Lactococcus lactis, an Attractive Cell Factory for the Expression of Functional Membrane Proteins
Abstract
:1. Introduction
2. Lactococcus lactis
2.1. The Nisin-Controlled Gene Expression System (NICE)
2.2. Host Strains Used for the NICE System
2.3. cDNA Cloning in Expression Vectors
2.3.1. Classical Cloning Using Restriction Enzymes
2.3.2. New Cloning Strategies
3. Expression of Membrane Proteins Using the NICE System
3.1. Expression of Prokaryotic MPs
3.2. Expression of Eukaryotic MPs
3.2.1. Membrane Proteins from Yeast (S. cerevisiae)
3.2.2. Membrane Proteins from Plants
3.2.3. Membrane Proteins from Humans
3.3. Comparison of Expression Levels between E. coli and L. lactis
4. Functional Expression of MPs
4.1. ABC Transporters
4.2. Secondary Transporters
4.3. MPs from Organelle
4.3.1. Mitochondrial MPs
4.3.2. Chloroplast MPs
4.4. Membrane Proteins from Other Families
5. Structures Resolved from MPs Expressed in L. lactis
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wallin, E.; von Heijne, G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998, 7, 1029–1038. [Google Scholar] [CrossRef]
- Lundstrom, K. Structural genomics and drug discovery. J. Cell Mol. Med. 2007, 11, 224–238. [Google Scholar] [CrossRef] [Green Version]
- Junge, F.; Schneider, B.; Reckel, S.; Schwarz, D.; Dötsch, V.; Bernhard, F. Large-scale production of functional membrane proteins. Cell Mol. Life Sci. 2008, 65, 1729–1755. [Google Scholar] [CrossRef]
- Kesidis, A.; Depping, P.; Lodé, A.; Vaitsopoulou, A.; Bill, R.M.; Goddard, A.D.; Rothnie, A.J. Expression of eukaryotic membrane proteins in eukaryotic and prokaryotic hosts. Methods 2020, 180, 3–18. [Google Scholar] [CrossRef]
- Lacapere, J.J.; Pebay-Peyroula, E.; Neumann, J.M.; Etchebest, C. Determining membrane protein structures: Still a challenge. Trends Biochem. Sci. 2007, 32, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Fogeron, M.L.; Lecoq, L.; Cole, L.; Harbers, M.; Böckmann, A. Easy Synthesis of Complex Biomolecular Assemblies: Wheat Germ Cell-Free Protein Expression in Structural Biology. Front. Mol. Biosci. 2021, 8, 639587. [Google Scholar] [CrossRef]
- Bernaudat, F.; Frelet-Barrand, A.; Pochon, N.; Dementin, S.; Hivin, P.; Boutigny, S.; Rioux, J.B.; Salvi, D.; Seigneurin-Berny, D.; Richaud, P.; et al. Heterologous expression of membrane proteins: Choosing the appropriate host. PLoS ONE 2011, 6, e29191. [Google Scholar] [CrossRef] [PubMed]
- Gordon, E.; Horsefield, R.; Swarts, H.G.; de Pont, J.J.; Neutze, R.; Snijder, A. Effective high-throughput overproduction of membrane proteins in Escherichia coli. Protein Expr. Purif. 2008, 62, 1–8. [Google Scholar] [CrossRef]
- Kaur, J.; Kumar, A.; Kaur, J. Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol. 2018, 106, 803–822. [Google Scholar] [CrossRef]
- Schlegel, S.; Klepsch, M.; Gialama, D.; Wickström, D.; Slotboom, D.J.; de Gier, J.W. Revolutionizing membrane protein overexpression in bacteria. Microb. Biotechnol. 2010, 3, 403–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunji, E.R.S.; Slotboom, D.J.; Poolman, B. Lactococcus lactis as host for overproduction of functional membrane proteins. Biochim. Biophys. Acta 2003, 1610, 97–108. [Google Scholar] [CrossRef] [Green Version]
- Bakari, S.; André, F.; Seigneurin-Berny, D.; Delaforge, M.; Rolland, N.; Frelet-Barrand, A. Lactococcus lactis, recent developments in functional expression of membrane proteins. In Membrane Proteins Production for Structural Analysis; Mus-Vuteau, I., Ed.; Springer eBook: Berlin/Heidelberg, Germany, 2014; pp. 107–132. [Google Scholar]
- Gasson, M.J.; de Vos, W.M. Genetics and Biotechnology of Lactic Acid Bacteria; Blackie: London, UK, 1994. [Google Scholar]
- Mierau, I.; Olieman, K.; Mond, J.; Smid, E.J. Optimization of the Lactococcus lactis nisin-controlled gene expression system NICE for industrial applications. Microb. Cell Fact. 2005, 4, 16. [Google Scholar] [CrossRef] [Green Version]
- Morello, E.; Bermúdez-Humarán, L.G.; Llull, D.; Solé, V.; Miraglio, N.; Langella, P.; Poquet, I. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J. Mol. Microbiol. Biotechnol. 2008, 14, 48–58. [Google Scholar] [CrossRef]
- Song, A.A.; In, L.L.A.; Lim, S.H.E.; Rahim, R.A. A review on Lactococcus lactis: From food to factory. Microb. Cell Fact. 2017, 16, 55. [Google Scholar] [CrossRef] [Green Version]
- Kunji, E.R.S.; Chan, K.W.; Slotboom, D.J.; Floyd, S.; O’Connor, R.; Monné, M. Eukaryotic membrane protein overproduction in Lactococcus lactis. Curr. Opin. Biotechnol. 2005, 16, 546–551. [Google Scholar] [CrossRef] [Green Version]
- Monné, M.; Chan, K.W.; Slotboom, D.J.; Kunji, E.R.S. Functional expression of eukaryotic membrane proteins in Lactococcus lactis. Protein Sci. 2005, 14, 3048–3056. [Google Scholar] [CrossRef] [Green Version]
- van Gijtenbeek, L.A.; Robinson, A.; van Oijen, A.M.; Poolman, B.; Kok, J. On the Spatial Organization of mRNA, Plasmids, and Ribosomes in a Bacterial Host Overexpressing Membrane Proteins. PLoS Genet. 2016, 12, e1006523. [Google Scholar] [CrossRef] [PubMed]
- Mierau, I.; Kleerebezem, M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 2005, 68, 705–717. [Google Scholar] [CrossRef]
- Oliveira, A.P.; Nielsen, J.; Förster, J. Modelling Lactococcus lactis using a genome-scale flux model. BMC Microbiol. 2005, 5, 39. [Google Scholar] [CrossRef] [Green Version]
- Ingram, L.O. Changes in lipid composition of Escherichia coli resulting from growth with organic solvents and with food additives. Appl. Environ. Microbiol. 1977, 33, 1233–1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opekarova, M.; Tanner, W. Specific lipid requirements of membrane proteins—A putative bottleneck in heterologous expression. Biochim. Biophys. Acta 2003, 1610, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Surade, S.; Klein, M.; Stolt-Bergner, P.C.; Muenke, C.; Roy, A.; Michel, H. Comparative analysis and “expression space” coverage of the production of prokaryotic membrane proteins for structural genomics. Protein Sci. 2006, 15, 2178–2189. [Google Scholar] [CrossRef] [Green Version]
- Pontes, D.S.; de Azevedo, M.S.; Chatel, J.M.; Langella, P.; Azevedo, V.; Miyoshi, A. Lactococcus lactis as a live vector: Heterologous protein production and DNA delivery systems. Protein Expr. Purif. 2011, 79, 165–175. [Google Scholar] [CrossRef]
- Lubelski, J.; Rink, R.; Khusainov, R.; Moll, G.N.; Kuipers, O.P. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol. Life Sci. 2008, 65, 455–476. [Google Scholar] [CrossRef] [Green Version]
- Delves-Broughton, J.; Blackburn, P.; Evans, R.J.; Hugenholtz, J. Applications of the bacteriocin, nisin. Antonie Van Leeuwenhoek 1996, 69, 193–202. [Google Scholar] [CrossRef]
- Gasson, M.J. Genetic transfer systems in lactic acid bacteria. Antonie Van Leeuwenhoek 1983, 49, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, O.P.; de Ruyter, P.G.G.A.; Kleerebezem, M.; de Vos, W.M. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 1998, 64, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Hasper, H.E.; de Kruijff, B.; Breukink, E. Assembly and stability of nisin-lipid II pores. Biochemistry 2004, 43, 11567–11575. [Google Scholar] [CrossRef]
- de Ruyter, P.G.; Kuipers, O.P.; Beerthuyzen, M.M.; Alen-Boerrigter, I.; de Vos, W.M. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 1996, 178, 3434–3439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Ruyter, P.G.; Kuipers, O.P.; de Vos, W.M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 1996, 62, 3662–3667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.X.; Li, W.F.; Ma, G.X.; Pan, Y.J. The nisin-controlled gene expression system: Construction, application and improvements. Biotechnol. Adv. 2006, 24, 285–295. [Google Scholar] [CrossRef]
- Mu, D.; Montalbán-López, M.; Masudaa, Y.; Kuipers, O.P. Zirex: A Novel Zinc-Regulated Expression System for Lactococcus lactis. Appl. Environ. Microbiol. 2013, 79, 4503–4508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linares, D.M.; Geertsma, E.R.; Poolman, B. Evolved Lactococcus lactis strains for enhanced expression of recombinant membrane proteins. J. Mol. Biol. 2010, 401, 45–55. [Google Scholar] [CrossRef] [Green Version]
- Poquet, I.; Saint, V.; Seznec, E.; Simoes, N.; Bolotin, A.; Gruss, A. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 2000, 35, 1042–1051. [Google Scholar] [CrossRef]
- Noreen, N.; Hooi, W.Y.; Baradaran, A.; Rosfarizan, M.; Sieo, C.C.; Rosli, M.I.; Yusoff, K.; Raha, A.R. Lactococcus lactis M4, a potential host for the expression of heterologous proteins. Microb. Cell Fact. 2011, 10, 28. [Google Scholar] [CrossRef] [Green Version]
- Pinto, J.P.; Kuipers, O.P.; Marreddy, R.K.; Poolman, B.; Kok, J. Efficient overproduction of membrane proteins in Lactococcus lactis requires the cell envelope stress sensor/regulator couple CesSR. PLoS ONE 2011, 6, e21873. [Google Scholar] [CrossRef] [Green Version]
- Kuipers, O.P.; Beerthuyzen, M.M.; Siezen, R.J.; de Vos, W.M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 1993, 216, 281–291. [Google Scholar] [CrossRef]
- de Vos, W.D. Gene cloning and expression in lactic streptococci. FEMS Microbiol. Lett. 1987, 46, 281–295. [Google Scholar] [CrossRef]
- Kok, J.; van der Vossen, J.M.; Venema, G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 1984, 48, 726–731. [Google Scholar] [CrossRef] [Green Version]
- de Vos, W.M.; Simons, G.F.M. Gene cloning and expression systems in Lactococci. In Genetics and Biotechnology of Lactic Acid Bacteria; Gasson, M.J., de Vos, W.M., Eds.; Blackie Academic and Professional: London, UK, 1994. [Google Scholar]
- Geertsma, E.R.; Poolman, B. High-throughput cloning and expression in recalcitrant bacteria. Nat. Methods 2007, 4, 705–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groeneveld, M.; Weme, R.G.; Duurkens, R.H.; Slotboom, D.J. Biochemical characterization of the C4-dicarboxylate transporter DctA from Bacillus subtilis. J. Bacteriol. 2010, 192, 2900–2907. [Google Scholar] [CrossRef] [Green Version]
- Erkens, G.B.; Slotboom, D.J. Biochemical characterization of ThiT from Lactococcus lactis: A thiamin transporter with picomolar substrate binding affinity. Biochemistry 2010, 49, 3203–3212. [Google Scholar] [CrossRef]
- Steen, A.; Wiederhold, E.; Gandhi, T.; Breitling, R.; Slotboom, D.J. Physiological adaptation of the bacterium Lactococcus lactis in response to the production of human CFTR. Mol. Cell Proteom. 2011, 10, M000052MCP200. [Google Scholar] [CrossRef]
- Hartley, J.L.; Temple, G.F.; Brasch, M.A. DNA cloning using in vitro site-specific recombination. Genome Res. 2000, 10, 1788–1795. [Google Scholar] [CrossRef] [Green Version]
- Eshaghi, S.; Hedrén, M.; Nasser, M.I.; Hammarberg, T.; Thornell, A.; Nordlund, P. An efficient strategy for high-throughput expression screening of recombinant integral membrane proteins. Protein Sci. 2005, 14, 676–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yashiroda, Y.; Matsuyama, A.; Yoshida, M. New insights into chemical biology from ORFeome libraries. Curr. Opin. Chem. Biol. 2008, 12, 55–59. [Google Scholar] [CrossRef]
- Frelet-Barrand, A.; Boutigny, S.; Moyet, L.; Deniaud, A.; Seigneurin-Berny, D.; Salvi, D.; Bernaudat, F.; Richaud, P.; Pebay-Peyroula, E.; Joyard, J.; et al. Lactococcus lactis, an alternative system for functional expression of peripheral and intrinsic Arabidopsis membrane proteins. PLoS ONE 2010, 5, e8746. [Google Scholar] [CrossRef]
- Bakari, S.; Lembrouk, M.; André, F.; Orlowski, S.; Delaforge, M.; Frelet-Barrand, A. Expression in Lactococcus lactis of two human membrane proteins involved in liver detoxification, cytochrome P450 3A4 and microsomal glutathione S-transferase MGST1. Mol. Biotechnol. 2016, 58, 299–310. [Google Scholar] [CrossRef]
- Douillard, F.P.; Mahony, J.; Campanacci, V.; Cambillau, C.; van Sinderen, D. Construction of two Lactococcus lactis expression vectors combining the Gateway and the NIsin Controlled Expression systems. Plasmid 2011, 66, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.; Klumpp, J.; Mahony, J.; O’Connell-Motherway, M.; Nauta, A.; van Sinderen, D. Methyltransferases acquired by lactococcal 936-type phage provide protection against restriction endonuclease activity. BMC Genom. 2014, 15, 831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berlec, A.; Štrukelj, B. Generating a custom TA-cloning expression plasmid for Lactococcus lactis. Biotechniques 2012, 52, 51–53. [Google Scholar] [CrossRef] [PubMed]
- Berlec, A.; Škrlec, K.; Kocjan, J.; Olenic, M.; Štrukelj, B. Single plasmid systems for inducible dual protein expression and for CRISPR-Cas9/CRISPRi gene regulation in lactic acid bacterium Lactococcus lactis. Sci. Rep. 2018, 8, 1009. [Google Scholar] [CrossRef] [PubMed]
- Plavec, T.V.; Mitrović, A.; Perišić Nanut, M.; Štrukelj, B.; Kos, J.; Berlec, A. Targeting of fluorescent Lactococcus lactis to colorectal cancer cells through surface display of tumour-antigen binding proteins. Microb. Biotechnol. 2021, 14, 2227–2240. [Google Scholar] [CrossRef]
- Noens, E.E.; Lolkema, J.S. Physiology and substrate specificity of two closely related amino acid transporters, SerP1 and SerP2, of Lactococcus lactis. J. Bacteriol. 2015, 197, 951–958. [Google Scholar] [CrossRef] [Green Version]
- Pols, T.; Singh, S.; Deelman-Driessen, C.; Gaastra, B.F.; Poolman, B. Enzymology of the pathway for ATP production by arginine breakdown. FEBS J. 2021, 288, 293–309. [Google Scholar] [CrossRef]
- Berntsson, R.P.; ter Beek, J.; Majsnerowska, M.; Duurkens, R.H.; Puri, P.; Poolman, B.; Slotboom, D.J. Structural divergence of paralogous S components from ECF-type ABC transporters. Proc. Natl. Acad. Sci. USA 2012, 109, 13990–13995. [Google Scholar] [CrossRef] [Green Version]
- Marreddy, R.K.R.; Geertsma, E.R.; Poolman, B. Recombinant Membrane Protein Production: Past, Present and Future. In Supramolecular Structure and Function; Brnjas-Kraljević, J., Pifat-Mrzljak, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Pudlik, A.M.; Lolkema, J.S. Rerouting citrate metabolism in Lactococcus lactis to citrate-driven transamination. Appl. Environ. Microbiol. 2012, 78, 6665–6673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filipic, B.; Golic, N.; Jovcic, B.; Tolinacki, M.; Bay, D.C.; Turner, R.J.; Antic-Stankovic, J.; Kojic, M.; Topisirovic, L. The cmbT gene encodes a novel major facilitator multidrug resistance transporter in Lactococcus lactis. Res. Microbiol. 2013, 164, 46–54. [Google Scholar] [CrossRef]
- Fulyani, F.; Schuurman-Wolters, G.K.; Slotboom, D.J.; Poolman, B. Relative Rates of Amino Acid Import via the ABC Transporter GlnPQ Determine the Growth Performance of Lactococcus lactis. J. Bacteriol. 2016, 198, 477–485. [Google Scholar] [CrossRef] [Green Version]
- Venter, H.; Shilling, R.A.; Velamakanni, S.; Balakrishnan, L.; Van Veen, H.W. An ABC transporter with a secondary-active multidrug translocator domain. Nature 2003, 426, 866–870. [Google Scholar] [CrossRef]
- Lubelski, J.; de Jong, A.; van Merkerk, R.; Agustiandari, H.; Kuipers, O.P.; Kok, J.; Driessen, A.J. LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol. Microbiol. 2006, 61, 771–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaedler, T.A.; Tong, Z.; van Veen, H.W. The multidrug transporter LmrP protein mediates selective calcium efflux. J. Biol. Chem. 2012, 287, 27682–27690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debruycker, V.; Hutchin, A.; Masureel, M.; Ficici, E.; Martens, C.; Legrand, P.; Stein, R.A.; Mchaourab, H.S.; Faraldo-Gómez, J.D.; Remaut, H.; et al. An embedded lipid in the multidrug transporter LmrP suggests a mechanism for polyspecificity. Nat. Struct. Mol. Biol. 2020, 27, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Swain, B.M.; Guo, D.; Singh, H.; Rawlins, P.B.; McAlister, M.; van Veen, H.W. Complexities of a protonatable substrate in measurements of Hoechst 33342 transport by multidrug transporter LmrP. Sci. Rep. 2020, 10, 20026. [Google Scholar] [CrossRef]
- Folgering, J.H.; Moe, P.C.; Schuurman-Wolters, G.K.; Blount, P.; Poolman, B. Lactococcus lactis uses MscL as its principal mechanosensitive channel. J. Biol. Chem. 2005, 280, 8784–8792. [Google Scholar] [CrossRef] [Green Version]
- Tassis, K.; Vietrov, R.; de Koning, M.; de Boer, M.; Gouridis, G.; Cordes, T. Single-molecule studies of conformational states and dynamics in the ABC importer OpuA. FEBS Lett. 2021, 595, 717–734. [Google Scholar] [CrossRef]
- Duurkens, R.H.; Tol, M.B.; Geertsma, E.R.; Permentier, H.P.; Slotboom, D.J. Flavin binding to the high affinity riboflavin transporter RibU. J. Biol. Chem. 2007, 282, 10380–10386. [Google Scholar] [CrossRef] [Green Version]
- Noens, E.E.; Kaczmarek, M.B.; Żygo, M.; Lolkema, J.S. ArcD1 and ArcD2 Arginine/Ornithine Exchangers Encoded in the Arginine Deiminase Pathway Gene Cluster of Lactococcus lactis. J. Bacteriol. 2015, 197, 3545–3553. [Google Scholar] [CrossRef] [Green Version]
- Margolles, A.; Flórez, A.B.; Moreno, J.A.; van Sinderen, D.; de los Reyes-Gavilán, C.G. Two membrane proteins from Bifidobacterium breve UCC2003 constitute an ABC-type multidrug transporter. Microbiology 2006, 152, 3497–3505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Liu, X.; O’Sullivan, D.J. Use of Lactococcus lactis as a production system for peptides and enzymes encoded by a Lantibiotic gene cluster from Bifidobacterium longum. Microbiology 2018, 164, 1481–1490. [Google Scholar] [CrossRef]
- Xu, Q.; Zhai, Z.; An, H.; Yang, Y.; Yin, J.; Wang, G.; Ren, F.; Hao, Y. The MarR Family Regulator BmrR Is Involved in Bile Tolerance of Bifidobacterium longum BBMN68 via Controlling the Expression of an ABC Transporter. Appl. Environ. Microbiol. 2019, 85, e02453-18. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; An, H.; Zhang, J.; Zhou, H.; Ren, F.; Hao, Y. Functional role of tlyC1 encoding a hemolysin-like protein from Bifidobacterium longum BBMN68 in bile tolerance. FEMS Microbiol. Lett. 2014, 360, 167–173. [Google Scholar] [CrossRef] [Green Version]
- Rezaei, M.; Rabbani Khorasgani, M.; Zarkesh Esfahani, S.H.; Emamzadeh, R.; Abtahi, H. Production of Brucella melitensis Omp16 protein fused to the human interleukin 2 in Lactococcus lactis MG1363 toward developing a Lactococcus-based vaccine against brucellosis. Can. J. Microbiol. 2020, 66, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Woebking, B.; Reuter, G.; Shilling, R.A.; Velamakanni, S.; Shahi, S.; Venter, H.; Balakrishnan, L.; van Veen, H.W. Drug-lipid A interactions on the Escherichia coli ABC transporter MsbA. J. Bacteriol. 2005, 187, 6363–6369. [Google Scholar] [CrossRef] [Green Version]
- Hürlimann, L.M.; Corradi, V.; Hohl, M.; Bloemberg, G.V.; Tieleman, D.P.; Seeger, M.A. The Heterodimeric ABC Transporter EfrCD Mediates Multidrug Efflux in Enterococcus faecalis. Antimicrob. Agents Chemother. 2016, 60, 5400–5411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Wang, C.; Cheng, W.; Duan, G.; Shi, Q.; Chen, S.; Fan, Q. Delivery of Helicobacter pylori HpaA to gastrointestinal mucosal immune sites using Lactococcus lactis and its immune efficacy in mice. Biotechnol. Lett. 2018, 40, 585–590. [Google Scholar] [CrossRef]
- Sakamoto, K.; Margolles, A.; van Veen, H.W.; Konings, W.N. Hop resistance in the beer spoilage bacterium Lactobacillus brevis is mediated by the ATP-binding cassette multidrug transporter HorA. J. Bacteriol. 2001, 183, 5371–5375. [Google Scholar] [CrossRef] [Green Version]
- Majsnerowska, M.; Hänelt, I.; Wunnicke, D.; Schäfer, L.V.; Steinhoff, H.J.; Slotboom, D.J. Substrate-induced conformational changes in the S-component ThiT from an energy coupling factor transporter. Structure 2013, 21, 861–867. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Zuo, F.; Yu, R.; Zeng, Z.; Ma, H.; Chen, S. Comparative genome-based identification of a cell wall-anchored protein from Lactobacillus plantarum increases adhesion of Lactococcus lactis to human epithelial cells. Sci. Rep. 2015, 5, 14109. [Google Scholar] [CrossRef]
- Martín, C.; Escobedo, S.; Pérez-Martínez, G.; Coll-Marqués, J.M.; Martín, R.; Suárez, J.E.; Quirós, L.M. Two alkaline motifs in the Lactobacillus salivarius Lv72 OppA surface are important to its adhesin function. Benef. Microbes 2019, 10, 101–109. [Google Scholar] [CrossRef]
- Hohl, M.; Remm, S.; Eskandarian, H.A.; Dal Molin, M.; Arnold, F.M.; Hürlimann, L.M.; Krügel, A.; Fantner, G.E.; Sander, P.; Seeger, M.A. Increased drug permeability of a stiffened mycobacterial outer membrane in cells lacking MFS transporter Rv1410 and lipoprotein LprG. Mol. Microbiol. 2019, 111, 1263–1282. [Google Scholar] [CrossRef] [PubMed]
- Rego, S.; Heal, T.J.; Pidwill, G.R.; Till, M.; Robson, A.; Lamont, R.J.; Sessions, R.B.; Jenkinson, H.F.; Race, P.R.; Nobbs, A.H. Structural and Functional Analysis of Cell Wall-anchored Polypeptide Adhesin BspA in Streptococcus agalactiae. J. Biol. Chem. 2016, 291, 15985–16000. [Google Scholar] [CrossRef] [Green Version]
- Velamakanni, S.; Yao, Y.; Gutmann, D.A.; van Veen, H.W. Multidrug transport by the ABC transporter Sav1866 from Staphylococcus aureus. Biochemistry 2008, 47, 9300–9308. [Google Scholar] [CrossRef] [PubMed]
- Freires, I.A.; Avilés-Reyes, A.; Kitten, T.; Simpson-Haidaris, P.J.; Swartz, M.; Knight, P.A.; Rosalen, P.L.; Lemos, J.A.; Abranches, J. Heterologous expression of Streptococcus mutans Cnm in Lactococcus lactis promotes intracellular invasion, adhesion to human cardiac tissues and virulence. Virulence 2017, 8, 18–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asmat, T.M.; Klingbeil, K.; Jensch, I.; Burchhardt, G.; Hammerschmidt, S. Heterologous expression of pneumococcal virulence factor PspC on the surface of Lactococcus lactis confers adhesive properties. Microbiology 2012, 158, 771–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, R.; Kim, B.J.; Paco, C.; Del Rosario, Y.; Courtney, H.S.; Doran, K.S. Identification of a group B streptococcal fibronectin binding protein, SfbA, that contributes to invasion of brain endothelium and development of meningitis. Infect. Immun. 2014, 82, 2276–2286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hohl, M.; Briand, C.; Grütter, M.G.; Seeger, M.A. Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat. Struct. Mol. Biol. 2012, 19, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Schaedler, T.A.; Thornton, J.D.; Kruse, I.; Schwarzländer, M.; Meyer, A.J.; van Veen, H.W.; Balk, J. A conserved mitochondrial ATP-binding cassette transporter exports glutathione polysulfide for cytosolic metal cofactor assembly. J. Biol. Chem. 2014, 289, 23264–23274. [Google Scholar] [CrossRef] [Green Version]
- Colinet, A.S.; Sengottaiyan, P.; Deschamps, A.; Colsoul, M.L.; Thines, L.; Demaegd, D.; Duchêne, M.C.; Foulquier, F.; Hols, P.; Morsomme, P. Yeast Gdt1 is a Golgi-localized calcium transporter required for stress-induced calcium signaling and protein glycosylation. Sci. Rep. 2016, 6, 24282. [Google Scholar] [CrossRef] [Green Version]
- Hofacker, M.; Gompf, S.; Zutz, A.; Presenti, C.; Haase, W.; van der Does, C.; Model, K.; Tampé, R. Structural and functional fingerprint of the mitochondrial ATP-binding cassette transporter Mdl1 from Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 3951–3961. [Google Scholar] [CrossRef]
- Vest, K.E.; Leary, S.C.; Winge, D.R.; Cobine, P.A. Copper Import into the Mitochondrial Matrix in Saccharomyces cerevisiae is Mediated by Pic2, a Mitochondrial Carrier Family Protein. J. Biol. Chem. 2013, 288, 23884–23892. [Google Scholar] [CrossRef] [Green Version]
- Furumoto, T. Pyruvate transport systems in organelles: Future directions in C4 biology research. Curr. Opin. Plant. Biol. 2016, 31, 143–148. [Google Scholar] [CrossRef]
- Herzig, S.; Raemy, E.; Montessuit, S.; Veuthey, J.L.; Zamboni, N.; Westermann, B.; Kunji, E.R.; Martinou, J.C. Identification and functional expression of the mitochondrial pyruvate carrier. Science 2012, 337, 93–96. [Google Scholar] [CrossRef]
- Taochy, C.; Gaillard, I.; Ipotesi, E.; Oomen, R.; Leonhardt, N.; Zimmermann, S.; Peltier, J.B.; Szponarski, W.; Simonneau, T.; Sentenac, H.; et al. The Arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress. Plant J. 2015, 83, 466–479. [Google Scholar] [CrossRef]
- Marreddy, R.K.; Pinto, J.P.; Wolters, J.C.; Geertsma, E.R.; Fusetti, F.; Permentier, H.P.; Kuipers, O.P.; Kok, J.; Poolman, B. The response of Lactococcus lactis to membrane protein production. PLoS ONE 2011, 6, e24060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monné, M.; Robinson, A.J.; Boes, C.; Harbour, M.E.; Fearnley, I.M.; Kunji, E.R. The mimivirus genome encodes a mitochondrial carrier that transports dATP and dTTP. J. Virol. 2007, 81, 3181–3186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janvilisri, T.; Venter, H.; Shahi, S.; Reuter, G.; Balakrishnan, L.; van Veen, H.W. Sterol transport by the human breast cancer resistance protein (ABCG2) expressed in Lactococcus lactis. J. Biol. Chem. 2003, 278, 20645–20651. [Google Scholar] [CrossRef] [Green Version]
- Stribny, J.; Thines, L.; Deschamps, A.; Goffin, P.; Morsomme, P. The human Golgi protein TMEM165 transports calcium and manganese in yeast and bacterial cells. J. Biol. Chem. 2020, 295, 3865–3874. [Google Scholar] [CrossRef]
- Mifsud, J.; Ravaud, S.; Krammer, E.M.; Chipot, C.; Kunji, E.R.; Pebay-Peyroula, E.; Dehez, F. The substrate specificity of the human ADP/ATP carrier AAC1. Mol. Membr. Biol. 2013, 30, 160–168. [Google Scholar] [CrossRef] [PubMed]
- King, M.S.; Thompson, K.; Hopton, S.; He, L.; Kunji, E.R.S.; Taylor, R.W.; Ortiz-Gonzalez, X.R. Expanding the phenotype of de novo SLC25A4-linked mitochondrial disease to include mild myopathy. Neurol. Genet. 2018, 4, e256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Tian, D.; Matsuyama, H.; Hamazaki, T.; Shiratsuchi, T.; Terada, N.; Hook, D.J.; Walters, M.A.; Georg, G.I.; Hawkinson, J.E. Human Adenine Nucleotide Translocase (ANT) Modulators Identified by High-Throughput Screening of Transgenic Yeast. J. Biomol. Screen. 2016, 21, 381–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulet, A.; Vest, K.E.; Maynard, M.K.; Gammon, M.G.; Russell, A.C.; Mathews, A.T.; Cole, S.E.; Zhu, X.; Phillips, C.B.; Kwong, J.Q.; et al. The mammalian phosphate carrier SLC25A3 is a mitochondrial copper transporter required for cytochrome c oxidase biogenesis. J. Biol. Chem. 2018, 293, 1887–1896. [Google Scholar] [CrossRef] [Green Version]
- Schleifer, K.H.; Kraus, J.; Dvorak, C.; Kilpper-Bälz, R.; Collins, M.D.; Fischer, W. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst. Appl. Microbiol. 1985, 6, 183–195. [Google Scholar] [CrossRef]
- Marreddy, R.K.; Geertsma, E.R.; Permentier, H.P.; Pinto, J.P.; Kok, J.; Poolman, B. Amino acid accumulation limits the overexpression of proteins in Lactococcus lactis. PLoS ONE 2010, 5, e10317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shilling, R.; Federici, L.; Walas, F.; Venter, H.; Velamakanni, S.; Woebking, B.; Balakrishnan, L.; Luisi, B.; van Veen, H.W. A critical role of a carboxylate in proton conduction by the ATP-binding cassette multidrug transporter LmrA. FASEB J. 2005, 19, 1698–1700. [Google Scholar] [CrossRef]
- Agboh, K.; Lau, C.H.F.; Khoo, Y.S.K.; Singh, H.; Raturi, S.; Nair, A.V.; Howard, J.; Chiapello, M.; Feret, R.; Deery, M.J.; et al. Powering the ABC multidrug exporter LmrA: How nucleotides embrace the ion-motive force. Sci. Adv. 2018, 4, eaas9365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellmich, U.A.; Glaubitz, C. NMR and EPR studies of membrane transporters. Biol. Chem. 2009, 390, 815–834. [Google Scholar] [CrossRef]
- Hellmich, U.A.; Lyubenova, S.; Kaltenborn, E.; Doshi, R.; van Veen, H.W.; Prisner, T.F.; Glaubitz, C. Probing the ATP hydrolysis cycle of the ABC multidrug transporter LmrA by pulsed EPR spectroscopy. J. Am. Chem. Soc. 2012, 134, 5857–5862. [Google Scholar] [CrossRef]
- Hellmich, U.A.; Mönkemeyer, L.; Velamakanni, S.; van Veen, H.W.; Glaubitz, C. Effects of nucleotide binding to LmrA: A combined MAS-NMR and solution NMR study. Biochim. Biophys. Acta 2015, 1848, 3158–3165. [Google Scholar] [CrossRef] [Green Version]
- Erkens, G.B.; Berntsson, R.P.; Fulyani, F.; Majsnerowska, M.; Vujičić-Žagar, A.; Ter Beek, J.; Poolman, B.; Slotboom, D.J. The structural basis of modularity in ECF-type ABC transporters. Nat. Struct. Mol. Biol. 2011, 18, 755–760. [Google Scholar] [CrossRef]
- Swier, L.J.; Guskov, A.; Slotboom, D.J. Structural insight in the toppling mechanism of an energy-coupling factor transporter. Nat. Commun. 2016, 7, 11072. [Google Scholar] [CrossRef]
- Woebking, B.; Velamakanni, S.; Federici, L.; Seeger, M.A.; Murakami, S.; van Veen, H.W. Functional role of transmembrane helix 6 in drug binding and transport by the ABC transporter MsbA. Biochemistry 2008, 47, 10904–10914. [Google Scholar] [CrossRef]
- Doshi, R.; Woebking, B.; van Veen, H.W. Dissection of the conformational cycle of the multidrug/lipidA ABC exporter MsbA. Proteins 2010, 78, 2867–2872. [Google Scholar] [CrossRef]
- Doshi, R.; van Veen, H.W. Substrate Binding Stabilizes a Pre-translocation Intermediate in the ATP-binding Cassette Transport Protein MsbA. J. Biol. Chem. 2013, 288, 21638–21647. [Google Scholar] [CrossRef] [Green Version]
- Trip, H.; Mulder, N.L.; Lolkema, J.S. Cloning, expression, and functional characterization of secondary amino acid transporters of Lactococcus lactis. J. Bacteriol. 2013, 195, 340–350. [Google Scholar] [CrossRef] [Green Version]
- Ter Horst, R.; Lolkema, J.S. Rapid screening of membrane topology of secondary transport proteins. Biochim. Biophys. Acta 2010, 1798, 672–680. [Google Scholar] [CrossRef] [Green Version]
- Halestrap, A.P. Stimulation of pyruvate transport in metabolizing mitochondria through changes in the transmembrane pH gradient induced by glucagon treatment of rats. Biochem. J. 1978, 172, 389–398. [Google Scholar] [CrossRef] [Green Version]
- Miras, S.; Salvi, D.; Ferro, M.; Grunwald, D.; Garin, J.; Joyard, J.; Rolland, N. Non-canonical transit peptide for import into the chloroplast. J. Biol. Chem. 2002, 277, 47770–47778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kühlbrandt, W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 2004, 5, 282–295. [Google Scholar] [CrossRef]
- Catty, P.; Boutigny, S.; Miras, R.; Joyard, J.; Rolland, N.; Seigneurin-Berny, D. Biochemical characterization of AtHMA6/PAA1, a chloroplast envelope Cu(I)-ATPase. J. Biol. Chem. 2011, 286, 36188–36197. [Google Scholar] [CrossRef] [Green Version]
- Neuhaus, H.E.; Thom, E.; Möhlmann, T.; Steup, M.; Kampfenkel, K. Characterization of a novel eukaryotic ATP/ADP translocator located in the plastid envelope of Arabidopsis thaliana L. Plant J. 1997, 11, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Tjaden, J.; Schwöppe, C.; Möhlmann, T.; Quick, P.W.; Neuhaus, H.E. Expression of a plastidic ATP/ADP transporter gene in Escherichia coli leads to a functional adenine nucleotide transport system in the bacterial cytoplasmic membrane. J. Biol. Chem. 1998, 273, 9630–9636. [Google Scholar] [CrossRef] [Green Version]
- Hostetler, K.Y.; Van den Bosch, H.; Van Deenen, L.L. Biosynthesis of cardiolipin in liver mitochondria. Biochim. Biophys. Acta 1971, 239, 113–119. [Google Scholar] [CrossRef] [Green Version]
- Block, M.A.; Douce, R.; Joyard, J.; Rolland, N. Chloroplast envelope membranes: A dynamic interface between plastids and the cytosol. Photosynth. Res. 2007, 92, 225–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolters, J.C.; Berntsson, R.P.; Gul, N.; Karasawa, A.; Thunnissen, A.M.; Slotboom, D.J.; Poolman, B. Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA. PLoS ONE 2010, 5, e10361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swier, L.J.; Monjas, L.; Guskov, A.; de Voogd, A.R.; Erkens, G.B.; Slotboom, D.J.; Hirsch, A.K. Structure-based design of potent small-molecule binders to the S-component of the ECF transporter for thiamine. ChemBioChem. 2015, 16, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Sikkema, H.R.; van den Noort, M.; Rheinberger, J.; de Boer, M.; Krepel, S.T.; Schuurman-Wolters, G.K.; Paulino, C.; Poolman, B. Gating by ionic strength and safety check by cyclic-di-AMP in the ABC transporter OpuA. Sci. Adv. 2020, 6, eabd7697. [Google Scholar] [CrossRef] [PubMed]
- Jäger, F.; Lamy, A.; Guerini, N.; Sun, W.S.; Berntsson, R.P.A. Structure of the enterococcal T4SS protein PrgL reveals unique dimerization interface in the VirB8 protein family. bioRxiv 2020. [Google Scholar] [CrossRef]
- Focht, D.; Neumann, C.; Lyons, J.; Eguskiza Bilbao, A.; Blunck, R.; Malinauskaite, L.; Schwarz, I.O.; Javitch, J.A.; Quick, M.; Nissen, P. A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition. EMBO J. 2021, 40, e105164. [Google Scholar] [CrossRef]
- Ploetz, E.; Schuurman-Wolters, G.K.; Zijlstra, N.; Jager, A.W.; Griffith, D.A.; Guskov, A.; Gouridis, G.; Poolman, B.; Cordes, T. Structural and biophysical characterization of the tandem substrate-binding domains of the ABC importer GlnPQ. Open Biol. 2021, 11, 200406. [Google Scholar] [CrossRef]
- Harborne, S.P.; Ruprecht, J.J.; Kunji, E.R. Calcium-induced conformational changes in the regulatory domain of the human mitochondrial ATP-Mg/Pi carrier. Biochim. Biophys. Acta 2015, 1847, 1245–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Januliene, D.; Moeller, A. Single-Particle Cryo-EM of Membrane Proteins. Methods Mol. Biol. 2021, 2302, 153–178. [Google Scholar] [CrossRef] [PubMed]
- Berntsson, R.P.; Alia Oktaviani, N.; Fusetti, F.; Thunnissen, A.M.; Poolman, B.; Slotboom, D.J. Selenomethionine incorporation in proteins expressed in Lactococcus lactis. Protein Sci. 2009, 18, 1121–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martens, C. Membrane Protein Production in Lactococcus lactis for Structural Studies. Methods Mol. Biol. 2020, 2127, 29–45. [Google Scholar] [CrossRef]

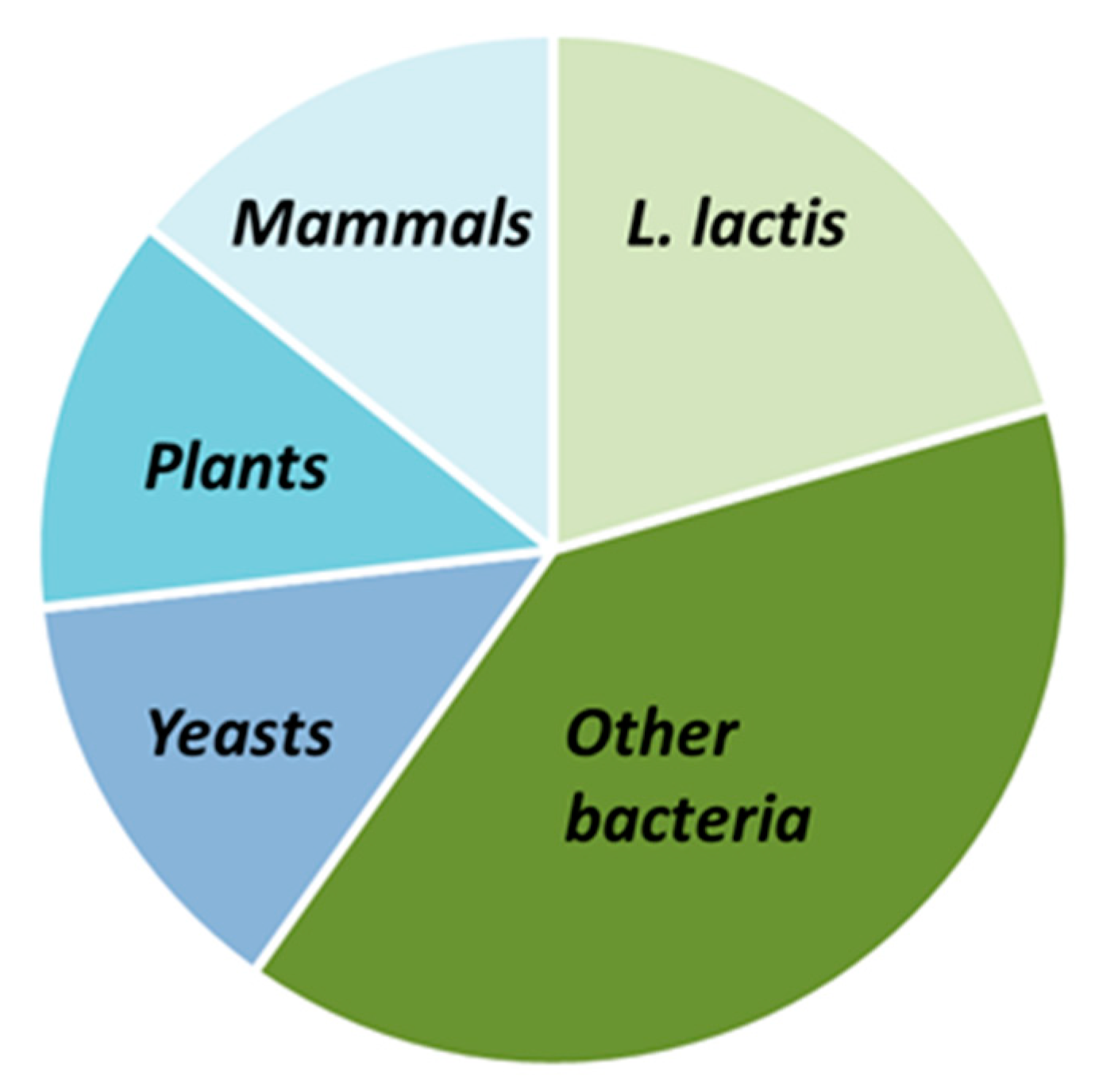


| Characteristics | References | ||
|---|---|---|---|
| Strains | |||
| L. lactis | NZ9700 | Progeny of the conjugation between nisin producer strain NIZO B8 and MG1614 (RifR StrpR derivative of MG1363). Nisin producer strain for nisin-induced gene expression. | [11,29,39] |
| NZ9800 | Derivative of NZ9700 with deletion of 4 bp in the nisA gene. No nisin production but nisRK-transcribed. Host of NICE system. | [29,39] | |
| NZ9000 | MG1363 strain with nisRK integrated into pepN gene. Most commonly used host for NICE system. | [29] | |
| NZ9100 | MG1363 strain with nisR and nisK integrated into a neutral locus. Standard host strain of NICE. | Mobitec Molecular Biotechnology | |
| DML1 | NZ9000 strain transformed with pNZ-X-GFP-EmrC and selected by increased concentration of erythromycin. | [35] | |
| Plasmids. | |||
| pNZ8048 | NcoI site used for translational fusions, CmR. | [29] | |
| pNZ8148 | pNZ8048 with deletion of 60 bp DNA from B. subtilis, CmR. | [20] | |
| pNZ8149 | pNZ8048 with NcoI site for translational fusions; lacF for food grade selection for growth on lactose. | Mobitec Molecular Biotechnology | |
| pNZ8150 | pNZ8148 with ScaI site used for translational fusions, CmR. | [20] | |
| pNZ8151 | pNZ8148 with ScaI site used for translational fusions, lacF. | Mobitec Molecular Biotechnology | |
| pNZ8152 | pNZ8148 with ScaI site used for translational fusions, alr gene for food grade selection. | Mobitec Molecular Biotechnology | |
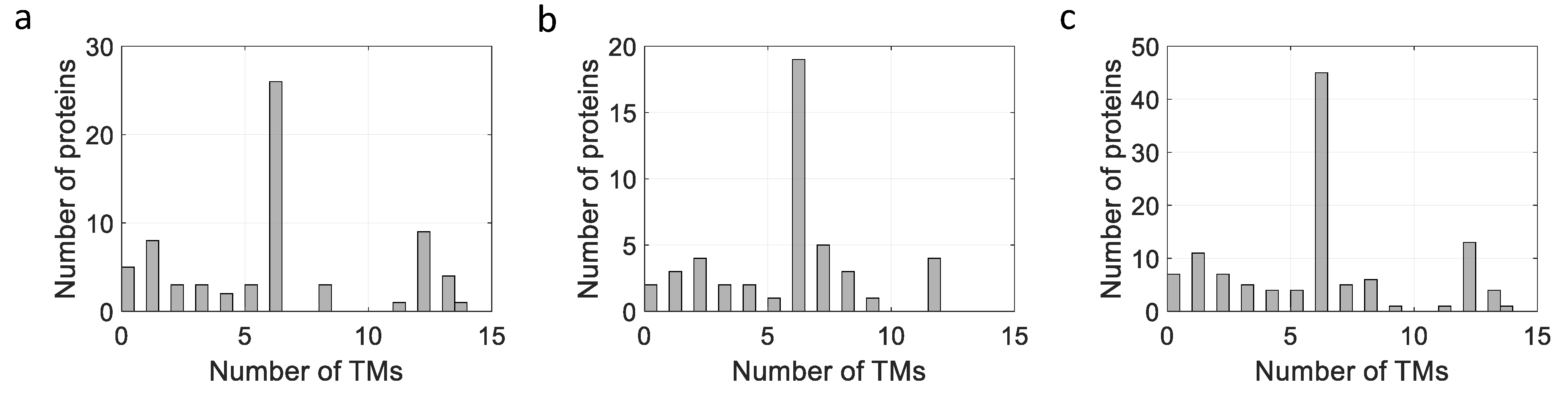
| Protein | Function | Size (kDa) a | TM Helices b | Expression Level c | References |
|---|---|---|---|---|---|
| ArcD1 | arginine/ornithine antiporter | 52.6 | 13 | - | [57] |
| ArcD2 | arginine/ornithine antiporter | 54 | 13 | - | [57,58] |
| BcaP | branched-chain amino acid permease | 50 | 12 | 20% | [38] |
| BioY | biotin transporter | 20.5 | 5 | 5% | [59] |
| ChoS | glycine betaine ABC transporter permease | 55.1 | 5 | 2% | [60] |
| CitP | citrate sodium symporter | 48.6 | 13 | 1–2% | [61] |
| CmbT | MFS transporter | 50 | 12 | <1% | [62] |
| DtpT | di-/tripeptide transporter | 54.8 | 12 | 10% | [11] |
| GlnP | ABC transporter | 78.5 | 3 | <1% | [60,63] |
| GlnQ | glutamine transport ATP- binding | 27 | 8 | 2–5% | |
| LmrA | ABC efflux pump | 65 | 6 | 30% | [64] |
| LmrCD | ABC transporter | 63 + 73.7 | 6 + 6 | 5–10% | [65] |
| LmrP | MFS efflux pump | 45 | 12 | 5% | [66,67,68] |
| MleP | MFS transporter | 46.7 | 11 | 1–2% | [11] |
| MscL | large-conductance mechanosensitive channel | 13.8 | 2 | 5–10% | [69] |
| OppB | ABC transporter with OpuC,D,F | 35.1 | 6 | <1% | [11] |
| OppC | ABC transporter with OpuB,D,F | 32.3 | 6 | <1% | |
| OpuABC | ABC transporter with OpuAA | 63 | 8 | 10% | [11,70] |
| RibU | riboflavin transporter | 23 | 6 | 5% | [71] |
| SerP1 | serine permease | 51.3 | 12 | - | [72] |
| SerP2 | DL-alanine permease | 51.5 | 12 | - | |
| ThiT | thiamine transporter | 20 | 6 | 2% | [45] |
| Protein | Function | Size (kDa) a | TM Helices b | Organism c | Expression Level d | References |
|---|---|---|---|---|---|---|
| abcA | ABC transporter | 70 | 6 | B. breve | 1% | [73] |
| abcB | ABC transporter | 66 | 6 | 5–10% | ||
| LanR1 | lantibiotic response regulator | 24 | 6 | B. longum | - | [74] |
| LanI | ABC transporter | 32.76 | - | - | ||
| LanT | lantibiotic transporter | 80.1 | 6 | - | ||
| BmrA | ABC transporter | 65.3 | 6 | 5–10% | [75] | |
| tlyC1 | hemolysin-like protein | 11.2 | 2 | - | [76] | |
| Omp16 | Peptidoglycan-associated lipoprotein | 18.2 | p | B. melitensis | - | [77] |
| DctA | C4-dicarboxylate transport | 45.4 | 8 | B. subtilis | 0.5–1% | [44] |
| CA_C2849 | proline/glycine betaine ABC-type transport system, permease | 57.6 | 6 | C. acetobutylicum | 2% | [60] |
| MsbA | lipid A export ATP-binding/ permease | 64.5 | 6 | E. coli | 20–30% | [78] |
| EfrA | ABC transporter | 56.3 | 4 | E. faecalis | - | [79] |
| EfrB | ABC transporter | 60.54 | 3 | - | ||
| Jhp0757 | putative osmoprotection binding protein | 62.6 | 6 | H. pylori | 1% | [60] |
| HpaA | neuraminyllactose-binding hemagglutinin | 29.1 | p | 25–30% | [80] | |
| HorA | Multidrug transporter | 64.2 | 5 | L. brevis | 30% | [81] |
| ArcD | arginine/ornithine exchangers | 51.9 | 13 | - | [82] | |
| Lin0840 | ABC transporter | 53.2 | 6 | L. innocua | <1% | [60] |
| Lin1461 | binding-protein-dependent transport system permease | 55.7 | 6 | 2% | ||
| Lin2352 | ABC transporter | 53.4 | 6 | 1% | ||
| Lmo1422 | binding-protein-dependent transport system permease | 55.7 | 6 | L. monocytogenes | 1% | [60] |
| Lmo2250 | ABC transporter | 53.1 | 6 | 2% | ||
| cwaA | cell wall-anchored adhesion-associated protein | 93.7 | 2 | L. plantarum | - | [83] |
| OppA | oligopeptide-binding protein | 59.7 | p | L. salivarius | - | [84] |
| XylP | xylose-proton symporter | 52.7 | 12 | Lb. pentosus | 20% | [11] |
| Rv1410 | MFS transporter | 54.7 | 14 | M. smegmatis | - | [85] |
| CYP201A2 | cytochrome-mono-oxygenase | 49.7 | p | R. palustris | 1.5% | [7] |
| TlcA,B,C | ATP/ADP translocator | 56.8 | 12 | R. prowazekii | 5–10% | [11] |
| NapC | cytochrome-electron transfer | 25.6 | 1 | R. sphaeroides | 0.5% | [7] |
| BspA | Gram+ anchoring domain containing protein | 101 | 1 | S. agalactiae | - | [86] |
| SAR1949 | putative extracellular glutamine-binding protein | 53.1 | 4 | S. aureus | 1% | [60] |
| Sav1866 | multidrug export ATP- binding/permease | 64.8 | 6 | 20–25% | [87] | |
| Cnm | collagen and laminin-binding glycoprotein | 58 | 1 | S. mutans | - | [88] |
| PspC | Choline-binding protein | 85.24 | 1 | S. pneumoniae | - | [89] |
| MreC | peptidoglycan synthesis | 32 | 1 | 1% | [7] | |
| ProWX | ABC transporter permease- choline transporter | 55.5 | 6 | 2–3% | [60] | |
| SP_0453 | ABC transporter, AA-binding protein/permease protein | 57.4 | 6 | <1% | ||
| SP_1241 | ABC transporter, AA-binding protein/permease protein | 78.4 | 3 | <1% | ||
| LacS | MFS transporter | 56.6 | 12 | S. thermophilus | 1–2% | [11] |
| SfbA/FbaA | streptococcal fibronectin-binding protein A | 37.8 | 1 | Streptococcus | - | [90] |
| SfbI | Fibronecting-binding protein | 67.3 | 1 | - | ||
| TM287/288 | ABC transporter | 60 + 60 | 6 + 6 | T. maritima | 0.5–1% | [91] |
| Protein | Function | Size (kDa) a | TM Helices b | Organism c | Expression Level d | References |
|---|---|---|---|---|---|---|
| ATM1 | mitochondrial iron-sulfur cluster transporter | 77.5 | 6 | S. cerevisiae | - | [92] |
| GDT1 | cation exchanger (homologous to TMEM) | 30.3 | 7 | S. cerevisiae | - | [93] |
| CTP1 | tricarboxylate transport protein | 32.9 | 6 | S. cerevisiae | 5% | [18] |
| SAM5 | mitochondrial S-adenosyl methionine carrier | 30.9 | 4 | S. cerevisiae | <1% | |
| Mdl1 | mitochondrial ATP-dependent permease | 76 | 5 | S. cerevisiae | <0.1% | [94] |
| MIR1 | mitochondrial phosphate carrier protein | 32.8 | 6 | S. cerevisiae | <1% | [18] |
| DIC1 | mitochondrial dicarboxylate transporter | 33 | 6 | S. cerevisiae | 10% | |
| GGC1 | mitochondrial GTP/GDP carrier protein | 33.2 | 6 | S. cerevisiae | 4% | |
| PIC2 | mitochondrial phosphate carrier protein 2 | 33.5 | 6 | S. cerevisiae | 1–2% | [95] |
| AAC3 | mitochondrial ADP/ATP carrier protein 3 | 33.7 | 6 | S. cerevisiae | 5% | [11] |
| ODC2 | mitochondrial 2-oxodicarboxylate carrier 2 | 34 | 6 | S. cerevisiae | 10% | [18] |
| AAC1 | mitochondrial ADP/ATP carrier protein 1 | 34.1 | 6 | S. cerevisiae | <1% | |
| ODC1 | mitochondrial 2-oxodicarboxylate carrier 1 | 34.2 | 6 | S. cerevisiae | 8% | |
| AAC2 | mitochondrial ADP/ATP carrier protein 2 | 34.4 | 6 | S. cerevisiae | <1% | |
| MPC1/2 | mitochondrial pyruvate carrier | 15 + 14.5 | 2 + 3 | S. cerevisiae | - | [96] |
| MPC1/2 | mitochondrial pyruvate carrier | 12.3 + 14.3 | 2 + 2 | M. musculus | <1% | [96,97] |
| MPC1/2 | mitochondrial pyruvate carrier | 12.4 + 12.2 | 2 + 3 | A. thaliana | - | [96] |
| ceQORH | quinone oxidoreductase-electron transfer | 33.1 | p | A. thaliana | 30% | [50] |
| LPR1 | multi-copper oxidase | 60.5 | p | A. thaliana | <0.1% | [7] |
| PHF | phosphate transport regulation | 42.4 | 1 | A. thaliana | 1.5% | |
| AtHMA1 | heavy metal transporter | 80.1 | 6 | A. thaliana | 3% | [50] |
| AtHMA3 | heavy metal transporter | 81.4 | 8 | A. thaliana | 1% | |
| AtHMA6 | heavy metal transporter | 100 | 8 | A. thaliana | 3% | |
| AtHMA4 | heavy metal transporter | 126.7 | 8 | A. thaliana | 0.75% | [7] |
| NTT1 | chloroplast ADP/ATP transporter | 57.5 | 12 | A. thaliana | 0.2% | [50] |
| NRT1 (NPF2.3) | nitrate excretion transporter | 61 | 12 | A. thaliana | - | [98] |
| ATM3 (ABCB25) | mitochondrial ABC transporter | 80 | 7 | A. thaliana | - | [92] |
| AAC hyd | hydrogenosomal carrier | 33.9 | 6 | N. patriciarum | <1% | [11] |
| SUT1 | sucrose transporter | 54.8 | 12 | S. tuberosum | 1–2% | [99] |
| L276 | mitochondrial carrier-like | 27.3 | 6 | A. polyphaga | 5% | [100] |
| Bcl-Xl | apoptosis regulation | 24.7 | 1 | H. sapiens | 1% | [7] |
| CYP3A4 | cytochrome-mono-oxygenase | 57.4 | 1 | H. sapiens | 5% | [51] |
| MGST1 | microsomal glutathione S-transferase 1 | 17.6 | 4 | H. sapiens | 3% | |
| ABCG2 | breast cancer resistance protein | 72 | 6 | H. sapiens | 0.5–1% | [101] |
| Erd2 | KDEL receptor | 24.4 | 7 | H. sapiens | <0.1% | [11] |
| CXCR4 | chemokine receptor type 4 | 37.9 | 7 | H. sapiens | <0.1% | [7] |
| CCR5 | chemokine receptor type 5 | 38.7 | 7 | H. sapiens | <0.1% | |
| PS1Δ9 | human alpha secretase component | 55 | 9 | H. sapiens | 0.1–0.2% | [100] |
| CFTR | cystic fibrosis transmembrane conductance regulator | 168 | 12 | H. sapiens | <0.1% | [46] |
| TMEM165 | cation transporter | 34.9 | 6 | H. sapiens | - | [102] |
| AAC1 | mitochondrial ADP/ATP carrier protein 1 | 34 | 6 | H. sapiens | 0.5–1% | [103,104,105] |
| ANT2(AAC2) | mitochondrial ADP/ATP carrier protein 2 | 32.8 | 6 | H. sapiens | - | [104] |
| ANT3(AAC3) | mitochondrial ADP/ATP carrier protein 3 | 32.8 | 6 | H. sapiens | - | [104] |
| SLC25A3 | mitochondrial pyruvate carrier (homologous to PIC) | 40.1 | 6 | H. sapiens | - | [106] |
| Protein | Organism | Code | Structure | References |
|---|---|---|---|---|
| OpuA | L. lactis | 7AHH | OpuA inhibited inward-facing, SBD docked | [131] |
| 7AHC | OpuA apo inward-facing | |||
| 7AHE | OpuA inhibited inward-facing | |||
| 7AHD | OpuA (E190Q) occluded | |||
| PrgL | E. faecalis | 7AED | VirB8 domain of PrgL from Enterococcus faecalis Pcf10 | [132] |
| MhsT | A. halodurans | 6YU2 | Crystal structure of MhsT in complex with L-isoleucine | [133] |
| 6YU3 | Crystal structure of MhsT in complex with L-phenylalanine | |||
| 6YU4 | Crystal structure of MhsT in complex with L-4F-phenylalanine | |||
| 6YU5 | Crystal structure of MhsT in complex with L-valine | |||
| 6YU6 | Crystal structure of MhsT in complex with L-leucine | |||
| 6YU7 | Crystal structure of MhsT in complex with L-tyrosine | |||
| GlnPQ | L. lactis | 6FXG | Crystal structure of substrate binding domain 1 (SBD1) OF ABC transporter GLNPQ in complex with Asparagine | [134] |
| ECF | L. delbrueckii subsp. Bulgaricus | 5D0Y | Substrate bound S-component of folate ECF transporter | [115] |
| ATP-Mg/Pi carrier (APC) | 4ZCU | Structure of calcium-bound regulatory domain of the human ATP-Mg/Pi carrier in the P2 form | [135] | |
| 4ZCV | Structure of calcium-bound regulatory domain of the human ATP-Mg/Pi carrier in the P212121 form | |||
| ThiT | L. lactis subsp. cremoris MG1363 | 4POP | ThiT with LMG139 bound | [130] |
| 4POV | ThiT with LMG135 bound | |||
| ECF | L. lactis subsp. cremoris MG1363 | 4DVE | Crystal structure at 2.1 A of the S-component for biotin from an ECF-type ABC transporter | [59] |
| OpuAC | L. lactis | 3L6G | Crystal structure of lactococcal OpuAC in its open conformation | [129] |
| 3L6H | Crystal structure of lactococcal OpuAC in its closed-liganded conformation complexed with glycine betaine |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frelet-Barrand, A. Lactococcus lactis, an Attractive Cell Factory for the Expression of Functional Membrane Proteins. Biomolecules 2022, 12, 180. https://doi.org/10.3390/biom12020180
Frelet-Barrand A. Lactococcus lactis, an Attractive Cell Factory for the Expression of Functional Membrane Proteins. Biomolecules. 2022; 12(2):180. https://doi.org/10.3390/biom12020180
Chicago/Turabian StyleFrelet-Barrand, Annie. 2022. "Lactococcus lactis, an Attractive Cell Factory for the Expression of Functional Membrane Proteins" Biomolecules 12, no. 2: 180. https://doi.org/10.3390/biom12020180
APA StyleFrelet-Barrand, A. (2022). Lactococcus lactis, an Attractive Cell Factory for the Expression of Functional Membrane Proteins. Biomolecules, 12(2), 180. https://doi.org/10.3390/biom12020180






