Insights into Non-Proteolytic Inhibitory Mechanisms of Polymorphic Early-Stage Amyloid β Oligomers by Insulin Degrading Enzyme
Abstract
1. Introduction
2. Methods and Materials
2.1. Construction of IDE Model
2.2. The Rational of the Choice of Aβ Dimer Models
2.3. Construction of IDE-Aβ Fibril-Like Dimer Complex Models
2.4. Construction of IDE-Aβ Random Coil/α-Helix Dimer Complex Models
2.5. Molecular Dynamics (MD) Simulations Protocol
2.6. Structural Analyses
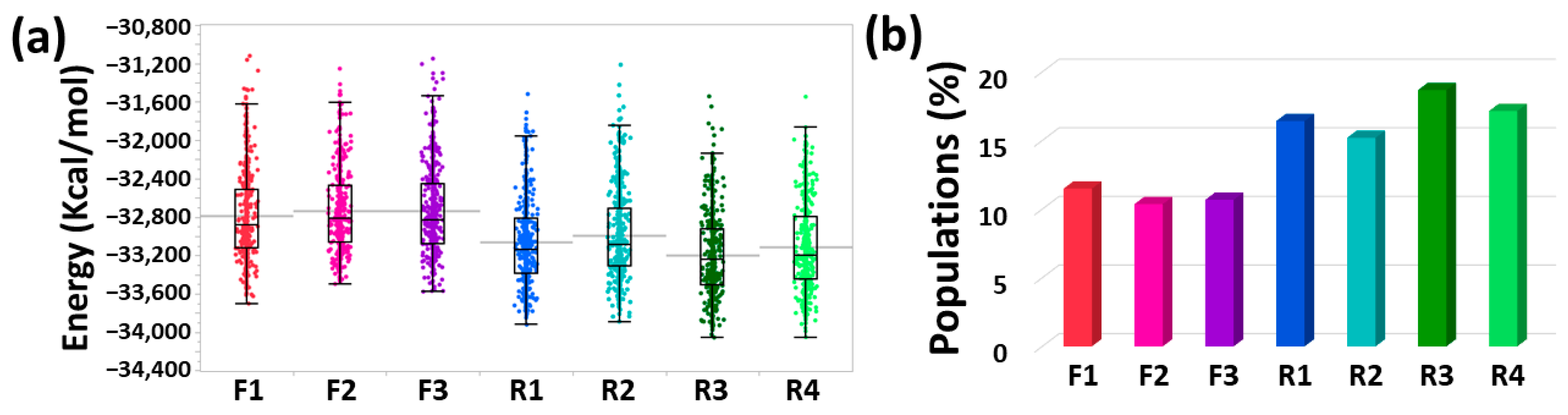
2.7. Determining the Conformational Energies and Populations for the Simulated IDE-Aβ Dimers
3. Results and Discussion
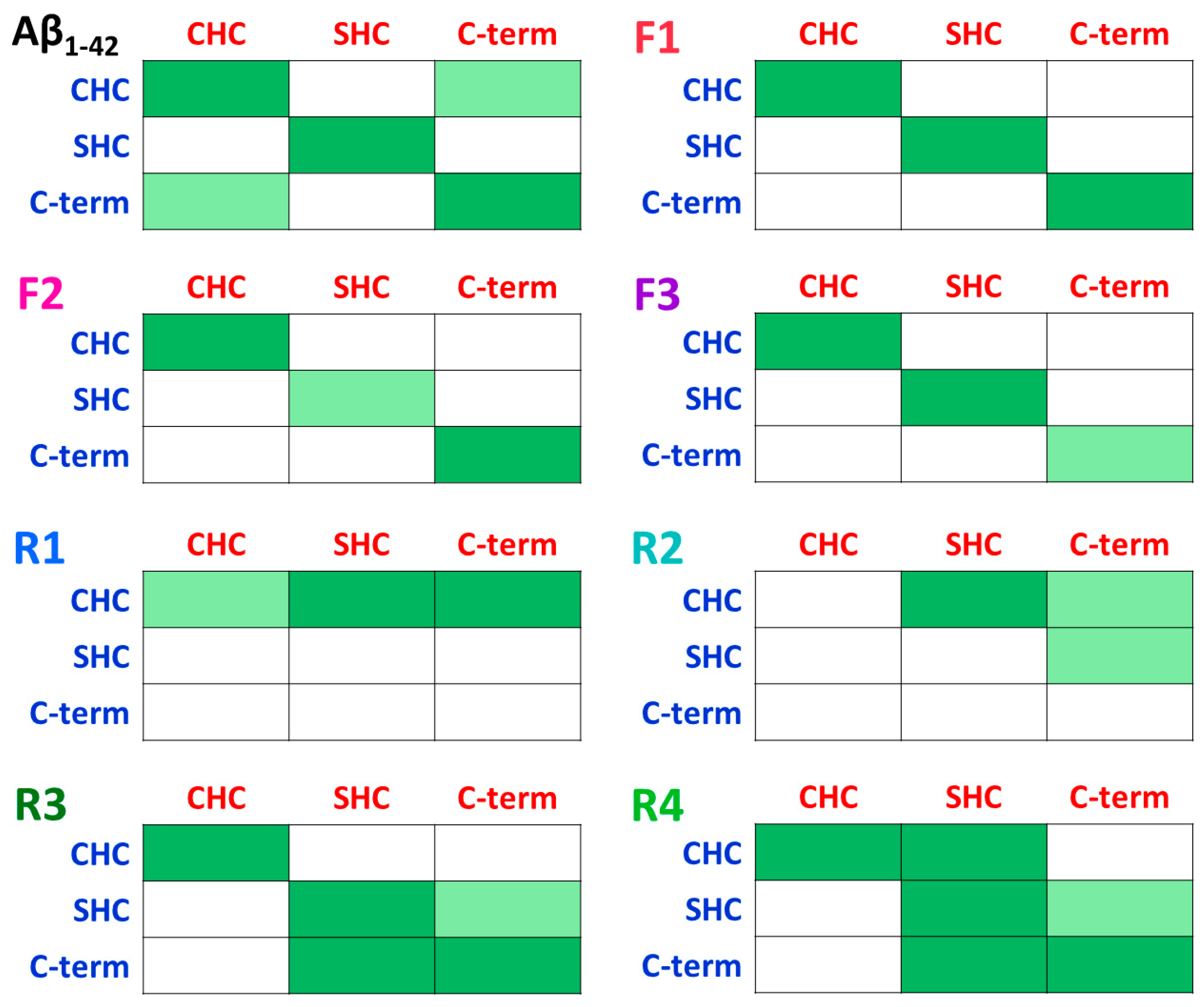
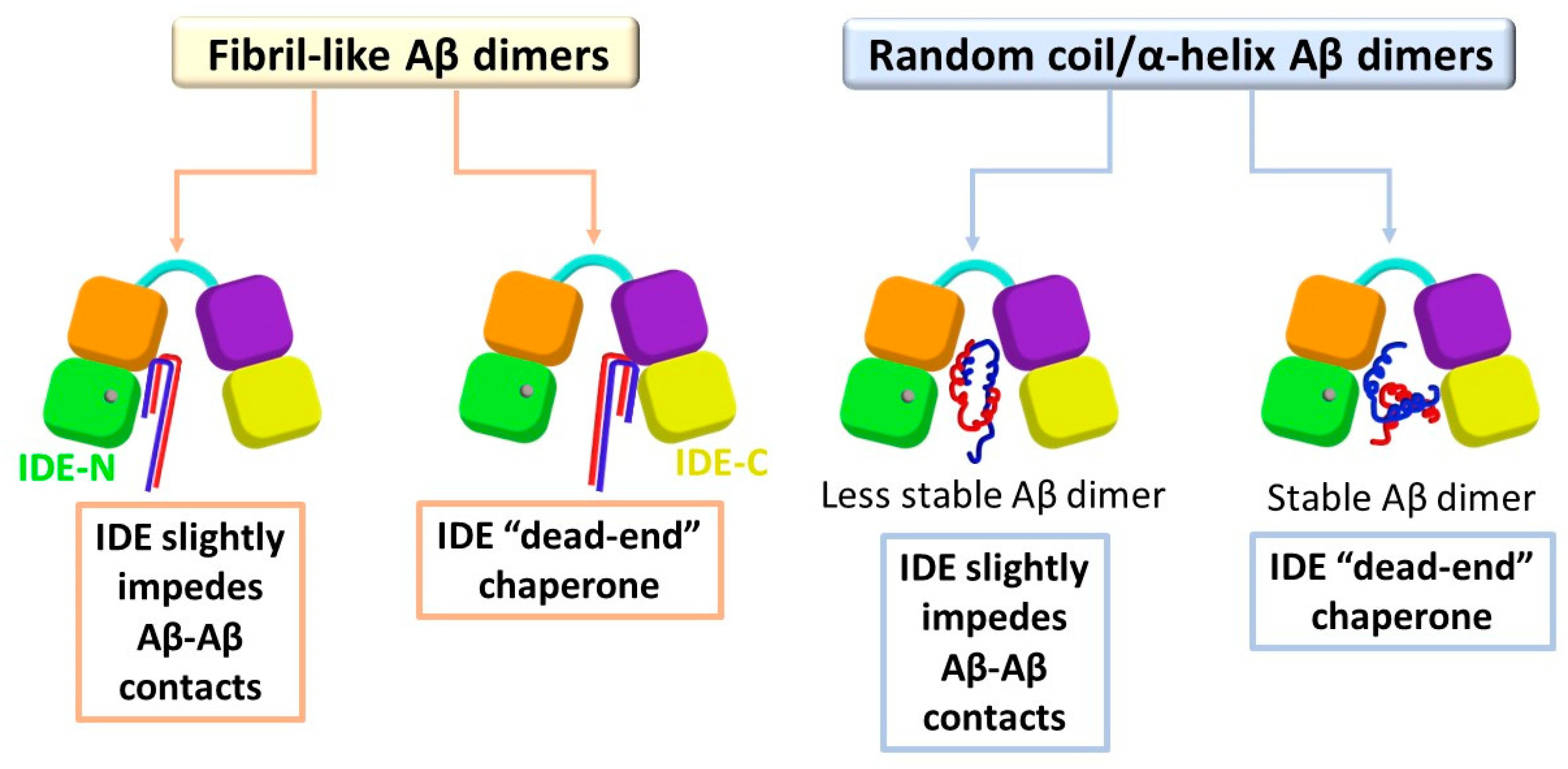
3.1. IDE Does Not Inhibit Contacts between Monomers in Fibril-Like Aβ Dimers
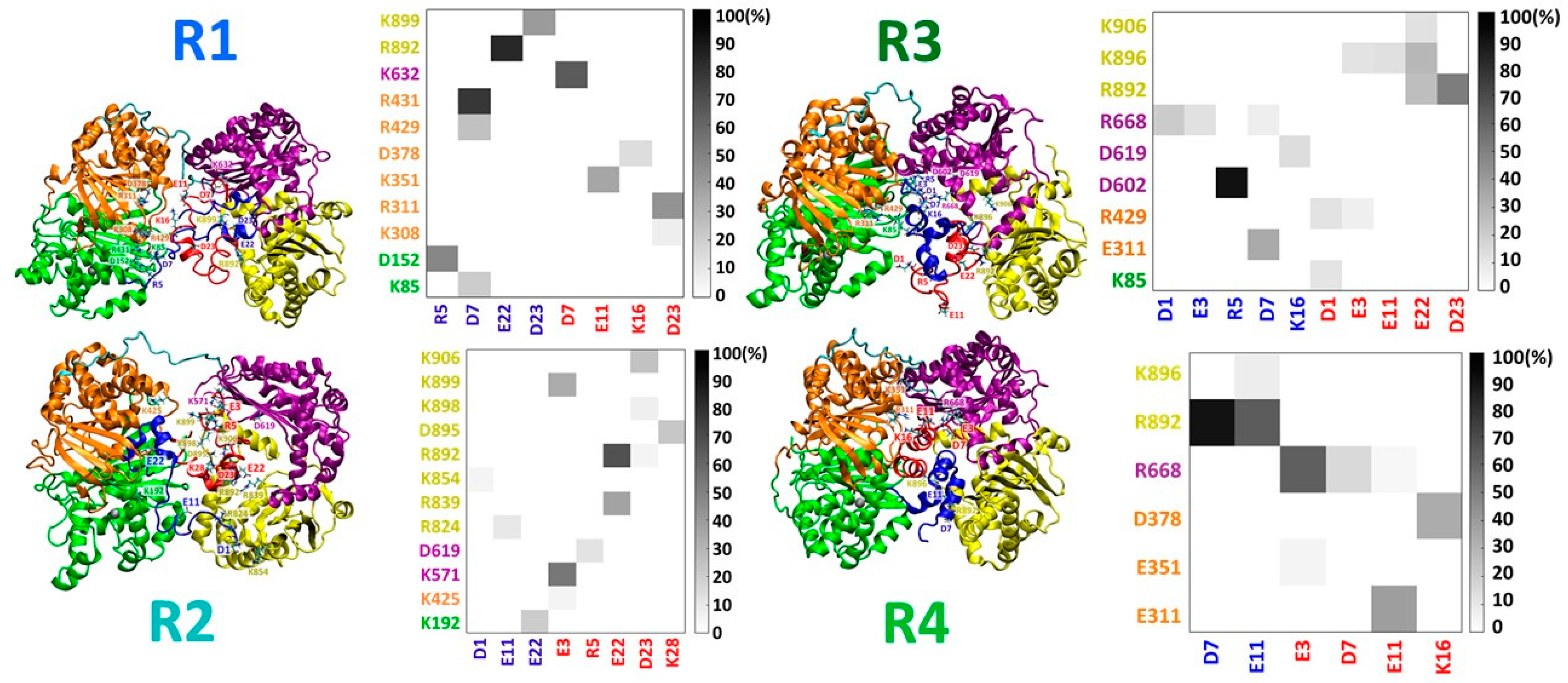
3.2. IDE Inhibitory Activity Depends on the Stability of the Random Coil/α-Helix Aβ Dimer
3.3. Distinct Effects of IDE on the Polymorphic Aβ Dimers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- 2021 Alzheimer’s disease facts and figures. Alzheimers Dement. 2021, 17, 327–406. [CrossRef] [PubMed]
- Sikanyika, N.L.; Parkington, H.C.; Smith, A.I.; Kuruppu, S. Powering Amyloid Beta Degrading Enzymes: A Possible Therapy for Alzheimer’s Disease. Neurochem. Res. 2019, 44, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Ramaraju, B.; Nelson, S.L.; Zheng, W.; Ghirlando, R.; Deshmukh, L. Quantitative NMR Study of Insulin-Degrading Enzyme Using Amyloid-beta and HIV-1 p6 Elucidates Its Chaperone Activity. Biochemistry 2021, 60, 2519–2523. [Google Scholar] [CrossRef]
- Authier, F.; Rachubinski, R.A.; Posner, B.I.; Bergeron, J.J. Endosomal proteolysis of insulin by an acidic thiol metalloprotease unrelated to insulin degrading enzyme. J. Biol. Chem. 1994, 269, 3010–3016. [Google Scholar] [CrossRef] [PubMed]
- Authier, F.; Bergeron, J.J.; Ou, W.J.; Rachubinski, R.A.; Posner, B.I.; Walton, P.A. Degradation of the cleaved leader peptide of thiolase by a peroxisomal proteinase. Proc. Natl. Acad. Sci. USA 1995, 92, 3859–3863. [Google Scholar] [CrossRef] [PubMed]
- Tundo, G.R.; Di Muzio, E.; Ciaccio, C.; Sbardella, D.; Di Pierro, D.; Polticelli, F.; Coletta, M.; Marini, S. Multiple allosteric sites are involved in the modulation of insulin-degrading-enzyme activity by somatostatin. FEBS J. 2016, 283, 3755–3770. [Google Scholar] [CrossRef]
- Perez, A.; Morelli, L.; Cresto, J.C.; Castano, E.M. Degradation of soluble amyloid beta-peptides 1-40, 1-42, and the Dutch variant 1-40Q by insulin degrading enzyme from Alzheimer disease and control brains. Neurochem. Res. 2000, 25, 247–255. [Google Scholar] [CrossRef]
- Cook, D.G.; Leverenz, J.B.; McMillan, P.J.; Kulstad, J.J.; Ericksen, S.; Roth, R.A.; Schellenberg, G.D.; Jin, L.W.; Kovacina, K.S.; Craft, S. Reduced hippocampal insulin-degrading enzyme in late-onset Alzheimer’s disease is associated with the apolipoprotein E-epsilon4 allele. Am. J. Pathol. 2003, 162, 313–319. [Google Scholar] [CrossRef]
- Farris, W.; Mansourian, S.; Chang, Y.; Lindsley, L.; Eckman, E.A.; Frosch, M.P.; Eckman, C.B.; Tanzi, R.E.; Selkoe, D.J.; Guenette, S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc. Natl. Acad. Sci. USA 2003, 100, 4162–4167. [Google Scholar] [CrossRef]
- Jha, N.K.; Jha, S.K.; Kumar, D.; Kejriwal, N.; Sharma, R.; Ambasta, R.K.; Kumar, P. Impact of Insulin Degrading Enzyme and Neprilysin in Alzheimer’s Disease Biology: Characterization of Putative Cognates for Therapeutic Applications. J. Alzheimers Dis. 2015, 48, 891–917. [Google Scholar] [CrossRef]
- Miners, J.S.; Barua, N.; Kehoe, P.G.; Gill, S.; Love, S. Abeta-degrading enzymes: Potential for treatment of Alzheimer disease. J. Neuropathol. Exp. Neurol. 2011, 70, 944–959. [Google Scholar] [CrossRef] [PubMed]
- Nalivaeva, N.N.; Fisk, L.R.; Belyaev, N.D.; Turner, A.J. Amyloid-degrading enzymes as therapeutic targets in Alzheimer’s disease. Curr. Alzheimer Res. 2008, 5, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Tundo, G.R.; Sbardella, D.; Ciaccio, C.; Grasso, G.; Gioia, M.; Coletta, A.; Polticelli, F.; Di Pierro, D.; Milardi, D.; Van Endert, P.; et al. Multiple functions of insulin-degrading enzyme: A metabolic crosslight? Crit. Rev. Biochem. Mol. Biol. 2017, 52, 554–582. [Google Scholar] [CrossRef] [PubMed]
- Kurochkin, I.V.; Goto, S. Alzheimer’s beta-amyloid peptide specifically interacts with and is degraded by insulin degrading enzyme. FEBS Lett. 1994, 345, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Morelli, L.; Llovera, R.E.; Mathov, I.; Lue, L.F.; Frangione, B.; Ghiso, J.; Castano, E.M. Insulin-degrading enzyme in brain microvessels: Proteolysis of amyloid {beta} vasculotropic variants and reduced activity in cerebral amyloid angiopathy. J. Biol. Chem. 2004, 279, 56004–56013. [Google Scholar] [CrossRef]
- Shen, Y.; Joachimiak, A.; Rosner, M.R.; Tang, W.J. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature 2006, 443, 870–874. [Google Scholar] [CrossRef]
- Ren, B.; Zhang, Y.; Zhang, M.; Liu, Y.; Zhang, D.; Gong, X.; Feng, Z.; Tang, J.; Chang, Y.; Zheng, J. Fundamentals of cross-seeding of amyloid proteins: An introduction. J. Mater. Chem. B 2019, 7, 7267–7282. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Tang, Y.J.; Zhang, D.; Liu, Y.L.; He, J.; Chang, Y.; Zheng, J. Amyloid cross-seeding between A beta and hIAPP in relation to the pathogenesis of Alzheimer and type 2 diabetes. Chin. J. Chem. Eng. 2021, 30, 225–235. [Google Scholar] [CrossRef]
- Atsmon-Raz, Y.; Miller, Y. Molecular Mechanisms of the Bindings between Non-Amyloid beta Component Oligomers and Amylin Oligomers. J. Phys. Chem. B 2016, 120, 10649–10659. [Google Scholar] [CrossRef]
- Atsmon-Raz, Y.; Miller, Y. Non-Amyloid-beta Component of Human alpha-Synuclein Oligomers Induces Formation of New Abeta Oligomers: Insight into the Mechanisms That Link Parkinson’s and Alzheimer’s Diseases. ACS Chem. Neurosci. 2016, 7, 46–55. [Google Scholar] [CrossRef]
- Baram, M.; Atsmon-Raz, Y.; Ma, B.; Nussinov, R.; Miller, Y. Amylin-Abeta oligomers at atomic resolution using molecular dynamics simulations: A link between Type 2 diabetes and Alzheimer’s disease. Phys. Chem. Chem. Phys. 2016, 18, 2330–2338. [Google Scholar] [CrossRef] [PubMed]
- Baram, M.; Gilead, S.; Gazit, E.; Miller, Y. Mechanistic perspective and functional activity of insulin in amylin aggregation. Chem. Sci. 2018, 9, 4244–4252. [Google Scholar] [CrossRef] [PubMed]
- Raz, Y.; Miller, Y. Interactions between Abeta and mutated Tau lead to polymorphism and induce aggregation of Abeta-mutated tau oligomeric complexes. PLoS ONE 2013, 8, e73303. [Google Scholar] [CrossRef] [PubMed]
- Hulse, R.E.; Ralat, L.A.; Wei-Jen, T. Structure, function, and regulation of insulin-degrading enzyme. Vitam. Horm. 2009, 80, 635–648. [Google Scholar] [PubMed]
- Llovera, R.E.; de Tullio, M.; Alonso, L.G.; Leissring, M.A.; Kaufman, S.B.; Roher, A.E.; de Prat Gay, G.; Morelli, L.; Castano, E.M. The catalytic domain of insulin-degrading enzyme forms a denaturant-resistant complex with amyloid beta peptide: Implications for Alzheimer disease pathogenesis. J. Biol. Chem. 2008, 283, 17039–17048. [Google Scholar] [CrossRef] [PubMed]
- de Tullio, M.B.; Castelletto, V.; Hamley, I.W.; Martino Adami, P.V.; Morelli, L.; Castano, E.M. Proteolytically inactive insulin-degrading enzyme inhibits amyloid formation yielding non-neurotoxic abeta peptide aggregates. PLoS ONE 2013, 8, e59113. [Google Scholar] [CrossRef]
- Sahoo, B.R.; Panda, P.K.; Liang, W.; Tang, W.J.; Ahuja, R.; Ramamoorthy, A. Degradation of Alzheimer’s Amyloid-beta by a Catalytically Inactive Insulin-Degrading Enzyme. J. Mol. Biol. 2021, 433, 166993. [Google Scholar] [CrossRef]
- Zou, K.; Gong, J.S.; Yanagisawa, K.; Michikawa, M. A novel function of monomeric amyloid beta-protein serving as an antioxidant molecule against metal-induced oxidative damage. J. Neurosci. 2002, 22, 4833–4841. [Google Scholar] [CrossRef]
- Giuffrida, M.L.; Caraci, F.; Pignataro, B.; Cataldo, S.; De Bona, P.; Bruno, V.; Molinaro, G.; Pappalardo, G.; Messina, A.; Palmigiano, A.; et al. Beta-amyloid monomers are neuroprotective. J. Neurosci. 2009, 29, 10582–10587. [Google Scholar] [CrossRef]
- Sharma, S.K.; Chorell, E.; Steneberg, P.; Vernersson-Lindahl, E.; Edlund, H.; Wittung-Stafshede, P. Insulin-degrading enzyme prevents alpha-synuclein fibril formation in a nonproteolytical manner. Sci. Rep. 2015, 5, 12531. [Google Scholar] [CrossRef]
- BIOVIA Software, Inc., San Diego, CA, United States. Available online: https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/ (accessed on 1 January 2021).
- Zhang, Z.; Liang, W.G.; Bailey, L.J.; Tan, Y.Z.; Wei, H.; Wang, A.; Farcasanu, M.; Woods, V.A.; McCord, L.A.; Lee, D.; et al. Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme. eLife 2018, 7, e33572. [Google Scholar] [CrossRef] [PubMed]
- Abramov-Harpaz, K.; Miller, Y. A zinc-dependent switching mechanism from an open to a new closed-state conformation of insulin-degrading enzyme. Inorg. Chem. Front. 2021, 8, 3205–3209. [Google Scholar] [CrossRef]
- Miller, Y.; Ma, B.; Nussinov, R. Polymorphism in Alzheimer Abeta amyloid organization reflects conformational selection in a rugged energy landscape. Chem. Rev. 2010, 110, 4820–4838. [Google Scholar] [CrossRef] [PubMed]
- Press-Sandler, O.; Miller, Y. Distinct Primary Nucleation of Polymorphic Abeta Dimers Yields to Distinguished Fibrillation Pathways. ACS Chem. Neurosci. 2019, 10, 4407–4413. [Google Scholar] [CrossRef]
- Kale, L.; Skeel, R.; Bhandarkar, M.; Brunner, R.; Gursoy, A.; Krawetz, N.; Phillips, J.; Shinozaki, A.; Varadarajan, K.; Schulten, K. NAMD2: Greater scalability for parallel molecular dynamics. J. Comput. Phys. 1999, 151, 283–312. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Mahoney, M.W.; Jorgensen, W.L. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J. Chem. Phys. 2000, 112, 8910–8922. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.H.; Pastor, R.W.; Brooks, B.R. Constant-Pressure Molecular-Dynamics Simulation—The Langevin Piston Method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Tu, K.; Tobias, D.J.; Klein, M.L. Constant pressure and temperature molecular dynamics simulation of a fully hydrated liquid crystal phase dipalmitoylphosphatidylcholine bilayer. Biophys. J. 1995, 69, 2558–2562. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald—An N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints—Molecular-Dynamics of N-Alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Israelachvili, J.; Pashley, R. The hydrophobic interaction is long range, decaying exponentially with distance. Nature 1982, 300, 341–342. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nussinov, R. Close-range electrostatic interactions in proteins. Chembiochem 2002, 3, 604–617. [Google Scholar] [CrossRef]
- Deloof, H.; Nilsson, L.; Rigler, R. Molecular-Dynamics Simulation of Galanin in Aqueous and Nonaqueous Solution. J. Am. Chem. Soc. 1992, 114, 4028–4035. [Google Scholar] [CrossRef]
- Lee, M.S.; Feig, M.; Salsbury, F.R.; Brooks, C.L. New analytic approximation to the standard molecular volume definition and its application to generalized born calculations. J. Comput. Chem. 2003, 24, 1348–1356. [Google Scholar] [CrossRef]
- Lee, M.S.; Salsbury, F.R.; Brooks, C.L. Novel generalized Born methods. J. Chem. Phys. 2002, 116, 10606–10614. [Google Scholar] [CrossRef]
- Press-Sandler, O.; Miller, Y. Molecular insights into the primary nucleation of polymorphic amyloid beta dimers in DOPC lipid bilayer membrane. Protein Sci. 2022, 31, e4283. [Google Scholar] [CrossRef]
- de Tullio, M.B.; Morelli, L.; Castano, E.M. The irreversible binding of amyloid peptide substrates to insulin-degrading enzyme: A biological perspective. Prion 2008, 2, 51–56. [Google Scholar] [CrossRef]
- Li, P.; Kuo, W.L.; Yousef, M.; Rosner, M.R.; Tang, W.J. The C-terminal domain of human insulin degrading enzyme is required for dimerization and substrate recognition. Biochem. Biophys. Res. Commun. 2006, 343, 1032–1037. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abramov-Harpaz, K.; Miller, Y. Insights into Non-Proteolytic Inhibitory Mechanisms of Polymorphic Early-Stage Amyloid β Oligomers by Insulin Degrading Enzyme. Biomolecules 2022, 12, 1886. https://doi.org/10.3390/biom12121886
Abramov-Harpaz K, Miller Y. Insights into Non-Proteolytic Inhibitory Mechanisms of Polymorphic Early-Stage Amyloid β Oligomers by Insulin Degrading Enzyme. Biomolecules. 2022; 12(12):1886. https://doi.org/10.3390/biom12121886
Chicago/Turabian StyleAbramov-Harpaz, Karina, and Yifat Miller. 2022. "Insights into Non-Proteolytic Inhibitory Mechanisms of Polymorphic Early-Stage Amyloid β Oligomers by Insulin Degrading Enzyme" Biomolecules 12, no. 12: 1886. https://doi.org/10.3390/biom12121886
APA StyleAbramov-Harpaz, K., & Miller, Y. (2022). Insights into Non-Proteolytic Inhibitory Mechanisms of Polymorphic Early-Stage Amyloid β Oligomers by Insulin Degrading Enzyme. Biomolecules, 12(12), 1886. https://doi.org/10.3390/biom12121886






