The Effects of Synthetic SREBP-1 and PPAR-γ Decoy Oligodeoxynucleotide on Acne-like Disease In Vivo and In Vitro via Lipogenic Regulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Bacteria
2.2. Synthesis of Decoy ODN
2.3. Animal Models and Transfection of Decoy ODN
2.4. Cell Culture and Treatment
2.5. Histological Analysis
2.6. Immunohistochemistry
2.7. Immunofluorescence Staining
2.8. Western Blot Analysis
2.9. Enzyme-Linked Immunosorbent Assay (ELISA)
2.10. Electrophoretic Mobility Shift Assay (EMSA)
2.11. Flow Cytometric Analysis
2.12. Nile Red Staining
2.13. Oil Red O Staining
2.14. Data and Statistical Analysis
3. Results
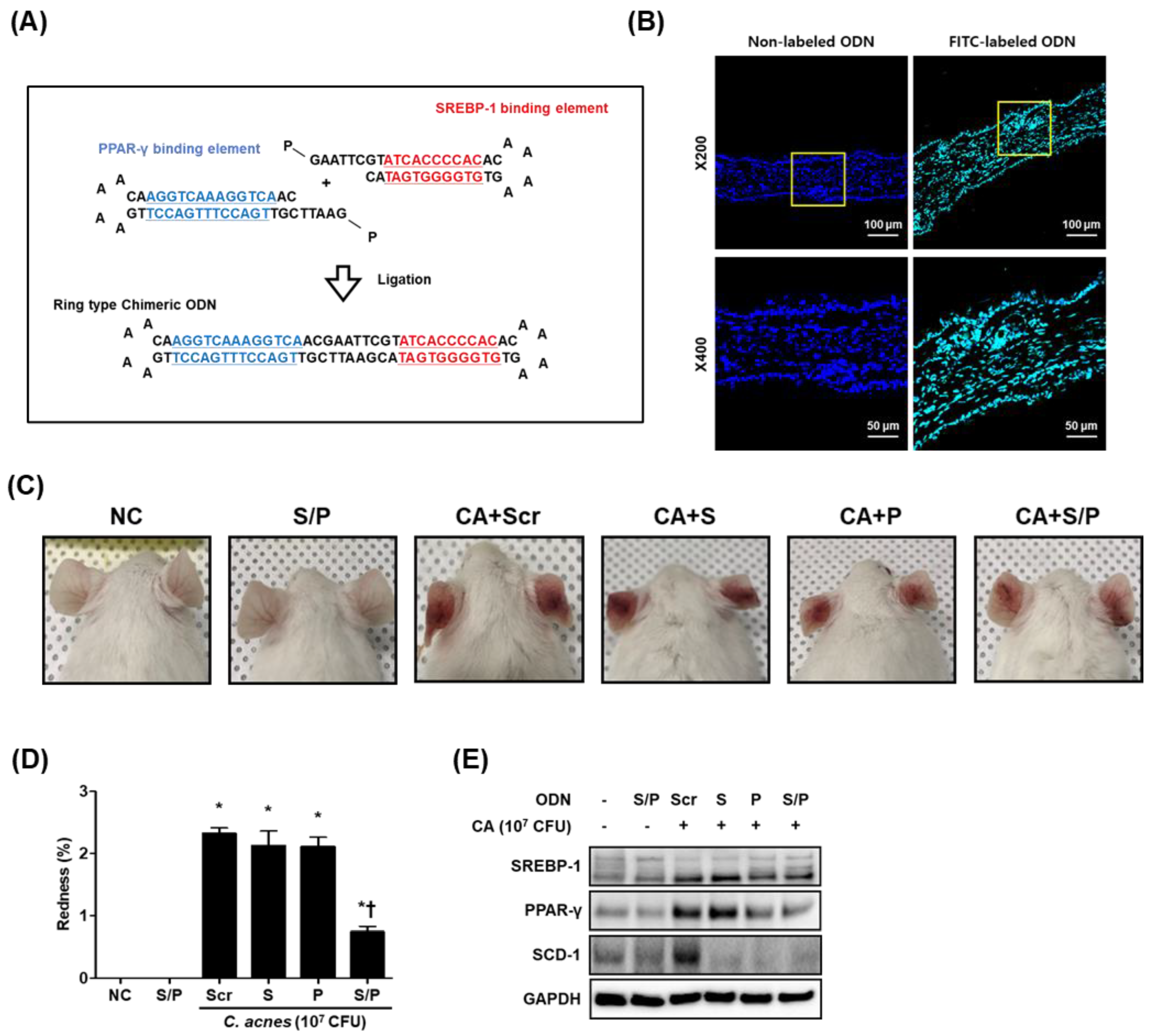
3.1. Construction of the Decoy ODNs
3.2. The SREBP/PPAR Chimeric Decoy ODN Alleviated C. acnes-Mediated Skin Lesions in the Mouse Model
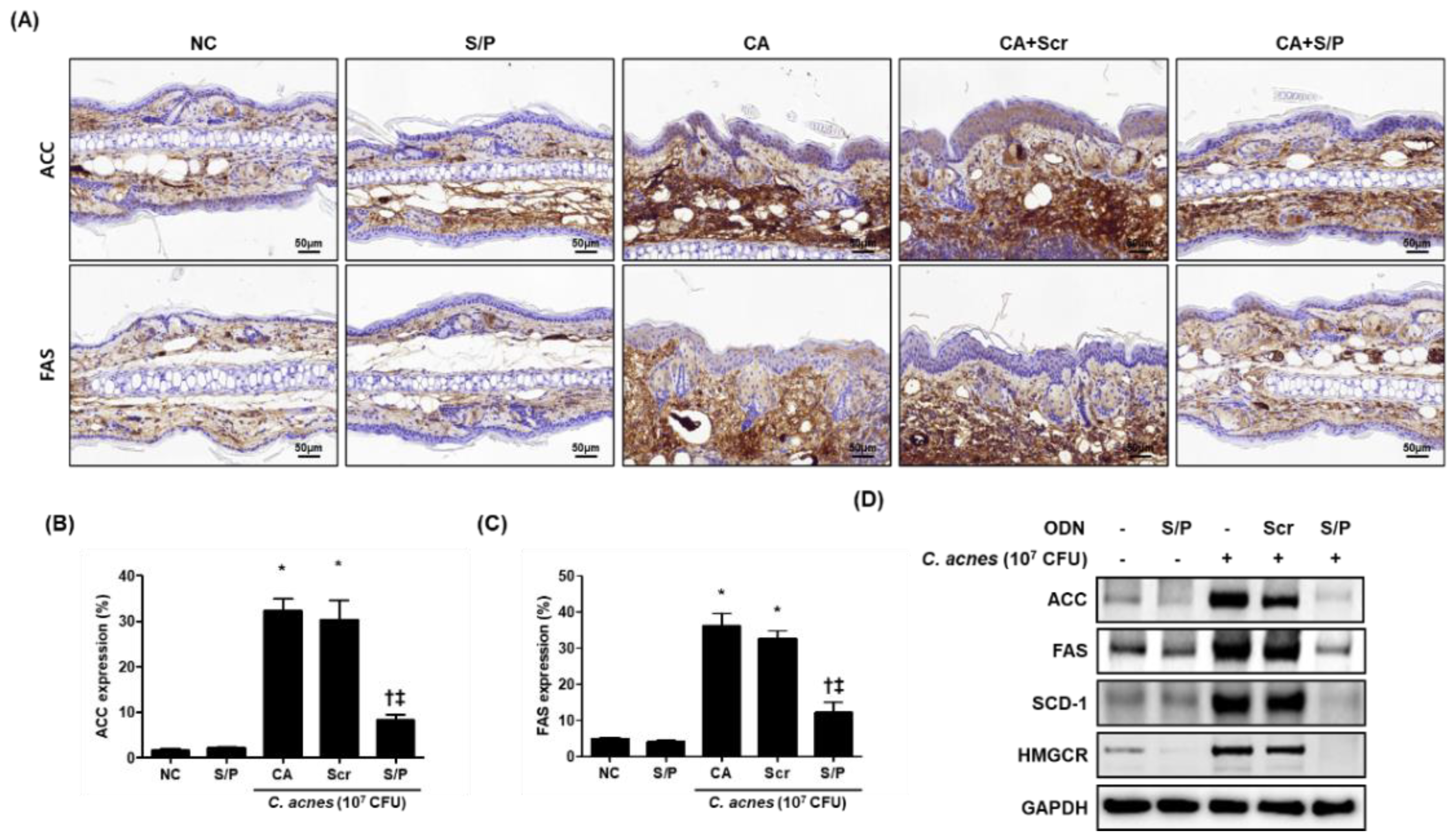
3.3. The SREBP/PPAR Chimeric Decoy Suppressed the Lipogenic Transcription Factors in the C. acnes-Mediated Skin Lesions
3.4. The SREBP/PPAR Chimeric Decoy Attenuated the Expression of Lipogenesis-Related Genes in the C. acnes -Mediatd Skin Lesions
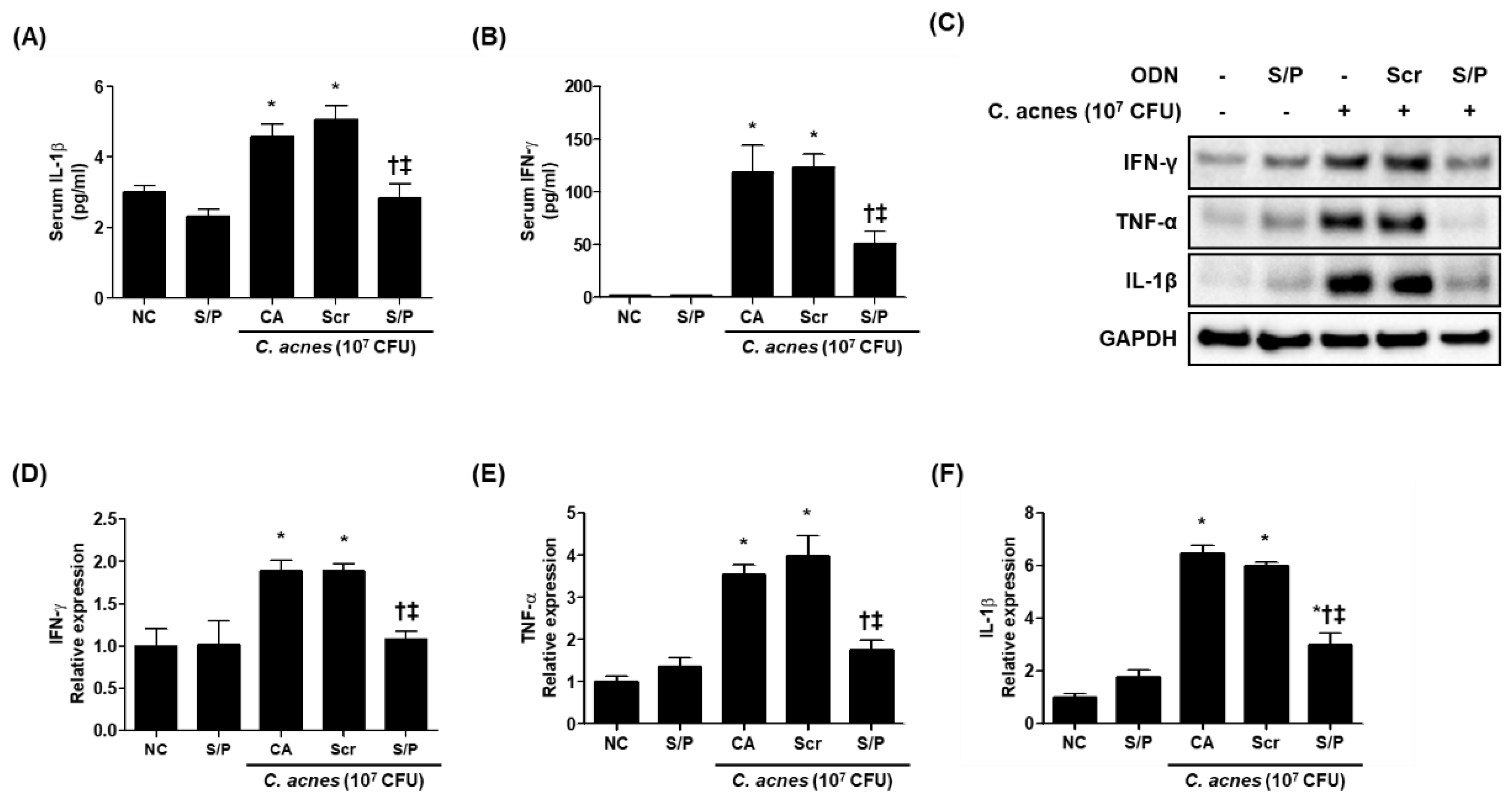
3.5. The SREBP/PPAR Chimeric Decoy ODN Reduced the Inflammatory Changes in the Animal Model of C. acnes-Like Lesions
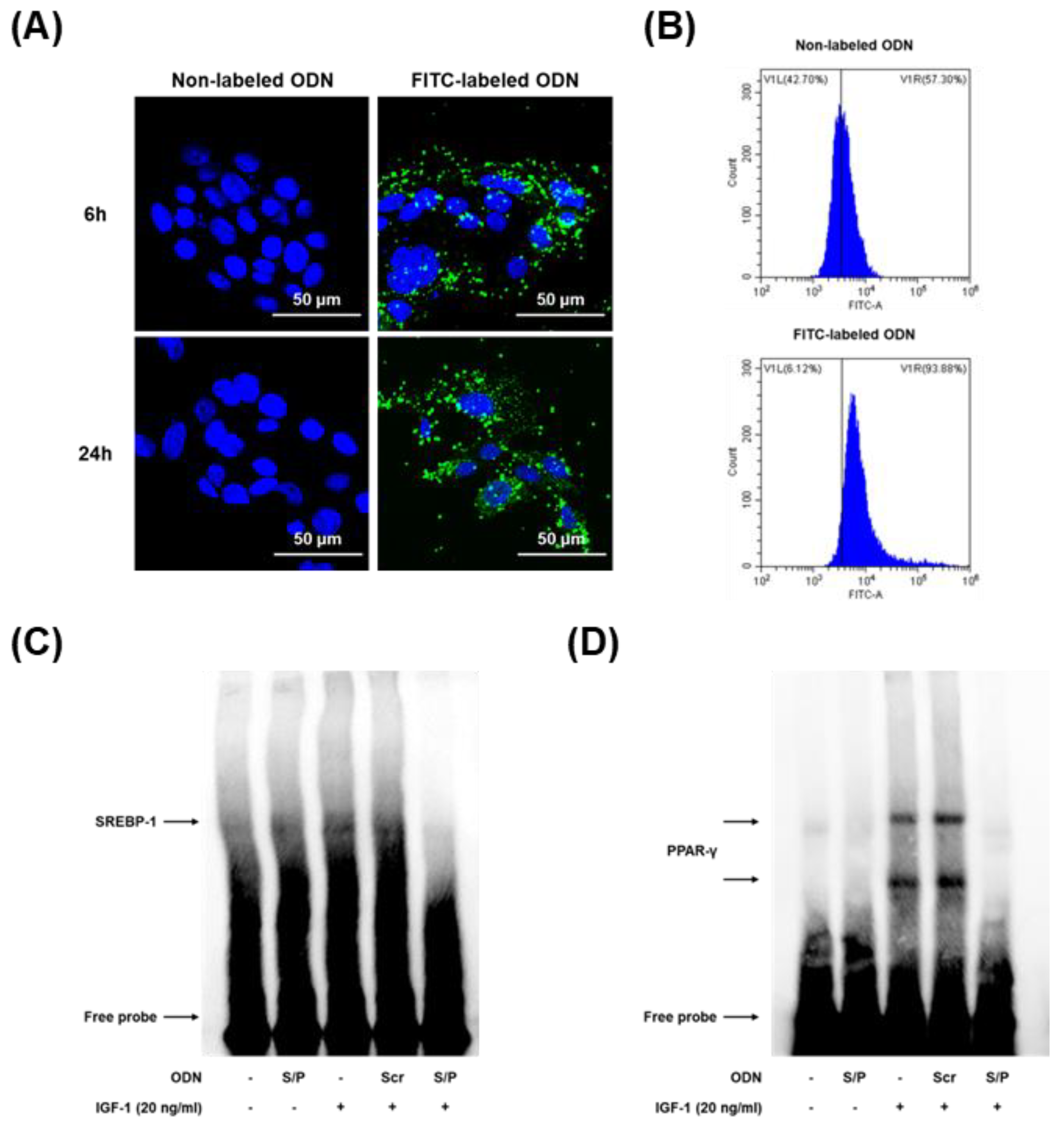
3.6. Effects of the SREBP/PPAR Chimeric Decoy ODN on the DNA-Binding Activity of SREBP and PPAR In Vitro
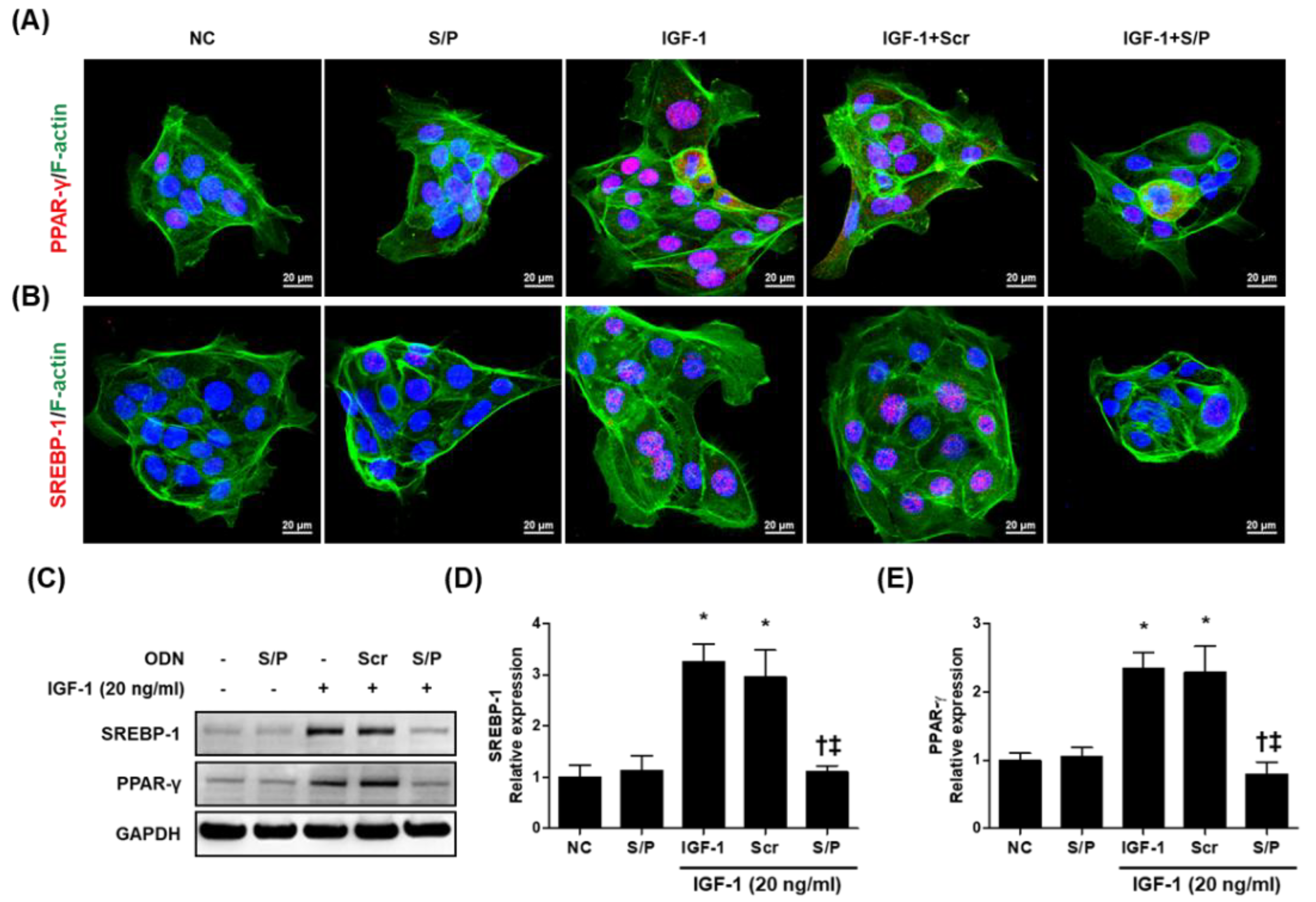
3.7. The SREBP/PPAR Chimeric Decoy ODN Decreased the Lipogenic Transcription Factors in IGF-1-Stimulated Human SZ95 Sebocytes
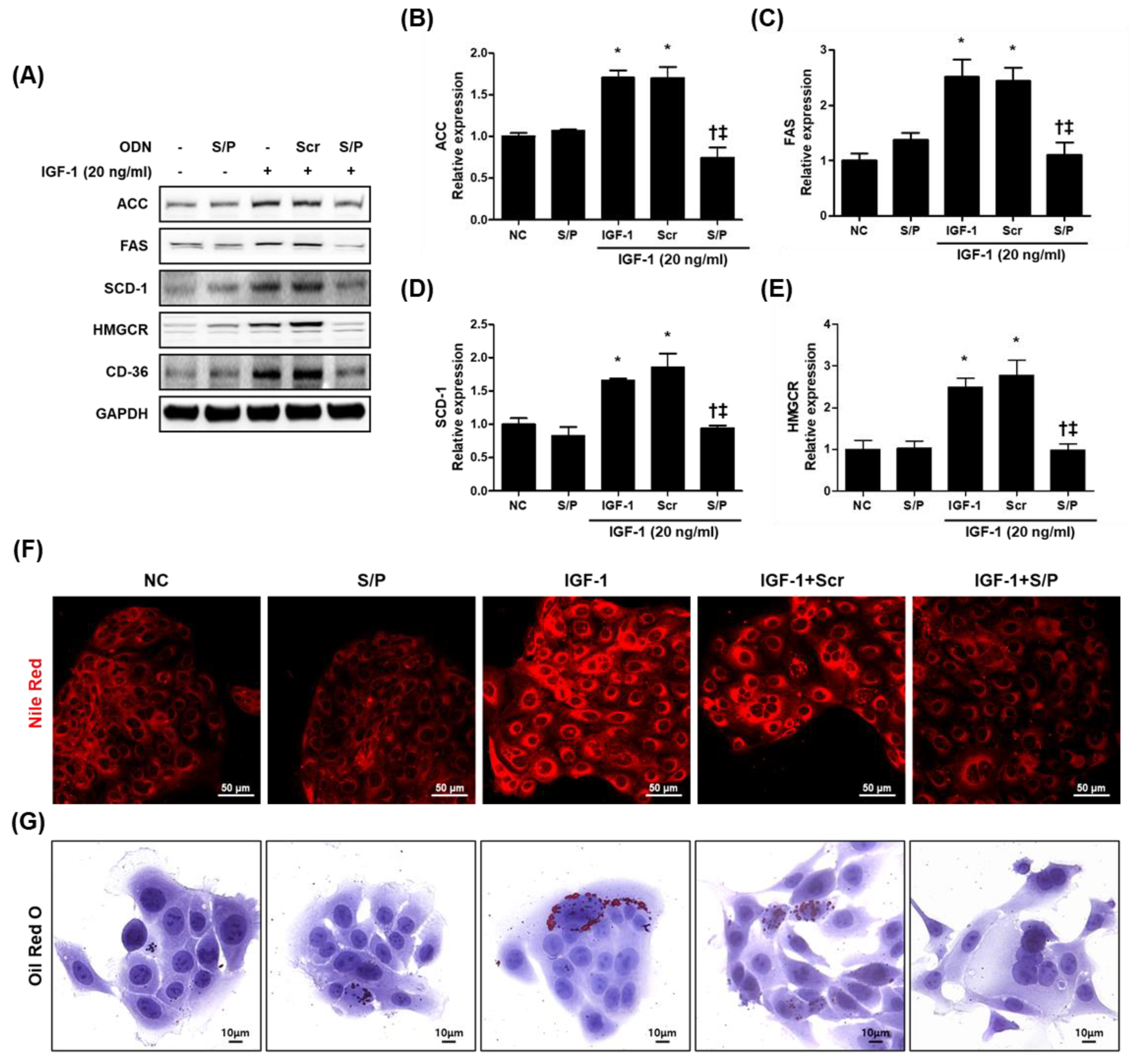
3.8. The SREBP/PPAR Chimeric Decoy ODN Impaired the Intracellular Lipid Accumulation in IGF-1-Treated SZ95 Cells
3.9. The SREBP/PPAR Chimeric Decoy ODN Suppressed the IGF-1-Induced Inflammatory Responses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bowe, W.P.; Leyden, J.J.; Crerand, C.E.; Sarwer, D.B.; Margolis, D.J. Body dysmorphic disorder symptoms among patients with acne vulgaris. J. Am. Acad. Dermatol. 2007, 57, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Esler, W.P.; Tesz, G.J.; Hellerstein, M.K.; Beysen, C.; Sivamani, R.; Turner, S.M.; Watkins, S.M.; Amor, P.A.; Carvajal-Gonzalez, S.; Geoly, F.J.; et al. Human sebum requires de novo lipogenesis, which is increased in acne vulgaris and suppressed by acetyl-CoA carboxylase inhibition. Sci. Transl. Med. 2019, 11, eaau8465. [Google Scholar] [CrossRef] [PubMed]
- Im, M.; Kim, S.Y.; Sohn, K.C.; Choi, D.K.; Lee, Y.; Seo, Y.J.; Kim, C.D.; Hwang, Y.L.; Zouboulis, C.C.; Lee, J.H. Epigallocatechin-3-gallate suppresses IGF-I-induced lipogenesis and cytokine expression in SZ95 sebocytes. J. Investig. Dermatol. 2012, 132, 2700–2708. [Google Scholar] [CrossRef]
- Melnik, B.C.; Schmitz, G. Role of insulin, insulin-like growth factor-1, hyperglycaemic food and milk consumption in the pathogenesis of acne vulgaris. Exp. Dermatol. 2009, 18, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Cong, T.-X.; Hao, D.; Wen, X.; Li, X.-H.; He, G.; Jiang, X. From pathogenesis of acne vulgaris to anti-acne agents. Arch. Dermatol. Res. 2019, 311, 337–349. [Google Scholar] [CrossRef]
- Melnik, B.C. Linking diet to acne metabolomics, inflammation, and comedogenesis: An update. Clin. Cosmet. Investig. Dermatol. 2015, 8, 371–388. [Google Scholar] [CrossRef]
- Mastrofrancesco, A.; Ottaviani, M.; Cardinali, G.; Flori, E.; Briganti, S.; Ludovici, M.; Zouboulis, C.; Lora, V.; Camera, E.; Picardo, M. Pharmacological PPARγ modulation regulates sebogenesis and inflammation in SZ95 human sebocytes. Biochem. Pharmacol. 2017, 138, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Dozsa, A.; Dezso, B.; Toth, B.I.; Bacsi, A.; Poliska, S.; Camera, E.; Picardo, M.; Zouboulis, C.C.; Bíró, T.; Schmitz, G.; et al. PPARγ-Mediated and Arachidonic Acid–Dependent Signaling Is Involved in Differentiation and Lipid Production of Human Sebocytes. J. Investig. Dermatol. 2014, 134, 910–920. [Google Scholar] [CrossRef]
- Smith, T.M.; Gilliland, K.; Clawson, G.A.; Thiboutot, D. IGF-1 induces SREBP-1 expression and lipogenesis in SEB-1 sebocytes via activation of the phosphoinositide 3-kinase/Akt pathway. J. Investig. Dermatol. 2008, 128, 1286–1293. [Google Scholar] [CrossRef]
- Melnik, B.C. Acne vulgaris: An inflammasomopathy of the sebaceous follicle induced by deviated FoxO1/mTORC1 signalling. Br. J. Dermatol. 2016, 174, 1186–1188. [Google Scholar] [CrossRef]
- Brown, N.F.; Stefanovic-Racic, M.; Sipula, I.J.; Perdomo, G. The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metabolism 2007, 56, 1500–1507. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Golub, T.R.; Sabatini, D.M. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol. Cell. Biol. 2002, 22, 5575–5584. [Google Scholar] [CrossRef]
- Mauvoisin, D.; Rocque, G.; Arfa, O.; Radenne, A.; Boissier, P.; Mounier, C. Role of the PI3-kinase/mTor pathway in the regulation of the stearoyl CoA desaturase (SCD1) gene expression by insulin in liver. J. Cell Commun. Signal. 2007, 1, 113–125. [Google Scholar] [CrossRef]
- An, H.J.; Kim, J.Y.; Gwon, M.G.; Gu, H.; Kim, H.J.; Leem, J.; Youn, S.W.; Park, K.K. Beneficial Effects of SREBP Decoy Oligodeoxynucleotide in an Animal Model of Hyperlipidemia. Int. J. Mol. Sci. 2020, 21, 552. [Google Scholar] [CrossRef] [PubMed]
- Gwon, M.G.; An, H.J.; Kim, J.Y.; Kim, W.H.; Gu, H.; Kim, H.J.; Leem, J.; Jung, H.J.; Park, K.K. Anti-fibrotic effects of synthetic TGF-β1 and Smad oligodeoxynucleotide on kidney fibrosis in vivo and in vitro through inhibition of both epithelial dedifferentiation and endothelial-mesenchymal transitions. FASEB J. 2020, 34, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; An, H.J.; Gwon, M.G.; Bae, S.; Leem, J.; Lee, S.J.; Han, S.M.; Zouboulis, C.C.; Park, K.K. Bee Venom and Its Major Component Melittin Attenuated Cutibacterium acnes- and IGF-1-Induced Acne Vulgaris via Inactivation of Akt/mTOR/SREBP Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 3152. [Google Scholar] [CrossRef] [PubMed]
- Zouboulis, C.C.; Seltmann, H.; Orfanos, C.E.; Neitzel, H. Establishment and characterization of an immortalized human sebaceous gland cell line (SZ95) 1. J. Investig. Dermatol. 1999, 113, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Kligman, A.M. An overview of acne. J. Investig. Dermatol. 1974, 62, 268–287. [Google Scholar] [CrossRef]
- Kwon, H.C.; Kim, T.Y.; Lee, C.M.; Lee, K.S.; Lee, K.K. Active compound chrysophanol of Cassia tora seeds suppresses heat-induced lipogenesis via inactivation of JNK/p38 MAPK signaling in human sebocytes. Lipids Health Dis. 2019, 18, 135. [Google Scholar] [CrossRef]
- Neves, J.R.; Francesconi, F. Propionibacterium acnes and bacterial resistance. Surg. Cosmet. Dermatol. 2015, 7, 27–38. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Picardo, M.; Ju, Q.; Kurokawa, I.; Torocsik, D.; Biro, T.; Schneider, M.R. Beyond acne: Current aspects of sebaceous gland biology and function. Rev. Endocr. Metab. Disord. 2016, 17, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Ben-Amitai, D.; Laron, Z. Effect of insulin-like growth factor-1 deficiency or administration on the occurrence of acne. J. Eur. Acad. Dermatol. Venereol. 2011, 25, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; John, S.M.; Schmitz, G. Over-stimulation of insulin/IGF-1 signaling by western diet may promote diseases of civilization: Lessons learnt from laron syndrome. Nutr. Metab. 2011, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.M.; Cong, Z.; Gilliland, K.L.; Clawson, G.A.; Thiboutot, D.M. Insulin-like growth factor-1 induces lipid production in human SEB-1 sebocytes via sterol response element-binding protein-1. J. Investig. Dermatol. 2006, 126, 1226–1232. [Google Scholar] [CrossRef]
- Zoete, V.; Grosdidier, A.; Michielin, O. Peroxisome proliferator-activated receptor structures: Ligand specificity, molecular switch and interactions with regulators. Biochim. Biophys. Acta 2007, 1771, 915–925. [Google Scholar] [CrossRef]
- Clayton, R.W.; Gobel, K.; Niessen, C.M.; Paus, R.; van Steensel, M.A.M.; Lim, X. Homeostasis of the sebaceous gland and mechanisms of acne pathogenesis. Br. J. Dermatol. 2019, 181, 677–690. [Google Scholar] [CrossRef]
- Bao, J.; Zhu, L.; Zhu, Q.; Su, J.; Liu, M.; Huang, W. SREBP-1 is an independent prognostic marker and promotes invasion and migration in breast cancer. Oncol. Lett. 2016, 12, 2409–2416. [Google Scholar]
- Zhang, B.; Wu, J.; Guo, P.; Wang, Y.; Fang, Z.; Tian, J.; Yu, Y.; Teng, W.; Luo, Y.; Li, Y. Down-Regulation of SREBP via PI3K/AKT/mTOR Pathway Inhibits the Proliferation and Invasion of Non-Small-Cell Lung Cancer Cells. OncoTargets Ther. 2020, 13, 8951–8961. [Google Scholar] [CrossRef]
- Rosignoli, C.; Nicolas, J.C.; Jomard, A.; Michel, S. Involvement of the SREBP pathway in the mode of action of androgens in sebaceous glands in vivo. Exp. Dermatol. 2003, 12, 480–489. [Google Scholar] [CrossRef]
- Hunt, D.W.; Winters, G.C.; Brownsey, R.W.; Kulpa, J.E.; Gilliland, K.L.; Thiboutot, D.M.; Hofland, H.E. Inhibition of sebum production with the acetyl coenzyme a carboxylase inhibitor olumacostat glasaretil. J. Investig. Dermatol. 2017, 137, 1415–1423. [Google Scholar] [CrossRef]
- Lenhard, J.M. Lipogenic enzymes as therapeutic targets for obesity and diabetes. Curr. Pharm. Des. 2011, 17, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Eilertsen, K.J.; Ge, L.; Zhang, L.; Sundberg, J.P.; Prouty, S.M.; Stenn, K.S.; Parimoo, S. Scd1 is expressed in sebaceous glands and is disrupted in the asebia mouse. Nat. Genet. 1999, 23, 268–270. [Google Scholar] [CrossRef] [PubMed]
- Gerondakis, S.; Grossmann, M.; Nakamura, Y.; Pohl, T.; Grumont, R. Genetic approaches in mice to understand Rel/NF-kappaB and IkappaB function: Transgenics and knockouts. Oncogene 1999, 18, 6888–6895. [Google Scholar] [CrossRef] [PubMed]
- Tomita, N.; Kashihara, N.; Morishita, R. Transcription factor decoy oligonucleotide-based therapeutic strategy for renal disease. Clin. Exp. Nephrol. 2007, 11, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.C.; Ding, X.Q.; Ou, Z.L.; Liu, C.F.; Li, P.; Wang, L.; Zhu, C.F. In vivo transfection of NF-kappaB decoy oligodeoxynucleotides attenuate renal ischemia/reperfusion injury in rats. Kidney Int. 2004, 65, 834–845. [Google Scholar] [CrossRef]
- Kim, K.H.; Lee, E.S.; Cha, S.H.; Park, J.H.; Park, J.S.; Chang, Y.C.; Park, K.K. Transcriptional regulation of NF-kappaB by ring type decoy oligodeoxynucleotide in an animal model of nephropathy. Exp. Mol. Pathol. 2009, 86, 114–120. [Google Scholar] [CrossRef]
- Yuan, H.F.; Huang, H.; Li, X.Y.; Guo, W.; Xing, W.; Sun, Z.Y.; Liang, H.P.; Yu, J.; Chen, D.F.; Wang, Z.G.; et al. A dual AP-1 and SMAD decoy ODN suppresses tissue fibrosis and scarring in mice. J. Investig. Dermatol. 2013, 133, 1080–1087. [Google Scholar] [CrossRef]
- Kim, K.H.; Park, J.H.; Lee, W.R.; Park, J.S.; Kim, H.C.; Park, K.K. The inhibitory effect of chimeric decoy oligodeoxynucleotide against NF-kappaB and Sp1 in renal interstitial fibrosis. J. Mol. Med. 2013, 91, 573–586. [Google Scholar] [CrossRef]


| Oligodeoxynucleotide | Sequence |
|---|---|
| SREBP-1 | 5′-GAATTCGTATCACCCCACACAAAAGTGTGGGGTGATAC-3′ |
| PPAR-γ | 5′-GAATTCGTTGACCTTTGACCTTGAAAACAAGGTCAAAGGTCAAC-3′ |
| Scramble | 5′-GAATTCAATTCAGGGTACGGCAAAAAATTGCCGTACCCTGAATT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, H.; An, H.-J.; Gwon, M.-G.; Bae, S.; Zouboulis, C.C.; Park, K.-K. The Effects of Synthetic SREBP-1 and PPAR-γ Decoy Oligodeoxynucleotide on Acne-like Disease In Vivo and In Vitro via Lipogenic Regulation. Biomolecules 2022, 12, 1858. https://doi.org/10.3390/biom12121858
Gu H, An H-J, Gwon M-G, Bae S, Zouboulis CC, Park K-K. The Effects of Synthetic SREBP-1 and PPAR-γ Decoy Oligodeoxynucleotide on Acne-like Disease In Vivo and In Vitro via Lipogenic Regulation. Biomolecules. 2022; 12(12):1858. https://doi.org/10.3390/biom12121858
Chicago/Turabian StyleGu, Hyemin, Hyun-Jin An, Mi-Gyeong Gwon, Seongjae Bae, Christos C. Zouboulis, and Kwan-Kyu Park. 2022. "The Effects of Synthetic SREBP-1 and PPAR-γ Decoy Oligodeoxynucleotide on Acne-like Disease In Vivo and In Vitro via Lipogenic Regulation" Biomolecules 12, no. 12: 1858. https://doi.org/10.3390/biom12121858
APA StyleGu, H., An, H.-J., Gwon, M.-G., Bae, S., Zouboulis, C. C., & Park, K.-K. (2022). The Effects of Synthetic SREBP-1 and PPAR-γ Decoy Oligodeoxynucleotide on Acne-like Disease In Vivo and In Vitro via Lipogenic Regulation. Biomolecules, 12(12), 1858. https://doi.org/10.3390/biom12121858







