Profiles of Metabolic Genes in Uncaria rhynchophylla and Characterization of the Critical Enzyme Involved in the Biosynthesis of Bioactive Compounds-(iso)Rhynchophylline
Abstract
1. Introduction
2. Results
2.1. Specific Indole Alkaloids in U. rhynchophylla
2.2. De Novo Transcriptome of U. rhynchophylla
2.3. Gene Expression Comparison among Tissues
2.4. Functional Annotation for U. rhynchophylla
2.5. GO and KEGG Classification
2.6. Transcription Factors
2.7. Phylogenetic Analysis
2.8. STR Activity
2.9. The Acceptance of Tryptamine and Analogues
2.10. Candidate Genes in Spiroindole Alkaloids Formation
3. Discussion
3.1. U. rhynchophylla Produces Specific Spiroindole Alkaloids
3.2. Candidate Genes Involve or Regulate Spiroindole Alkaloids Biosynthesis
3.3. N-Methyltryptamines Are Not the Natural Substrates for N-Methylstrictosidine Formation
3.4. Biosynthesis of Spiroindole Alkaloids in U. rhynchophylla
3.5. Global Transcriptome Provides a Valuable Genetic Resource for the Biosynthetic Pathway Elucidation of Indole Alkaloids
4. Materials and Methods
4.1. Plant Materials
4.2. Non-Targeted Metabolites Analysis
4.3. Transcriptome Sequencing
4.4. De novo Transcriptome Assembly and Gene Expression Comparison among Tissues
4.5. Functional Annotation for U. rhynchophylla
4.6. Phylogenetic Analysis
4.7. Plasmids Construction, Protein Purification
4.8. Enzymatic Assay and LCMS Analysis
4.9. Structural and Docking Analyses for UrSTR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heitzman, M.E.; Neto, C.C.; Winiarz, E.; Vaisberg, A.J.; Hammond, G.B. Ethnobotany, phytochemistry and pharmacology of Uncaria (Rubiaceae). Phytochemistry 2005, 66, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhou, S. Antihypertensive and neuroprotective activities of rhynchophylline: The role of rhynchophylline in neurotransmission and ion channel activity. J. Ethnopharmacol. 2010, 132, 15–27. [Google Scholar] [CrossRef]
- Xian, Y.F.; Lin, Z.X.; Zhao, M.; Mao, Q.Q.; Ip, S.P.; Che, C.T. Uncaria rhynchophylla ameliorates cognitive deficits induced by D-galactose in mice. Planta Med. 2011, 77, 1977–1983. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Ma, B.; Yang, J.Y.; Xie, Y.Y.; Wang, L.; Zhang, L.J.; Kano, Y.; Wu, C.F. Anti-inflammatory effects of rhynchophylline and isorhynchophylline in mouse N9 microglial cells and the molecular mechanism. Int. Immunopharmacol. 2009, 9, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Zhang, S.; Wang, H.; Chen, Z.; Sun, M.; Sun, L.; Gong, L.; Li, Y.; Jiang, H. Intervention of Uncaria and Its Components on Liver Lipid Metabolism in Spontaneously Hypertensive Rats. Front. Pharmacol. 2020, 11, 910. [Google Scholar] [CrossRef]
- Xian, Y.F.; Lin, Z.X.; Mao, Q.Q.; Hu, Z.; Zhao, M.; Che, C.T.; Ip, S.P. Bioassay-Guided Isolation of Neuroprotective Compounds from Uncaria rhynchophylla against Beta-Amyloid-Induced Neurotoxicity. Evid. Based Complement Alternat. Med. 2012, 2012, 802625. [Google Scholar] [CrossRef]
- Xian, Y.F.; Lin, Z.X.; Mao, Q.Q.; Ip, S.P.; Su, Z.R.; Lai, X.P. Protective effect of isorhynchophylline against beta-amyloid-induced neurotoxicity in PC12 cells. Cell Mol. Neurobiol. 2012, 32, 353–360. [Google Scholar] [CrossRef]
- Shi, J.S.; Yu, J.X.; Chen, X.P.; Xu, R.X. Pharmacological actions of Uncaria alkaloids, rhynchophylline and isorhynchophylline. Acta. Pharmacol. Sin. 2003, 24, 97–101. [Google Scholar]
- Wu, S.W.; Yang, M.Q.; Xiao, Y.L. Synthetic Biology Studies of Monoterpene Indole Alkaloids. Chin. J. Org. Chem. 2018, 38, 2243–2258. [Google Scholar] [CrossRef]
- O’Connor, S.E.; Maresh, J.J. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat. Prod. Rep. 2006, 23, 532–547. [Google Scholar] [CrossRef]
- Vera-Reyes, I.; Huerta-Heredia, A.A.; Ponce-Noyola, T.; Flores-Sanchez, I.J.; Esparza-Garcia, F.; Cerda-Garcia-Rojas, C.M.; Trejo-Tapia, G.; Ramos-Valdivia, A.C. Strictosidine-related enzymes involved in the alkaloid biosynthesis of Uncaria tomentosa root cultures grown under oxidative stress. Biotechnol. Prog. 2013, 29, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Ma, X.; Wei, S.; Qiu, D.; Wilson, I.W.; Wu, P.; Tang, Q.; Liu, L.; Dong, S.; Zu, W. De novo transcriptome sequencing and digital gene expression analysis predict biosynthetic pathway of rhynchophylline and isorhynchophylline from Uncaria rhynchophylla, a non-model plant with potent anti-alzheimer’s properties. BMC Genom. 2014, 15, 676. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.H.; Qiang, W.; Zheng, H.J.; Shangguan, L.Y.; Zhang, M.S. Transcriptome revealing the dual regulatory mechanism of ethylene on the rhynchophylline and isorhynchophylline in Uncaria rhynchophylla. J. Plant. Res. 2022, 135, 485–500. [Google Scholar] [CrossRef] [PubMed]
- Dai, G.; Shen, Q.; Zhang, Y.; Bian, X. Biosynthesis of Fungal Natural Products Involving Two Separate Pathway Crosstalk. J. Fungi 2022, 8, 320. [Google Scholar] [CrossRef]
- Kishimoto, S.; Tsunematsu, Y.; Sato, M.; Watanabe, K. Elucidation of Biosynthetic Pathways of Natural Products. Chem. Rec. 2017, 17, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; You, W.; Wu, S.; Fan, Z.; Xu, B.; Zhu, M.; Li, X.; Xiao, Y. Global transcriptome analysis of Huperzia serrata and identification of critical genes involved in the biosynthesis of huperzine A. BMC Genom. 2017, 18, 245. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Wu, S.; You, W.; Jaisi, A.; Xiao, Y. Selection of Reference Genes for Expression Analysis in Chinese Medicinal Herb Huperzia serrata. Front. Pharmacol. 2019, 10, 44. [Google Scholar] [CrossRef]
- Liang, J.H.; Wang, C.; Huo, X.K.; Tian, X.G.; Zhao, W.Y.; Wang, X.; Sun, C.P.; Ma, X.C. The genus Uncaria: A review on phytochemical metabolites and biological aspects. Fitoterapia 2020, 147, 104772. [Google Scholar] [CrossRef]
- Ndagijimana, A.; Wang, X.M.; Pan, G.X.; Zhang, F.; Feng, H.; Olaleye, O. A review on indole alkaloids isolated from Uncaria rhynchophylla and their pharmacological studies. Fitoterapia 2013, 86, 35–47. [Google Scholar] [CrossRef]
- Leclercq, J.; Angenot, L. Dolichantoside, Main Alkaloid from Stem Bark of Strychnos tricalysioides. Planta Med. 1984, 50, 457–458. [Google Scholar] [CrossRef]
- Zhu, H.; Kercmar, P.; Wu, F.; Rajendran, C.; Sun, L.; Wang, M.; Stockigt, J. Using Strictosidine Synthase to Prepare Novel Alkaloids. Curr. Med. Chem. 2015, in press. [Google Scholar] [CrossRef]
- Caputi, L.; Franke, J.; Farrow, S.C.; Chung, K.; Payne, R.M.E.; Nguyen, T.D.; Dang, T.T.; Soares Teto Carqueijeiro, I.; Koudounas, K.; Duge de Bernonville, T.; et al. Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science 2018, 360, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hansen, L.G.; Gudich, O.; Viehrig, K.; Lassen, L.M.M.; Schrubbers, L.; Adhikari, K.B.; Rubaszka, P.; Carrasquer-Alvarez, E.; Chen, L.; et al. A microbial supply chain for production of the anti-cancer drug vinblastine. Nature 2022, 609, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Rohani, E.R.; Chiba, M.; Kawaharada, M.; Asano, T.; Oshima, Y.; Mitsuda, N.; Ohme-Takagi, M.; Fukushima, A.; Rai, A.; Saito, K.; et al. An MYB transcription factor regulating specialized metabolisms in Ophiorrhiza pumila. Plant Biotechnol. 2016, 33, 1–9. [Google Scholar] [CrossRef]
- Hao, X.L.; Xie, C.H.; Ruan, Q.Y.; Zhang, X.C.; Wu, C.; Han, B.; Qian, J.; Zhou, W.; Nutzmann, H.W.; Kai, G.Y. The transcription factor OpWRKY2 positively regulates the biosynthesis of the anticancer drug camptothecin in Ophiorrhiza pumila. Hortic Res-Engl. 2021, 8, 7. [Google Scholar] [CrossRef]
- Udomsom, N.; Rai, A.; Suzuki, H.; Okuyama, J.; Imai, R.; Mori, T.; Nakabayashi, R.; Saito, K.; Yamazaki, M. Function of AP2/ERF Transcription Factors Involved in the Regulation of Specialized Metabolism in Ophiorrhiza pumila Revealed by Transcriptomics and Metabolomics. Front. Plant Sci. 2016, 7, 1861. [Google Scholar] [CrossRef]
- Patra, B.; Pattanaik, S.; Schluttenhofer, C.; Yuan, L. A network of jasmonate-responsive bHLH factors modulate monoterpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol. 2018, 217, 1566–1581. [Google Scholar] [CrossRef]
- Pu, X.; Gao, H.C.; Wang, M.J.; Zhang, J.H.; Shan, J.H.; Chen, M.H.; Zhang, L.; Wang, H.G.; Wen, A.X.; Luo, Y.G.; et al. Integrative Analysis of Elicitor-Induced Camptothecin Biosynthesis in Camptotheca acuminata Plantlets Through a Combined Omics Approach. Front. Plant Sci. 2022, 13, 851077. [Google Scholar] [CrossRef]
- Hong, B.K.; Grzech, D.; Caputi, L.; Sonawane, P.; Lopez, C.E.R.; Kamileen, M.O.; Lozada, N.J.H.; Grabe, V.; O’Connor, S.E. Biosynthesis of strychnine. Nature 2022, 607, 617. [Google Scholar] [CrossRef]
- Farrow, S.C.; Kamileen, M.O.; Caputi, L.; Bussey, K.; Mundy, J.E.A.; McAtee, R.C.; Stephenson, C.R.J.; O’Connor, S.E. Biosynthesis of an Anti-Addiction Agent from the Iboga Plant. J. Am. Chem. Soc. 2019, 141, 12979–12983. [Google Scholar] [CrossRef]
- Farrow, S.C.; Kamileen, M.O.; Meades, J.; Ameyaw, B.; Xiao, Y.; O’Connor, S.E. Cytochrome P450 and O-methyltransferase catalyze the final steps in the biosynthesis of the anti-addictive alkaloid ibogaine from Tabernanthe iboga. J. Biol. Chem. 2018, 293, 13821–13833. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.Q.; Wang, Q.; Liu, Y.N.; Hao, X.L.; Wang, C.; Liang, Y.C.; Chen, J.B.; Xiao, Y.L.; Kai, G.Y. Divergent camptothecin biosynthetic pathway in Ophiorrhiza pumila. BMC Biol. 2021, 19, 122. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nature Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]

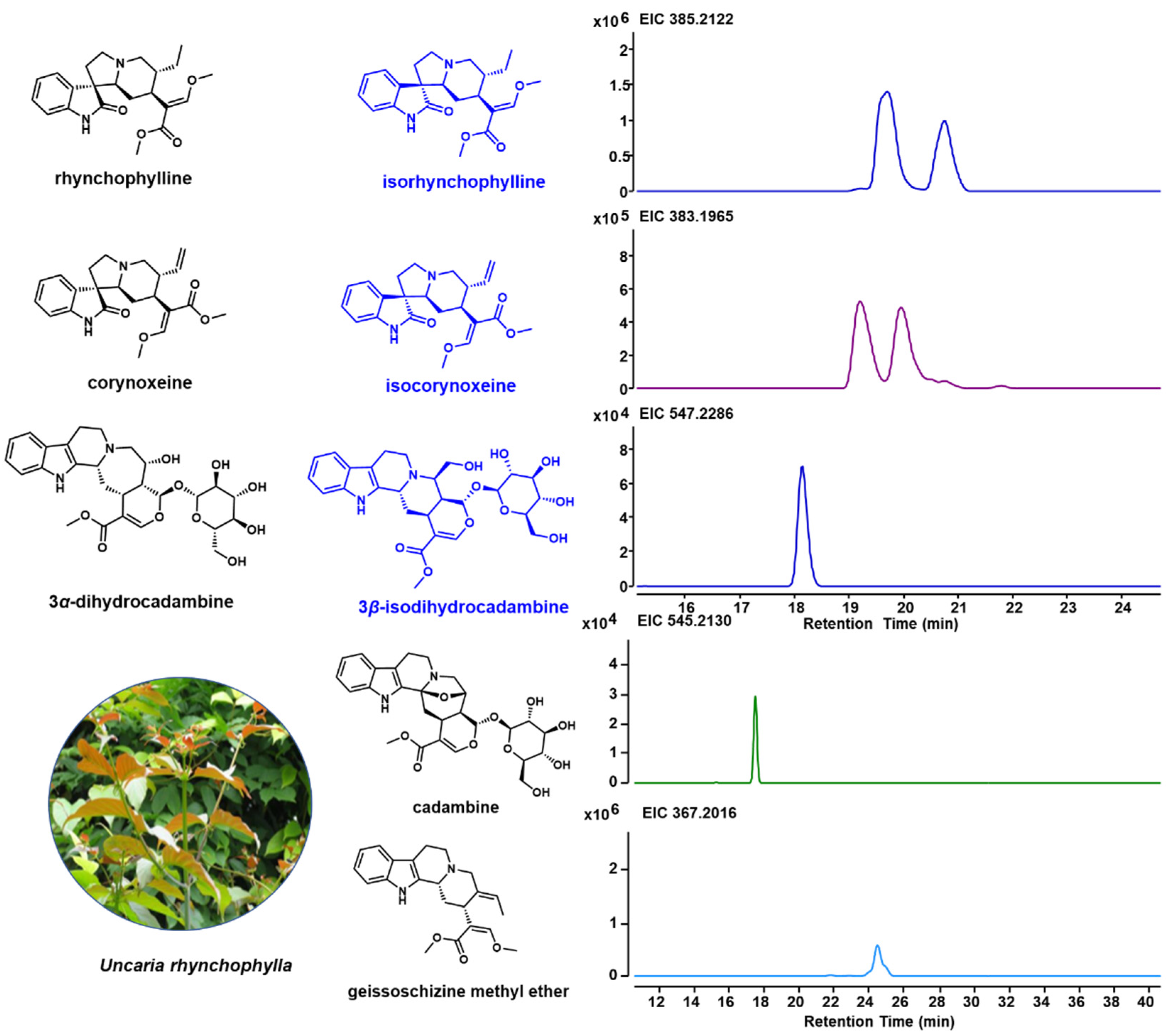
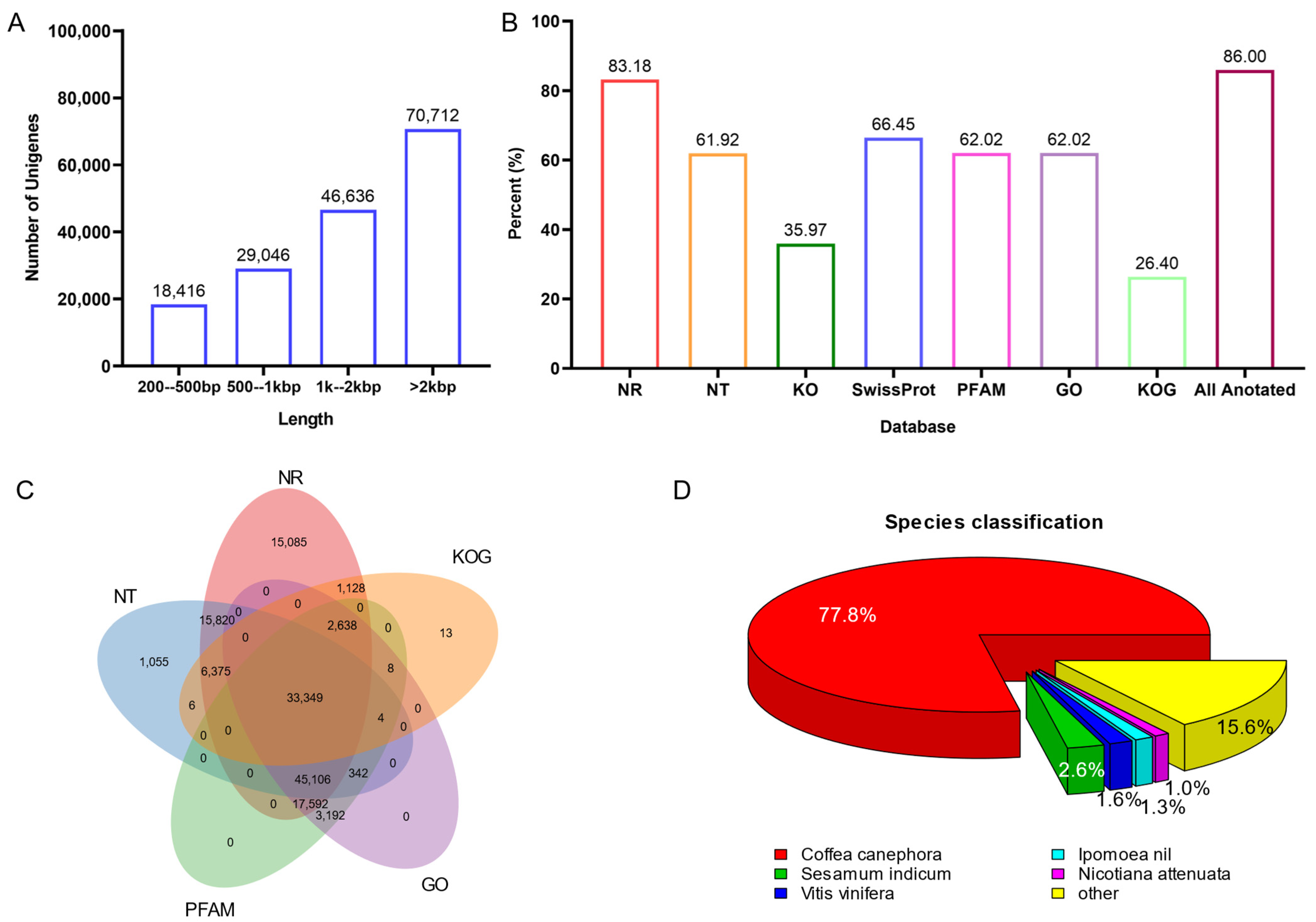
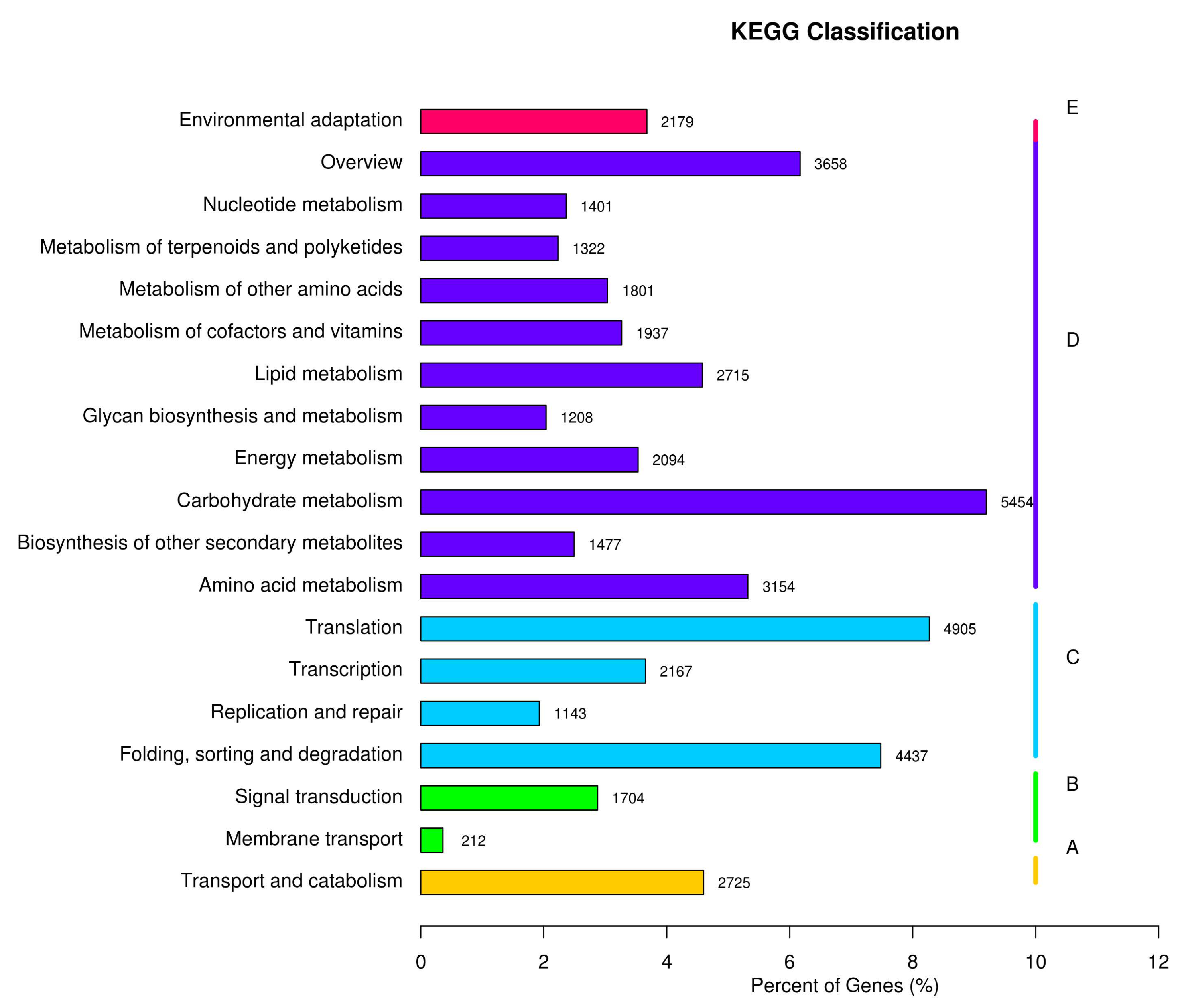





| Items | RAW READS | Clean Reads | Clean Bases |
|---|---|---|---|
| Leaf | 71,187,462 | 69,670,550 | 10.45 G |
| Stem Bark | 56,685,480 | 55,496,154 | 8.32 G |
| Root | 55,949,240 | 54,721,630 | 8.21 G |
| Bud | 57,681,414 | 56,408,860 | 8.46 G |
| Total data | 241,503,596 | 236,297,194 | 35.44 G |
| Unigenes ≥ 500 bp | 311,204 | ||
| N50 (bp) | 2887 | ||
| Number of Unigenes | Percentage (%) | |
|---|---|---|
| Annotated in NR | 137,093 | 83.18 |
| Annotated in NT | 102,057 | 61.92 |
| Annotated in KO | 59,289 | 35.97 |
| Annotated in SwissProt | 109,519 | 66.45 |
| Annotated in PFAM | 102,231 | 62.02 |
| Annotated in GO | 102,231 | 62.02 |
| Annotated in KOG | 43,521 | 26.4 |
| Annotated in Total | 141,747 | 86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Yao, B.; Lin, R. Profiles of Metabolic Genes in Uncaria rhynchophylla and Characterization of the Critical Enzyme Involved in the Biosynthesis of Bioactive Compounds-(iso)Rhynchophylline. Biomolecules 2022, 12, 1790. https://doi.org/10.3390/biom12121790
Yang M, Yao B, Lin R. Profiles of Metabolic Genes in Uncaria rhynchophylla and Characterization of the Critical Enzyme Involved in the Biosynthesis of Bioactive Compounds-(iso)Rhynchophylline. Biomolecules. 2022; 12(12):1790. https://doi.org/10.3390/biom12121790
Chicago/Turabian StyleYang, Mengquan, Bowen Yao, and Rongmei Lin. 2022. "Profiles of Metabolic Genes in Uncaria rhynchophylla and Characterization of the Critical Enzyme Involved in the Biosynthesis of Bioactive Compounds-(iso)Rhynchophylline" Biomolecules 12, no. 12: 1790. https://doi.org/10.3390/biom12121790
APA StyleYang, M., Yao, B., & Lin, R. (2022). Profiles of Metabolic Genes in Uncaria rhynchophylla and Characterization of the Critical Enzyme Involved in the Biosynthesis of Bioactive Compounds-(iso)Rhynchophylline. Biomolecules, 12(12), 1790. https://doi.org/10.3390/biom12121790







