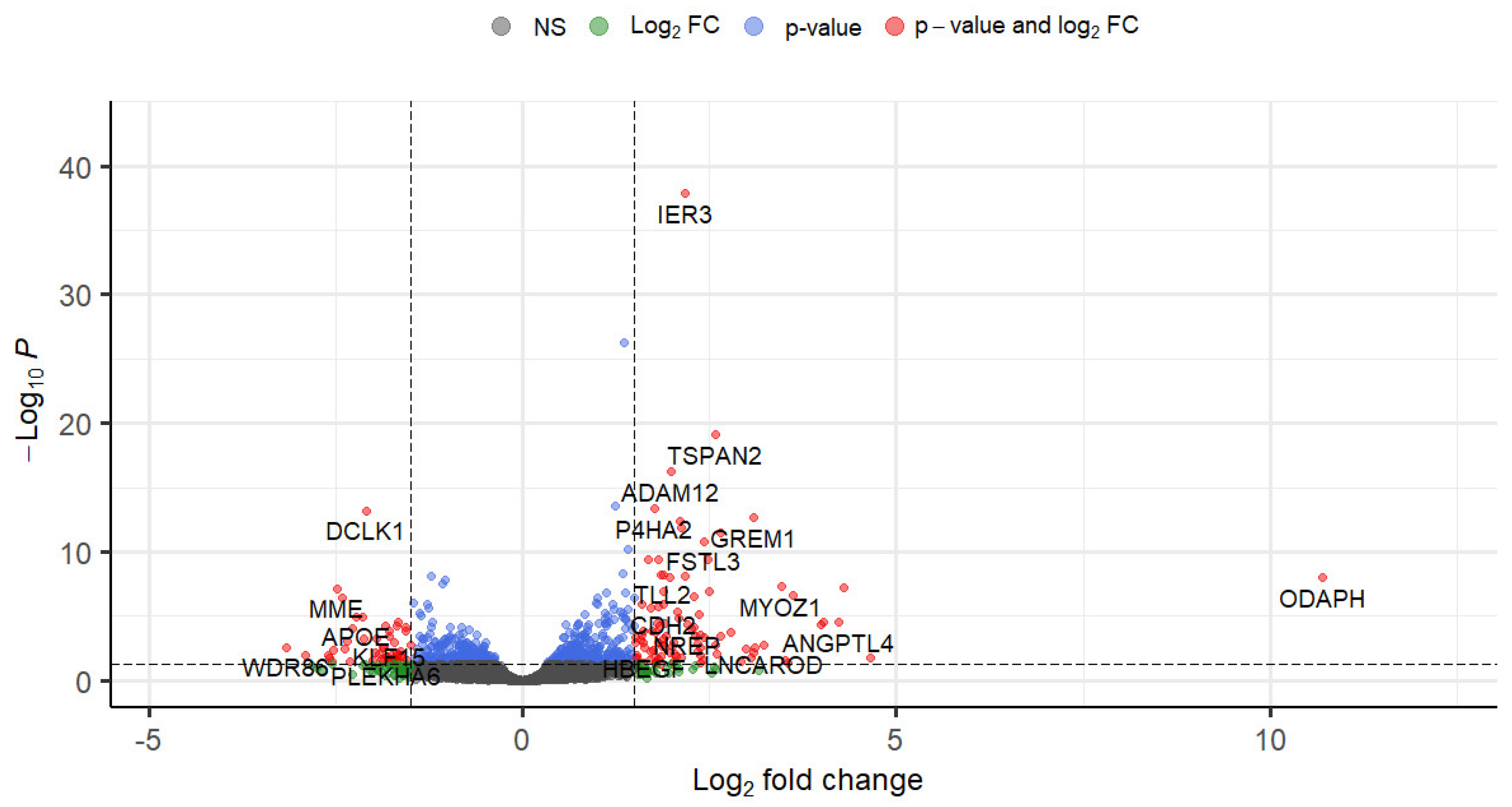
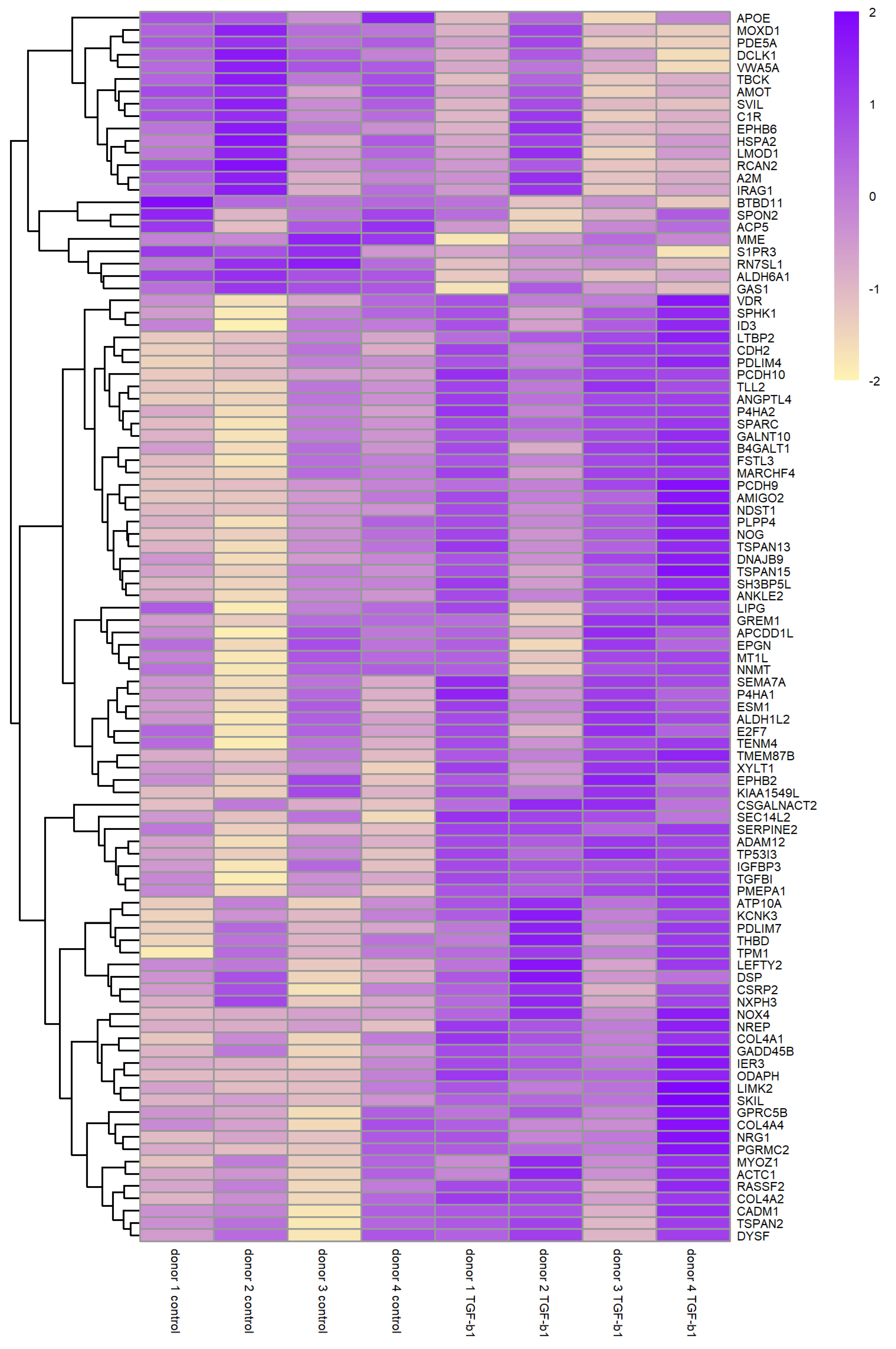
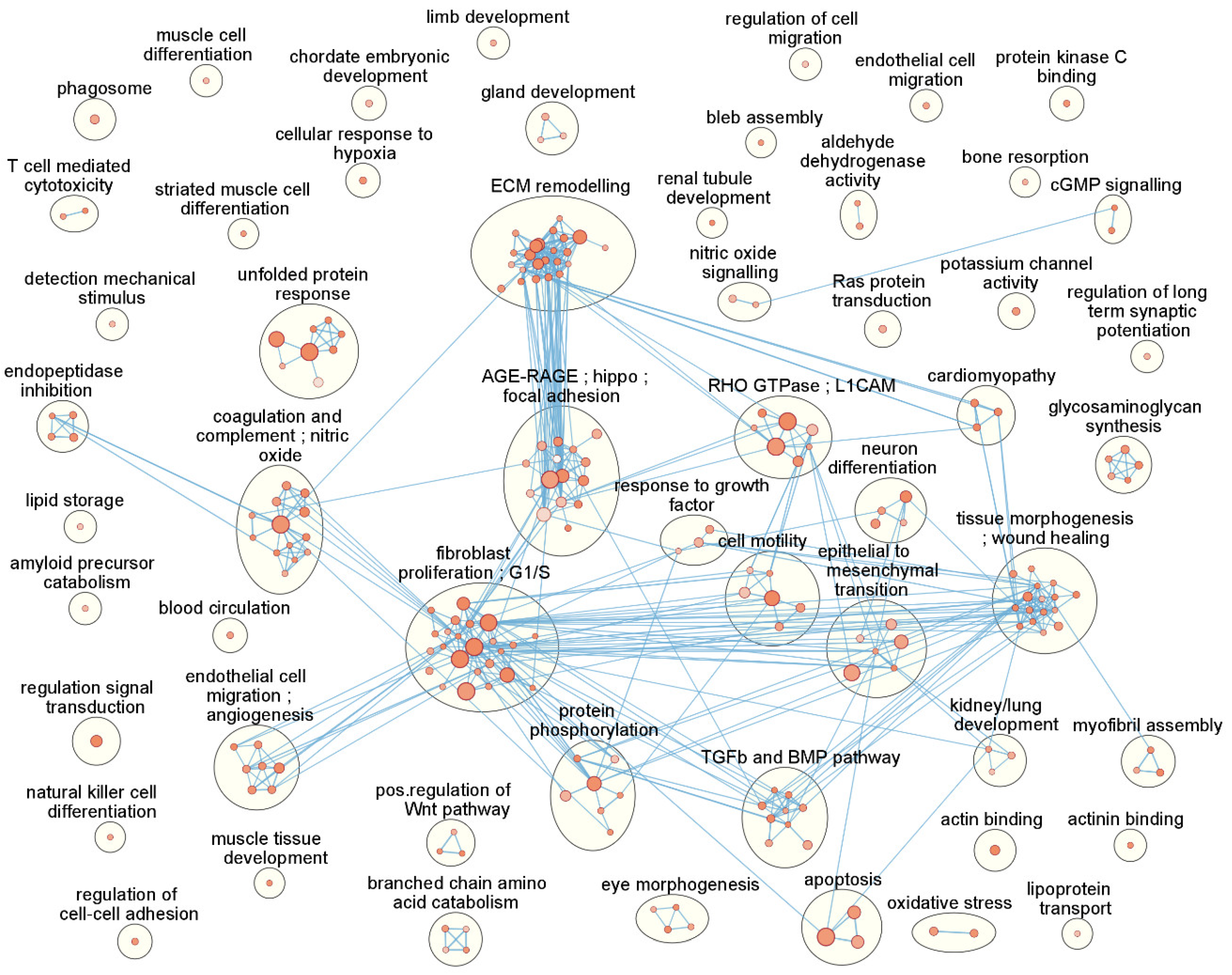




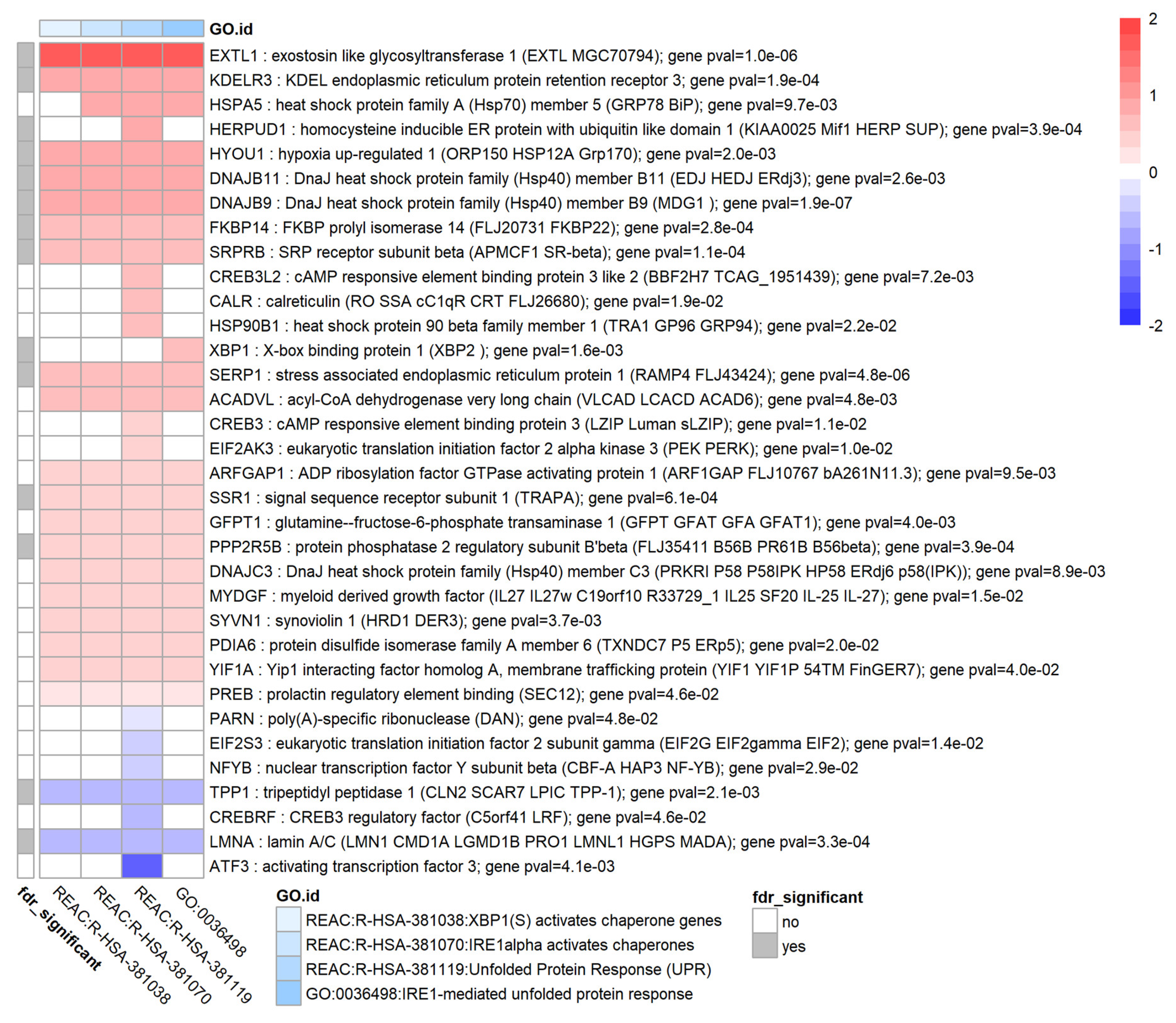

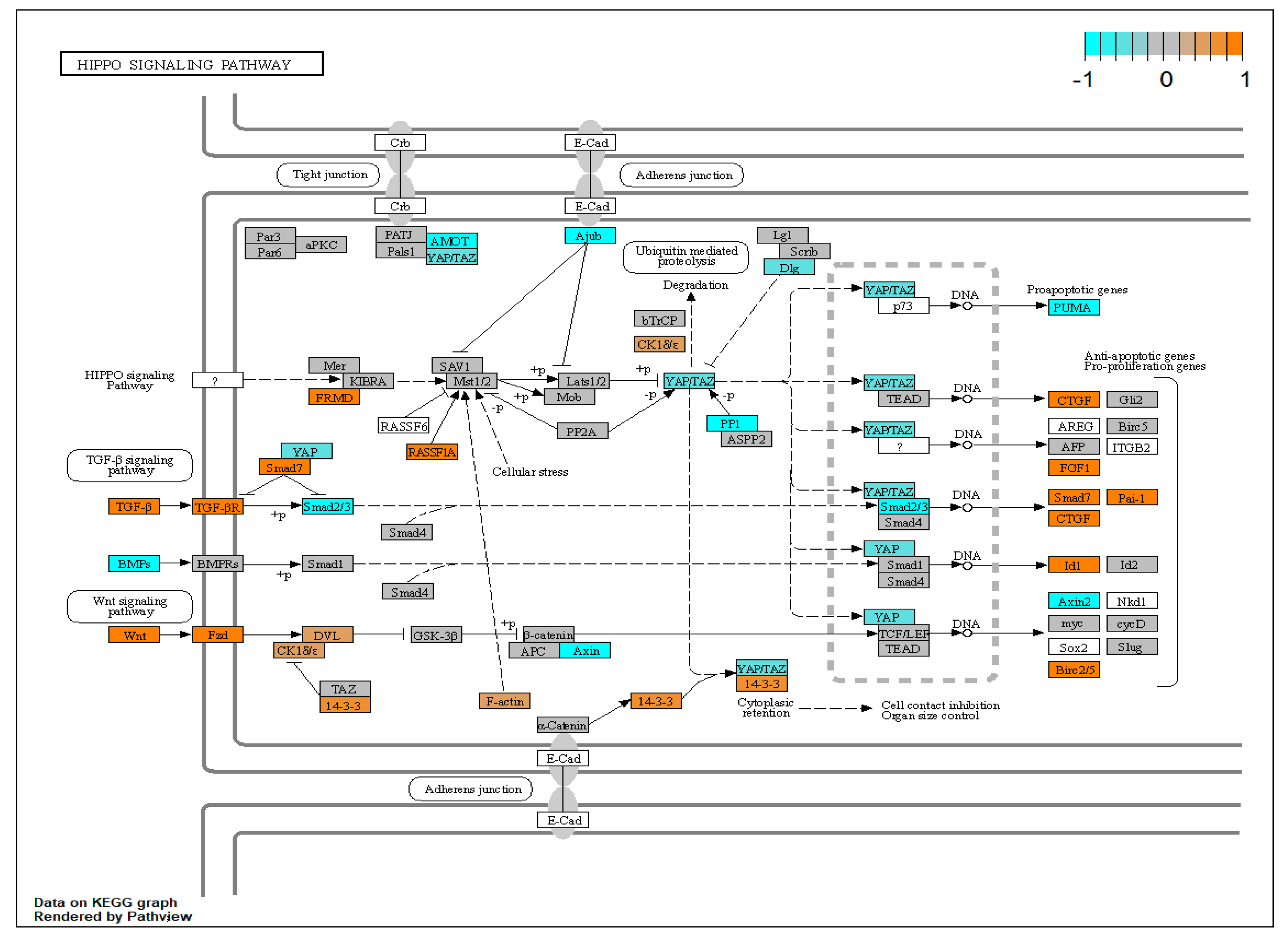

| Gene Symbol | Gene Name | log2FC | p-Value | FDR |
|---|---|---|---|---|
| IER3 | immediate early response 3 | 2.17 | 1.05 × 10−42 | 1.36 × 10−38 |
| DSP | desmoplakin | 1.36 | 7.75 × 10−31 | 4.99 × 10−27 |
| TSPAN2 | tetraspanin 2 | 2.59 | 1.90 × 10−23 | 8.16 × 10−20 |
| ADAM12 | ADAM metallopeptidase domain 12 | 1.98 | 1.80 × 10−20 | 5.80 × 10−17 |
| TP53I3 | tumor protein p53 inducible protein 3 | 1.24 | 1.14 × 10−17 | 2.94 × 10−14 |
| P4HA2 | prolyl 4-hydroxylase subunit alpha 2 | 1.76 | 2.14 × 10−17 | 4.59 × 10−14 |
| DCLK1 (DCAMKL1) | doublecortin like kinase 1 | –2.10 | 3.47 × 10−17 | 6.39 × 10−14 |
| GREM1 (CKTSF1B1) | gremlin 1, DAN family BMP antagonist | 3.08 | 1.42 × 10−16 | 2.29 × 10−13 |
| PCDH10 | protocadherin 10 | 2.10 | 3.02 × 10−16 | 4.32 × 10−13 |
| ATP10A (ATP10C) | ATPase phospholipid transporting 10A | 2.13 | 1.03 × 10−15 | 1.33 × 10−12 |
| SEMA7A (SEMAL) | semaphorin 7A | 2.65 | 2.78 × 10−15 | 3.26 × 10−12 |
| FSTL3 | follistatin like 3 | 2.43 | 1.43 × 10−14 | 1.54 × 10−11 |
| SPHK1 | sphingosine kinase 1 | 1.41 | 6.41 × 10−14 | 6.35 × 10−11 |
| TGFBI | transforming growth factor beta induced | 2.48 | 4.71 × 10−13 | 3.79 × 10−10 |
| LTBP2 (LTBP3 C14orf141) | latent transforming growth factor beta binding protein 2 | 1.81 | 4.45 × 10−13 | 3.79 × 10−10 |
| CADM1 (TSLC1 IGSF4) | cell adhesion molecule 1 | 1.68 | 4.40 × 10−13 | 3.79 × 10−10 |
| PLPP4 (PPAPDC1) | phospholipid phosphatase 4 | 1.33 | 6.69 × 10−12 | 5.07 × 10−9 |
| TLL2 | tolloid like 2 | 1.88 | 7.94 × 10−12 | 5.69 × 10−9 |
| AMIGO2 | adhesion molecule with Ig like domain 2 | 1.85 | 9.79 × 10−12 | 6.64 × 10−9 |
| SERPINE2 (PI7) | serpin family E member 2 | 2.16 | 1.35 × 10−11 | 8.31 × 10−9 |
| ALDH6A1 (MMSDH) | aldehyde dehydrogenase 6 family member A1 | –1.22 | 1.33 × 10−11 | 8.31 × 10−9 |
| ODAPH (C4orf26) | odontogenesis associated phosphoprotein | 10.70 | 1.87 × 10−11 | 1.07 × 10−8 |
| NOG (SYNS1 SYM1) | noggin | 1.97 | 1.91 × 10−11 | 1.07 × 10−8 |
| RCAN2 (DSCR1L1) | regulator of calcineurin 2 | –1.04 | 2.71 × 10−11 | 1.46 × 10−8 |
| RN7SL1 (RN7SL) | RNA component of signal recognition particle 7SL1 | –1.08 | 6.59 × 10−11 | 3.39 × 10−8 |
| MYOZ1 (MYOZ) | myozenin 1 | 3.47 | 1.03 × 10−10 | 5.12 × 10−8 |
| ESM1 | endothelial cell specific molecule 1 | 4.29 | 1.21 × 10−10 | 5.76 × 10−8 |
| MME | membrane metalloendopeptidase | –2.49 | 1.53 × 10−10 | 7.03 × 10−8 |
| PMEPA1 (TMEPAI) | prostate transmembrane protein, androgen induced 1 | 2.50 | 2.85 × 10−10 | 1.22 × 10−7 |
| COL4A1 | collagen type IV alpha 1 chain | 1.89 | 2.76 × 10−10 | 1.22 × 10−7 |
| SPARC (ON) | secreted protein acidic and cysteine rich | 1.38 | 3.34 × 10−10 | 1.39 × 10−7 |
| GALNT10 | polypeptide N-acetylgalactosaminyltransferase 10 | 1.11 | 3.98 × 10−10 | 1.60 × 10−7 |
| ACTC1 (ACTC) | actin alpha cardiac muscle 1 | 3.62 | 5.83 × 10−10 | 2.28 × 10−7 |
| TSPAN13 (TM4SF13) | tetraspanin 13 | 2.29 | 7.79 × 10−10 | 2.95 × 10−7 |
| EPHB2 (DRT EPHT3) | EPH receptor B2 | 1.48 | 9.65 × 10−10 | 3.55 × 10−7 |
| LIMK2 | LIM domain kinase 2 | 1.01 | 1.03 × 10−9 | 3.68 × 10−7 |
| A2M | Alpha-2-macroglobulin | –2.41 | 1.06 × 10−9 | 3.68 × 10−7 |
| SVIL | supervillin | –1.46 | 2.75 × 10−9 | 9.33 × 10−7 |
| CSRP2 | cysteine and glycine rich protein 2 | 0.99 | 3.19 × 10−9 | 1.05 × 10−6 |
| PCDH9 | protocadherin 9 | 1.00 | 3.60 × 10−9 | 1.16 × 10−6 |
| CDH2 (NCAD) | cadherin 2 | 1.88 | 3.84 × 10−9 | 1.21 × 10−6 |
| S1PR3 (EDG3) | sphingosine-1-phosphate receptor 3 | –1.27 | 4.15 × 10−9 | 1.27 × 10−6 |
| MT1L (MT1) | metallothionein 1L, pseudogene | 1.60 | 4.25 × 10−9 | 1.27 × 10−6 |
| NNMT | nicotinamide N-methyltransferase | 1.20 | 4.51 × 10−9 | 1.32 × 10−6 |
| THBD | thrombomodulin | 1.81 | 7.09 × 10−9 | 2.03 × 10−6 |
| MOXD1 | monooxygenase DBH like 1 | –1.26 | 8.46 × 10−9 | 2.37 × 10−6 |
| TMEM87B | transmembrane protein 87B | 1.71 | 9.06 × 10−9 | 2.48 × 10−6 |
| GPRC5B | G protein-coupled receptor class C group 5 member B | 1.41 | 1.21 × 10−8 | 3.19 × 10−6 |
| EPGN | epithelial mitogen | 1.22 | 1.21 × 10−8 | 3.19 × 10−6 |
| VDR | vitamin D receptor | 1.30 | 1.80 × 10−8 | 4.64 × 10−6 |
| Enrichment Result | p Value |
|---|---|
| BP-extracellular matrix organization (GO:0030198) | 3.34 × 10−22 |
| BP-extracellular structure organization (GO:0043062) | 5.75 × 10−16 |
| BP-external encapsulating structure organization (GO:0045229) | 4.04 × 10−15 |
| Extracellular matrix organization Homo sapiens (REAC:R-HAS-1474244) | 3.11 × 10−14 |
| BP-collagen fibril organization (GO:0030199) | 1.80 × 10−11 |
| Elastic fibre formation Homo sapiens (REAC:R-HSA-1566948) | 8.02 × 10−9 |
| BP-supramolecular fiber organization (GO:0097435) | 1.95 × 10−8 |
| XBP1(S) activates chaperone genes Homo sapiens (REAC:R-HSA-381038) | 2.03 × 10−8 |
| BP-IRE1-mediated unfolded protein response (GO:0036498) | 2.03 × 10−8 |
| Collagen formation Homo sapiens (REAC:R-HSA-1474290) | 2.85 × 10−8 |
| IRE1alpha activates chaperones Homo sapiens (REAC:R-HSA-381070) | 3.29 × 10−8 |
| MF-receptor ligand activity (GO:0048018) | 9.49 × 10−8 |
| BP-negative regulation of pathway-restricted SMAD protein phosphor. (GO:0060394) | 1.26 × 10−7 |
| Collagen biosynthesis and modifying enzymes Homo sapiens (REAC:R-HSA-1650814) | 1.84 × 10−7 |
| BP-regulation of cell population proliferation (GO:0042127) | 4.90 × 10−7 |
| Unfolded Protein Response (UPR) Homo sapiens (REAC:R-HSA-381119) | 1.33 × 10−6 |
| BP-ventricular cardiac muscle tissue morphogenesis (GO:0055010) | 1.57 × 10−6 |
| BP-regulation of pathway-restricted SMAD protein phosphorylation (GO:0060393) | 3.90 × 10−6 |
| Dilated cardiomyopathy—Homo sapiens (human) (KEGG:hsa05414) | 5.11 × 10−6 |
| MF-growth factor activity (GO:0008083) | 8.42 × 10−6 |
| BP-negative regulation of cell population proliferation (GO:0008285) | 8.55 × 10−6 |
| BP-positive regulation of cell population proliferation (GO:0008284) | 8.70 × 10−6 |
| BP-actomyosin structure organization (GO:0031032) | 1.05 × 10−5 |
| BP-positive regulation of cell differentiation (GO:0045597) | 1.09 × 10−5 |
| Glycosaminoglycan biosynthesis—heparan sulfate/heparin—(KEGG:hsa00534) | 1.13 × 10−5 |
| Arrhythmogenic right ventricular cardiomyopathy—(KEGG:hsa05412) | 1.20 × 10−5 |
| Hypertrophic cardiomyopathy—Homo sapiens (human) (KEGG:hsa05410) | 1.23 × 10−5 |
| Assembly of collagen fibrils and other multimeric structures (REAC:R-HSA-2022090) | 1.34 × 10−5 |
| BP-heart development (GO:0007507) | 1.50 × 10−5 |
| ECM proteoglycans Homo sapiens (REAC:R-HSA-3000178) | 1.59 × 10−5 |
| Integrin cell surface interactions Homo sapiens (REAC:R-HSA-216083) | 1.66 × 10−5 |
| BP-positive regulation of non-canonical Wnt signaling pathway (GO:2000052) | 1.86 × 10−5 |
| BP-blood vessel morphogenesis (GO:0048514) | 1.87 × 10−5 |
| BP-ventricular compact myocardium morphogenesis (GO:0003223) | 1.98 × 10−5 |
| L1CAM interactions Homo sapiens (REAC:R-HSA-373760) | 2.50 × 10−5 |
| BP-regulation of angiogenesis (GO:0045765) | 2.54 × 10−5 |
| BP-positive regulation of cellular process (GO:0048522) | 3.09 × 10−5 |
| MF-vascular endothelial growth factor receptor binding (GO:0005172) | 3.10 × 10−5 |
| BP-ventricular septum morphogenesis (GO:0060412) | 3.51 × 10−5 |
| BP-regulation of cell migration (GO:0030334) | 3.60 × 10−5 |
| Cluster Name | # | Top 10 Genes Ranked by FDR |
|---|---|---|
| fibroblast proliferation; G1/S | 24 | GREM1,SEMA7A,FSTL3,SPHK1,TGFBI,SERPINE2,NOG,ESM1,EPHB2,S1PR3 |
| ECM remodelling | 23 | DSP,ADAM12,P4HA2,GREM1,SEMA7A,TGFBI,LTBP2,TLL2,MYOZ1,COL4A1 |
| morphogenesis; wound healing | 16 | DSP,NOG,MYOZ1,COL4A1,ACTC1,CDH2,DYSF,NRG1,TPM1,PDLIM7 |
| AGE-RAGE;hippo;focal adhesion | 15 | COL4A1,THBD,DYSF,KCNK3,COL4A2,NOX4,GADD45B,TPM1,LEFTY2,ID3 |
| coagulation; complement; NO | 13 | SERPINE2,MYOZ1,SPARC,EPHB2,A2M,THBD,APOE,LEFTY2,C1R,PDE5A |
| TGFb and BMP pathway | 9 | GREM1,FSTL3,NOG,PMEPA1,EPHB2,RASSF2,LEFTY2,IGFBP3,ID3,SKIL |
| unfolded protein response | 8 | FSTL3,SPHK1,TGFBI,MME,GALNT10,CDH2,VDR,APOE,SPON2,HSPA2 |
| protein phosphorylation | 7 | GREM1,SPHK1,NOG,PMEPA1,LIMK2,NNMT,RASSF2,LEFTY2,ANKLE2,IGFBP3 |
| cell migration; angiogenesis | 7 | ADAM12,GREM1,SPHK1,ESM1,SPARC,APOE,COL4A2,ANGPTL4,E2F7,AMOT |
| RHO GTPase; L1CAM | 7 | DSP,SEMA7A,COL4A1,EPHB2,LIMK2,CDH2,COL4A2,ANGPTL4,EPHB6,LEFTY2 |
| epithelial to mesenchymal trans. | 6 | DCLK1,GREM1,CADM1,ODAPH,NOG,EPHB2,CDH2,VDR,APOE,NRG1 |
| cell motility | 6 | SEMA7A,SPHK1,SERPINE2,NOG,EPHB2,CDH2,NRG1,TPM1,IGFBP3,AMOT |
| glycosaminoglycan synthesis | 5 | XYLT1,CSGALNACT2, NDST1, B4GALT1, EXTL1, HS3ST2, HS3ST3B1, HAS1 |
| branched chain amino acid catab. | 4 | ALDH6A1,ALDH3A2,PPM1K,OXCT1,HIBADH,MCCC2,ALDH7A1,ACADSB |
| endopeptidase inhibition | 4 | SERPINE2,A2M,FURIN,ATP13A2,SERPINE1,PROS1,PLAUR,NGF,AGT,RECK |
| eye morphogenesis | 4 | VEGFA,STRA6,COL5A1,FBN1,SALL2,LAMA1,COL5A2,SMOC1,FBN2,PBX1 |
| neuron differentiation | 4 | GREM1,CDH2,GPRC5B,NREP,IGFBP3,DDR2,DKK1,SCIN,PPARG,PTN |
| response to growth factor | 3 | SPHK1,LTBP2,GAS1,LEFTY2,VEGFA,FURIN,ITGB5,BMP4,TGFBR3,HAS1 |
| cardiomyopathy | 3 | DSP,ACTC1,CDH2,TPM1,SGCG,LAMA1,ITGB5,ITGA5,ITGAV,TPM2 |
| gland development | 3 | NRG1,E2F7,STRA6,TGFBR3,TGFBR1,CITED2,XBP1,PKD2,TG |
| myofibril assembly | 3 | MYOZ1,ACTC1,CSRP2,TPM1,LMOD1,EPB41L2,ITGB5,CDC42BPA,ANKRD1,MYH10 |
| pos.regulation of Wnt pathway | 3 | DKK1,WNT5B,SFRP1,PLEKHA4, RSPO3 |
| apoptosis | 3 | TP53I3, GREM1,SPHK1,RASSF2,GAS1,ANGPTL4,GADD45B,ANKLE2, IGFBP3 |
| kidney/lung development | 3 | MME,VEGFA,STRA6,FOXD1,BMP4,TGFBR1,KANK2,WNT7B,AGT,CTSH |
| aldehyde dehydrogenase activity | 2 | P4HA2,P4HA1,ALDH1L2,ALDH3A2,ALDH3B1,SAT1,PYCR1,SRM,ALDH7A1 |
| cGMP signalling | 2 | APOE,IRAG1,PDE3A,HTR2B,PDE7B,AQP1,AKAP6 |
| nitric oxide signalling | 2 | SPHK1,RCAN2,APOE,FPR1,CHRM3,HTR2B,AGT,LAT2,GUCY1A1 |
| oxidative stress | 2 | NOX4,TPM1,ALDH3B1,ATP13A2,SOD3,GPX3,PYCR1,GPX8,CPEB2,PPARGC1A |
| T cell mediated cytotoxicity | 2 | CADM1,EMP2,MICA,CTSH |
| regulation signal transduction | 1 | ESM1,TPD52L1,TGFBR1,EMP2,ERFE,ITGA5,ITGAV,MYORG,CITED2,SFRP1 |
| actinin binding | 1 | CSRP2,PDLIM4,PPARG,CSRP1,PKD2 |
| lipid storage | 1 | PPARG,ITGAV,TTC39B,ITGB3 |
| Ras protein transduction | 1 | EPHB2,EPS8,ARHGEF2,KANK2,NGF,RAPGEF1,OPHN1,DENND4C,LRRC59 |
| regulation of cell–cell adhesion | 1 | FSTL3,ADAM19,PLPP3,PIEZO1,CITED2,PLAUR,SMAD7 |
| endothelial cell migration | 1 | VEGFA,EMP2,DPP4,PLEKHG5,LOXL2,ID1 |
| bleb assembly | 1 | EMP1,EMP2,P2RX7 |
| actin binding | 1 | MYOZ1,TPM1,PDLIM7,PDLIM4,SCIN,EPS8,SPTBN5,UTRN,TPM2,CALD1 |
| striated muscle cell differentiation | 1 | NRG1,HDAC5,HDAC9 |
| lipoprotein transport | 1 | PPARG,ZDHHC17,VMP1 |
| renal tubule development | 1 | COL4A1,PKD2,MTSS1 |
| regulation of cell migration | 1 | VEGFA,CARD10,HDAC5,PTGS2,HDAC9 |
| blood circulation | 1 | ACTC1,SGCG,ELN,HTR7,SERPING1,TBC1D8,GUCY1A1 |
| protein kinase C binding | 1 | DSP,CAVIN2,HDAC5,ITGAV,PKP2,HDAC9,IRS1 |
| muscle tissue development | 1 | CSRP2,STRA6,BMP4,CSRP1 |
| potassium channel activity | 1 | KCNK3,GRIK2,KCNK6,KCNMB1,KCNT2,KCNC4,AQP1,KCNA1,PKD2,KCND1 |
| bone resorption | 1 | ACP5,RAB3D,TPP1 |
| natural killer cell differentiation | 1 | TOX,PBX1,NFIL3 |
| amyloid precursor catabolism | 1 | APOE,FLOT2,PICALM,BIN1 |
| phagosome | 1 | C1R,DYNC2H1,ITGB5,ATP6V1G1,SEC61G,ITGA5,ITGAV,ITGA2,ITGB3,THBS2 |
| limb development | 1 | GREM1,NOG,DKK1,MAP3K20,SMOC1 |
| detection of a mechanical stimulus | 1 | CDH2,PIEZO1,PKD1L2,PKD2 |
| muscle cell differentiation | 1 | DKK1,HDAC5,XBP1,FZD7 |
| long term synaptic potentiation | 1 | EPHB2,APOE,CALB2,PTN,LGMN |
| chordate embryonic development | 1 | NOG,XYLT1,VEGFA,TGFBR1,ARNT2,WNT7B,PKD2 |
| cellular response to hypoxia | 1 | VEGFA,PTGS2,LMNA,SFRP1,CPEB2,SCN2A,TBL2,AQP1,HYOU1 |
| Pathway/Gene Ontology | This Work | Reported in XFG Literature |
|---|---|---|
| exfoliation material XFM gene expression | VCAN, ELN, LTBP2, ADAM19, LDHA, FBN1, MYL6, LOXL1, MYH9 ↑ | LTBP2, APOE, IRAG1, ADAM19, LDHA, FBN1 ↑ [5], LAMA1, ELN ↑ [5], VCAN, VTN, C4B, TKT, BFSP1, LOXL1, MYL6, MYH9, C4A, S100A6, LAMB1, SASH1, HSPB1, YWHAB, PKM, ENO1, TRRAP, FN1, CLU, ADAM21, ANXA7, FBLN2, SDC3, CRYAB, HEATR1, ARHGAP42, LAMC1, COL18A1, C3, LTBP1, PRDX2, NID1, TIMP3, RASD1, EMILIN1, VIM, FTH1, ANXA1, MFAP2, SDC2, ALDH3A1 |
| APOE, LAMA1, C4B, VTN, IRAG1, C4A, TKT, BFSP1, S100A6 ↓ | ||
| XFS GWAS genes | LOLX1 ↑ POMP ↑ RBMS3 ↑ | LOXL1 ↑ and ↓, may be a temporal effect [97] POMP ↑ [4] and ↓ [36] |
| extracellular signal molecules | CCN2 (CTGF) ↑ | CCN2 ↑ [41] |
| TGF-β1 ↑ | TGF-β1 ↑ [14] | |
| THBS1 ↑, THBS2 ↓ | THBS1 ↑ in POAG [61] | |
| EDN1 ↑ | EDN1 in POAG ↑ [102] | |
| VEGFA ↑ | VEGFA ↑ [42] | |
| interleukins, complement | IL-6, IL-8 == | IL-6↑ IL-8 ↑ |
| IL11↑ IL12A ↑ | IL-11, IL-12 ↑ in POAG [69,70] | |
| TGF-β pathway regulators | GREM1,2 ↑ | GREM1,2 ↑ [64] |
| Wnt pathway | WNT5B ↑ WNT2B ↓ WNT7B ↑ DKK1 ↑ | Wnt pathway implicated [74] |
| unfolded protein response | UPR pathway ↑ KDELR3 ↑ HSPA5 (GRP78) ↑ HSP90B1 (TRA1) ↑ XBP1 ↑ EIF2AK3 ↑ ERO1A ↑ | UPR pathway implicated [40] |
| oxidative stress & antioxidant system | GPX3 ↑, GPX7 ↑, GPX8 ↑, GSR ↓ SOD2 ↓, SOD3 ↓ GSTM4 ↓ GSTM3 ↓ GSTA4 ↓ NOX4 ↑ | SOD2 ↑ [17] SOD3 ↑ [4] GST1 ↓ [18] NOX4 ? |
| retinoic acid signalling | STRA6 ↑ ALDH1A1 ↓ RORB ↓ AKR1C3 ↓ RDH5 ↓ | STRA ↓ [12] ALDH1A1↓ RORB ↓ [12] AKR1C3 ? RDH5 ? |
| ECM remodelling | TIMP1..4 == | TIMP1..4 all ↑ [38] |
| MMP2 == MMP15 ↑, MMP24 ↑ | MMP2 ↑ [38] MMP15 ? MMP24 ? | |
| ADAM12 ↑ ADAM19 ↑ | ADAM12 ↑ [47] | |
| SERPINE1 ↑, SERPINE2 ↑ | SERPINE1 ↑ [39], SERPINE2 ? | |
| SPARC ↑ | SPARC ? | |
| TSPAN2 ↑, TSPAN13 ↑, TSPAN15 ↑ | TSPAN2 ? TSPAN13 ? TSPAN15 ? | |
| LOXL1 ↑ LOXL2 ↑ | LOXL1 ↑ | |
| ECM components | COL1A1 ↑ COL4A1 ↑ COL4A2 ↑ COL4A4 ↑ COL5A2 ↑ COL14A1 ↓ | COL4A1 ↓ COL4A2 ↓ [4] (lens tissue) |
| homocysteine metabolism | not enriched | - |
| mitochondrial dysfunction | not enriched | - |
| impaired autophagy | not enriched | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roodnat, A.W.; Callaghan, B.; Doyle, C.; Henry, M.; Goljanek-Whysall, K.; Simpson, D.A.; Sheridan, C.; Atkinson, S.D.; Willoughby, C.E. Genome-Wide RNA Sequencing of Human Trabecular Meshwork Cells Treated with TGF-β1: Relevance to Pseudoexfoliation Glaucoma. Biomolecules 2022, 12, 1693. https://doi.org/10.3390/biom12111693
Roodnat AW, Callaghan B, Doyle C, Henry M, Goljanek-Whysall K, Simpson DA, Sheridan C, Atkinson SD, Willoughby CE. Genome-Wide RNA Sequencing of Human Trabecular Meshwork Cells Treated with TGF-β1: Relevance to Pseudoexfoliation Glaucoma. Biomolecules. 2022; 12(11):1693. https://doi.org/10.3390/biom12111693
Chicago/Turabian StyleRoodnat, Anton W., Breedge Callaghan, Chelsey Doyle, Megan Henry, Katarzyna Goljanek-Whysall, David A. Simpson, Carl Sheridan, Sarah D. Atkinson, and Colin E. Willoughby. 2022. "Genome-Wide RNA Sequencing of Human Trabecular Meshwork Cells Treated with TGF-β1: Relevance to Pseudoexfoliation Glaucoma" Biomolecules 12, no. 11: 1693. https://doi.org/10.3390/biom12111693
APA StyleRoodnat, A. W., Callaghan, B., Doyle, C., Henry, M., Goljanek-Whysall, K., Simpson, D. A., Sheridan, C., Atkinson, S. D., & Willoughby, C. E. (2022). Genome-Wide RNA Sequencing of Human Trabecular Meshwork Cells Treated with TGF-β1: Relevance to Pseudoexfoliation Glaucoma. Biomolecules, 12(11), 1693. https://doi.org/10.3390/biom12111693