Interaction of TOR and PKA Signaling in S. cerevisiae
Abstract
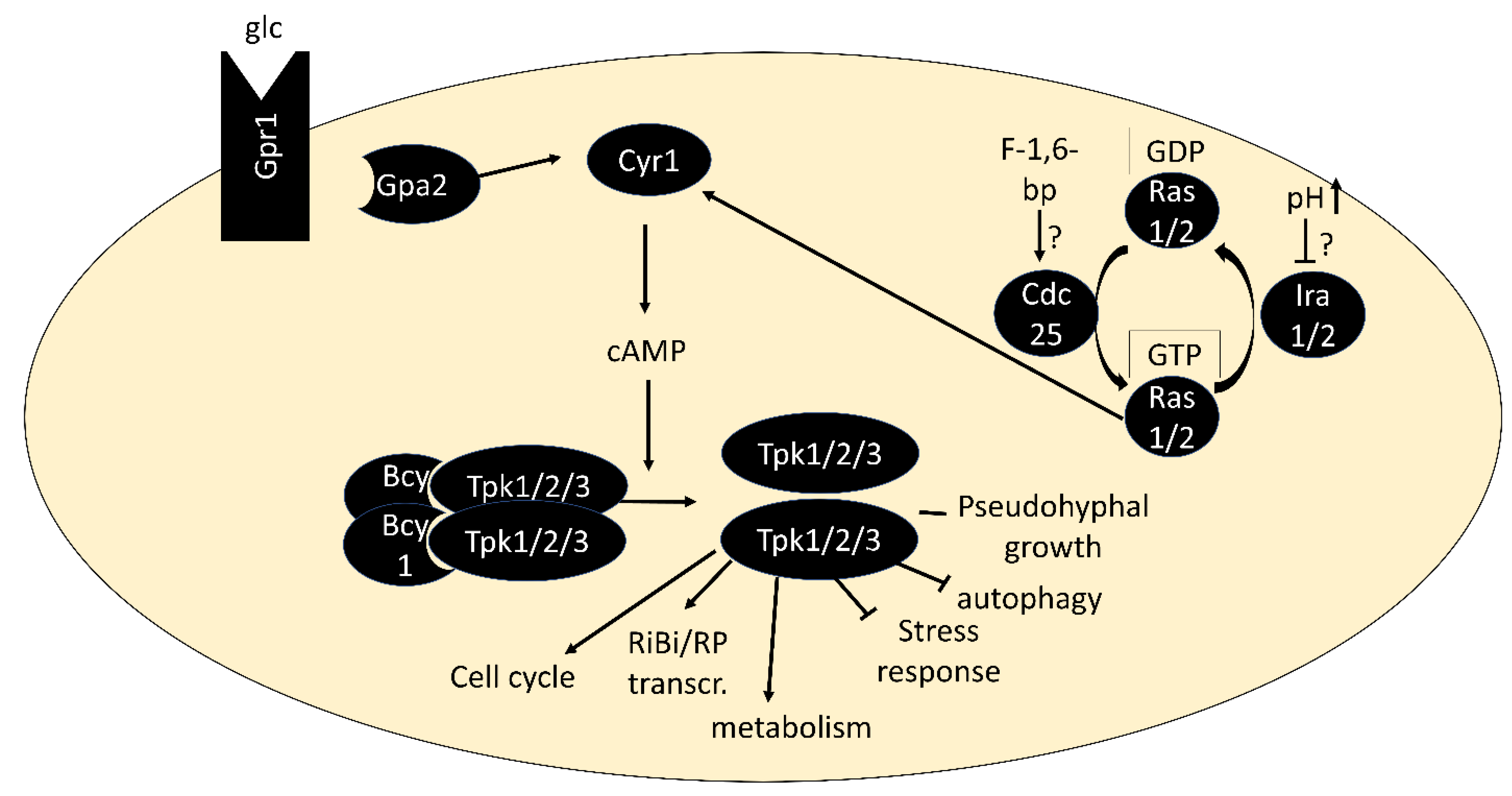
:1. Introduction
1.1. TOR Signaling
1.2. PKA Signaling
2. TOR–PKA Genetic Interactions
3. Shared Targets
3.1. Ribosome Production
3.1.1. Ribosome Biogenesis: Dot6/Tod6 and Stb3
3.1.2. Ribosomal Protein Production: Ifh1, Crf1 and Spf1
3.1.3. RNA Pol III Transcription: Maf1
3.2. Autophagy
3.3. Stress Response
3.3.1. Rim15
3.3.2. Msn2/4
3.4. Summary of Shared PKA and TOR Targets
4. Substrate Specificity of PKA and Sch9
5. Potential Mechanisms of TOR–PKA Interplay
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zaman, S.; Lippman, S.I.; Zhao, X.; Broach, J.R. How Saccharomyces Responds to Nutrients. Annu. Rev. Genet. 2008, 42, 27–81. [Google Scholar] [CrossRef]
- Broach, J.R. Nutritional Control of Growth and Development in Yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef] [Green Version]
- Loewith, R.; Hall, M.N. Target of Rapamycin (TOR) in Nutrient Signaling and Growth Control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef] [Green Version]
- Eltschinger, S.; Loewith, R. TOR Complexes and the Maintenance of Cellular Homeostasis. Trends Cell Biol. 2016, 26, 148–159. [Google Scholar] [CrossRef]
- De Virgilio, C. The Essence of Yeast Quiescence. FEMS Microbiol. Rev. 2012, 36, 306–339. [Google Scholar] [CrossRef] [Green Version]
- González, A.; Hall, M.N. Nutrient Sensing and TOR Signaling in Yeast and Mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef] [Green Version]
- De Virgilio, C.; Loewith, R. The TOR Signalling Network from Yeast to Man. Int. J. Biochem. Cell Biol. 2006, 38, 1476–1481. [Google Scholar] [CrossRef] [Green Version]
- Inoki, K.; Guan, K.-L. Complexity of the TOR Signaling Network. Trends Cell Biol. 2006, 16, 206–212. [Google Scholar] [CrossRef]
- Soulard, A.; Cohen, A.; Hall, M.N. TOR Signaling in Invertebrates. Curr. Opin. Cell Biol. 2009, 21, 825–836. [Google Scholar] [CrossRef] [Green Version]
- De Virgilio, C.; Loewith, R. Cell Growth Control: Little Eukaryotes Make Big Contributions. Oncogene 2006, 25, 6392–6415. [Google Scholar] [CrossRef] [Green Version]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient Sensing and Signaling in the Yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef] [Green Version]
- Healy, A.M.; Zolnierowicz, S.; Stapleton, A.E.; Goebl, M.; DePaoli-Roach, A.A.; Pringle, J.R. CDC55, a Saccharomyces cerevisiae Gene Involved in Cellular Morphogenesis: Identification, Characterization, and Homology to the B Subunit of Mammalian Type 2A Protein Phosphatase. Mol. Cell. Biol. 1991, 11, 5767–5780. [Google Scholar] [CrossRef] [Green Version]
- Van Zyl, W.; Huang, W.; Sneddon, A.A.; Stark, M.; Camier, S.; Werner, M.; Marck, C.; Sentenac, A.; Broach, J.R. Inactivation of the Protein Phosphatase 2A Regulatory Subunit A Results in Morphological and Transcriptional Defects in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992, 12, 4946–4959. [Google Scholar] [CrossRef] [Green Version]
- Shu, Y.; Yang, H.; Hallberg, E.; Hallberg, R. Molecular Genetic Analysis of Rts1p, a B’ Regulatory Subunit of Saccharomyces cerevisiae Protein Phosphatase 2A. Mol. Cell. Biol. 1997, 17, 3242–3253. [Google Scholar] [CrossRef] [Green Version]
- Sneddon, A.A.; Cohen, P.T.; Stark, M.J. Saccharomyces cerevisiae Protein Phosphatase 2A Performs an Essential Cellular Function and Is Encoded by Two Genes. EMBO J. 1990, 9, 4339–4346. [Google Scholar] [CrossRef]
- Sutton, A.; Immanuel, D.; Arndt, K.T. The SIT4 Protein Phosphatase Functions in Late G1 for Progression into S Phase. Mol. Cell. Biol. 1991, 11, 2133–2148. [Google Scholar]
- Luke, M.M.; Della Seta, F.; Di Como, C.J.; Sugimoto, H.; Kobayashi, R.; Arndt, K.T. The SAP, a New Family of Proteins, Associate and Function Positively with the SIT4 Phosphatase. Mol. Cell. Biol. 1996, 16, 2744–2755. [Google Scholar] [CrossRef] [Green Version]
- Di Como, C.J.; Arndt, K.T. Nutrients, via the Tor Proteins, Stimulate the Association of Tap42 with Type 2A Phosphatases. Genes Dev. 1996, 10, 1904–1916. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Broach, J.R. Tor Proteins and Protein Phosphatase 2A Reciprocally Regulate Tap42 in Controlling Cell Growth in Yeast. EMBO J. 1999, 18, 2782–2792. [Google Scholar] [CrossRef] [Green Version]
- Cooper, T.G. Transmitting the Signal of Excess Nitrogen in Saccharomyces cerevisiae from the Tor Proteins to the GATA Factors: Connecting the Dots. FEMS Microbiol. Rev. 2002, 26, 223–238. [Google Scholar] [CrossRef] [Green Version]
- Cardenas, M.E.; Cutler, N.S.; Lorenz, M.C.; Di Como, C.J.; Heitman, J. The TOR Signaling Cascade Regulates Gene Expression in Response to Nutrients. Genes Dev. 1999, 13, 3271–3279. [Google Scholar] [CrossRef] [Green Version]
- Beck, T.; Hall, M.N. The TOR Signalling Pathway Controls Nuclear Localization of Nutrient-Regulated Transcription Factors. Nature 1999, 402, 689–692. [Google Scholar] [CrossRef]
- Schmidt, A.; Beck, T.; Koller, A.; Kunz, J.; Hall, M.N. The TOR Nutrient Signalling Pathway Phosphorylates NPR1 and Inhibits Turnover of the Tryptophan Permease. EMBO J. 1998, 17, 6924–6931. [Google Scholar] [CrossRef] [Green Version]
- Huber, A.; French, S.L.; Tekotte, H.; Yerlikaya, S.; Stahl, M.; Perepelkina, M.P.; Tyers, M.; Rougemont, J.; Beyer, A.L.; Loewith, R. Sch9 Regulates Ribosome Biogenesis via Stb3, Dot6 and Tod6 and the Histone Deacetylase Complex RPD3L. EMBO J. 2011, 30, 3052–3064. [Google Scholar] [CrossRef] [Green Version]
- Soulard, A.; Cremonesi, A.; Moes, S.; Schütz, F.; Jenö, P.; Hall, M.N. The Rapamycin-Sensitive Phosphoproteome Reveals That TOR Controls Protein Kinase A toward Some but Not All Substrates. Mol. Biol. Cell 2010, 21, 3475–3486. [Google Scholar] [CrossRef] [Green Version]
- Urban, J.; Soulard, A.; Huber, A.; Lippman, S.; Mukhopadhyay, D.; Deloche, O.; Wanke, V.; Anrather, D.; Ammerer, G.; Riezman, H.; et al. Sch9 Is a Major Target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 2007, 26, 663–674. [Google Scholar] [CrossRef]
- Pearce, L.R.; Komander, D.; Alessi, D.R. The Nuts and Bolts of AGC Protein Kinases. Nat. Rev. Mol. Cell Biol. 2010, 11, 9–22. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, X.; Lester, R.L.; Dickson, R.C. The Sphingoid Long Chain Base Phytosphingosine Activates AGC-Type Protein Kinases in Saccharomyces cerevisiae Including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 2005, 280, 22679–22687. [Google Scholar] [CrossRef] [Green Version]
- Casamayor, A.; Torrance, P.D.; Kobayashi, T.; Thorner, J.; Alessi, D.R. Functional Counterparts of Mammalian Protein Kinases PDK1 and SGK in Budding Yeast. Curr. Biol. CB 1999, 9, 186–197. [Google Scholar] [CrossRef] [Green Version]
- Van Dam, T.J.P.; Zwartkruis, F.J.T.; Bos, J.L.; Snel, B. Evolution of the TOR Pathway. J. Mol. Evol. 2011, 73, 209–220. [Google Scholar] [CrossRef] [Green Version]
- Toda, T.; Cameron, S.; Sass, P.; Zoller, M.; Wigler, M. Three Different Genes in S. Cerevisiae Encode the Catalytic Subunits of the CAMP-Dependent Protein Kinase. Cell 1987, 50, 277–287. [Google Scholar] [CrossRef]
- Toda, T.; Cameron, S.; Sass, P.; Zoller, M.; Scott, J.D.; McMullen, B.; Hurwitz, M.; Krebs, E.G.; Wigler, M. Cloning and Characterization of BCY1, a Locus Encoding a Regulatory Subunit of the Cyclic AMP-Dependent Protein Kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987, 7, 1371–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kemp, B.E.; Bylund, D.B.; Huang, T.S.; Krebs, E.G. Substrate Specificity of the Cyclic AMP-Dependent Protein Kinase. Proc. Natl. Acad. Sci. USA 1975, 72, 3448–3452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabb, J.B. Physiological Substrates of CAMP-Dependent Protein Kinase. Chem. Rev. 2001, 101, 2381–2412. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Uno, I.; Oshima, Y.; Ishikawa, T. Isolation and Characterization of Yeast Mutants Deficient in Adenylate Cyclase and CAMP-Dependent Protein Kinase. Proc. Natl. Acad. Sci. USA 1982, 79, 2355–2359. [Google Scholar] [CrossRef] [Green Version]
- Kraakman, L.; Lemaire, K.; Ma, P.; Teunissen, A.W.; Donaton, M.C.; Van Dijck, P.; Winderickx, J.; de Winde, J.H.; Thevelein, J.M. A Saccharomyces cerevisiae G-Protein Coupled Receptor, Gpr1, Is Specifically Required for Glucose Activation of the CAMP Pathway during the Transition to Growth on Glucose. Mol. Microbiol. 1999, 32, 1002–1012. [Google Scholar] [CrossRef]
- Tatchell, K.; Chaleff, D.T.; DeFeo-Jones, D.; Scolnick, E.M. Requirement of Either of a Pair of Ras-Related Genes of Saccharomyces cerevisiae for Spore Viability. Nature 1984, 309, 523–527. [Google Scholar] [CrossRef]
- Mazón, M.J.; Gancedo, J.M.; Gancedo, C. Inactivation of Yeast Fructose-1,6-Bisphosphatase. In Vivo Phosphorylation of the Enzyme. J. Biol. Chem. 1982, 257, 1128–1130. [Google Scholar] [CrossRef]
- Purwin, C.; Leidig, F.; Holzer, H. Cyclic AMP-Dependent Phosphorylation of Fructose-1,6-Bisphosphatase in Yeast. Biochem. Biophys. Res. Commun. 1982, 107, 1482–1489. [Google Scholar] [CrossRef]
- Thevelein, J.M. Fermentable Sugars and Intracellular Acidification as Specific Activators of the RAS-Adenylate Cyclase Signalling Pathway in Yeast: The Relationship to Nutrient-Induced Cell Cycle Control. Mol. Microbiol. 1991, 5, 1301–1307. [Google Scholar] [CrossRef]
- Nikawa, J.; Sass, P.; Wigler, M. Cloning and Characterization of the Low-Affinity Cyclic AMP Phosphodiesterase Gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 1987, 7, 3629–3636. [Google Scholar] [CrossRef] [PubMed]
- Sass, P.; Field, J.; Nikawa, J.; Toda, T.; Wigler, M. Cloning and Characterization of the High-Affinity CAMP Phosphodiesterase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1986, 83, 9303–9307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, P.; Wera, S.; Van Dijck, P.; Thevelein, J.M. The PDE1-Encoded Low-Affinity Phosphodiesterase in the Yeast Saccharomyces cerevisiae Has a Specific Function in Controlling Agonist-Induced CAMP Signaling. Mol. Biol. Cell 1999, 10, 91–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peeters, T.; Louwet, W.; Gelade, R.; Nauwelaers, D.; Thevelein, J.M.; Versele, M. Kelch-Repeat Proteins Interacting with the G Protein Gpa2 Bypass Adenylate Cyclase for Direct Regulation of Protein Kinase A in Yeast. Proc. Natl. Acad. Sci. USA 2006, 103, 13034–13039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conrad, M.; Kankipati, H.N.; Kimpe, M.; Van Zeebroeck, G.; Zhang, Z.; Thevelein, J.M. The Nutrient Transceptor/PKA Pathway Functions Independently of TOR and Responds to Leucine and Gcn2 in a TOR-Independent Manner. FEMS Yeast Res. 2017, 17, fox048. [Google Scholar] [CrossRef] [Green Version]
- Thevelein, J.M.; Beullens, M. Cyclic AMP and the Stimulation of Trehalase Activity in the Yeast Saccharomyces cerevisiae by Carbon Sources, Nitrogen Sources and Inhibitors of Protein Synthesis. J. Gen. Microbiol. 1985, 131, 3199–3209. [Google Scholar] [CrossRef] [Green Version]
- Steyfkens, F.; Zhang, Z.; Van Zeebroeck, G.; Thevelein, J.M. Multiple Transceptors for Macro- and Micro-Nutrients Control Diverse Cellular Properties Through the PKA Pathway in Yeast: A Paradigm for the Rapidly Expanding World of Eukaryotic Nutrient Transceptors up to Those in Human Cells. Front. Pharmacol. 2018, 9, 191. [Google Scholar] [CrossRef]
- Haesendonckx, S.; Tudisca, V.; Voordeckers, K.; Moreno, S.; Thevelein, J.M.; Portela, P. The Activation Loop of PKA Catalytic Isoforms Is Differentially Phosphorylated by Pkh Protein Kinases in Saccharomyces cerevisiae. Biochem. J. 2012, 448, 307–320. [Google Scholar] [CrossRef] [Green Version]
- Steichen, J.M.; Iyer, G.H.; Li, S.; Saldanha, S.A.; Deal, M.S.; Woods, V.L.; Taylor, S.S. Global Consequences of Activation Loop Phosphorylation on Protein Kinase A. J. Biol. Chem. 2010, 285, 3825–3832. [Google Scholar] [CrossRef] [Green Version]
- Marshall, M.S.; Gibbs, J.B.; Scolnick, E.M.; Sigal, I.S. Regulatory Function of the Saccharomyces cerevisiae RAS C-Terminus. Mol. Cell. Biol. 1987, 7, 2309–2315. [Google Scholar] [CrossRef] [Green Version]
- Cannon, J.F.; Tatchell, K. Characterization of Saccharomyces cerevisiae Genes Encoding Subunits of Cyclic AMP-Dependent Protein Kinase. Mol. Cell. Biol. 1987, 7, 2653–2663. [Google Scholar] [PubMed] [Green Version]
- Matsumoto, K.; Uno, I.; Ishikawa, T. Control of Cell Division in Saccharomyces cerevisiae Mutants Defective in Adenylate Cyclase and CAMP-Dependent Protein Kinase. Exp. Cell Res. 1983, 146, 151–161. [Google Scholar] [CrossRef]
- Cameron, S.; Levin, L.; Zoller, M.; Wigler, M. CAMP-Independent Control of Sporulation, Glycogen Metabolism, and Heat Shock Resistance in S. Cerevisiae. Cell 1988, 53, 555–566. [Google Scholar] [CrossRef]
- Uno, I.; Matsumoto, K.; Ishikawa, T. Characterization of a Cyclic Nucleotide Phosphodiesterase-Deficient Mutant in Yeast. J. Biol. Chem. 1983, 258, 3539–3542. [Google Scholar] [CrossRef]
- Santangelo, G.M. Glucose Signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 253–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dihazi, H.; Kessler, R.; Eschrich, K. Glucose-Induced Stimulation of the Ras-CAMP Pathway in Yeast Leads to Multiple Phosphorylations and Activation of 6-Phosphofructo-2-Kinase. Biochemistry 2003, 42, 6275–6282. [Google Scholar] [CrossRef]
- Portela, P.; Howell, S.; Moreno, S.; Rossi, S. In Vivo and in Vitro Phosphorylation of Two Isoforms of Yeast Pyruvate Kinase by Protein Kinase A. J. Biol. Chem. 2002, 277, 30477–30487. [Google Scholar] [CrossRef] [Green Version]
- Schepers, W.; Van Zeebroeck, G.; Pinkse, M.; Verhaert, P.; Thevelein, J.M. In Vivo Phosphorylation of Ser21 and Ser83 during Nutrient-Induced Activation of the Yeast Protein Kinase A (PKA) Target Trehalase. J. Biol. Chem. 2012, 287, 44130–44142. [Google Scholar] [CrossRef] [Green Version]
- Rittenhouse, J.; Moberly, L.; Marcus, F. Phosphorylation in Vivo of Yeast (Saccharomyces cerevisiae) Fructose-1,6-Bisphosphatase at the Cyclic AMP-Dependent Site. J. Biol. Chem. 1987, 262, 10114–10119. [Google Scholar] [CrossRef]
- Garrett, S.; Broach, J. Loss of Ras Activity in Saccharomyces cerevisiae Is Suppressed by Disruptions of a New Kinase Gene, YAKI, Whose Product May Act Downstream of the CAMP-Dependent Protein Kinase. Genes Dev. 1989, 3, 1336–1348. [Google Scholar] [CrossRef] [Green Version]
- Reinders, A.; Bürckert, N.; Boller, T.; Wiemken, A.; De Virgilio, C. Saccharomyces cerevisiae CAMP-Dependent Protein Kinase Controls Entry into Stationary Phase through the Rim15p Protein Kinase. Genes Dev. 1998, 12, 2943–2955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.; Ward, M.P.; Garrett, S. Yeast PKA Represses Msn2p/Msn4p-Dependent Gene Expression to Regulate Growth, Stress Response and Glycogen Accumulation. EMBO J. 1998, 17, 3556–3564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrett, S.; Menold, M.M.; Broach, J.R. The Saccharomyces cerevisiae YAK1 Gene Encodes a Protein Kinase That Is Induced by Arrest Early in the Cell Cycle. Mol. Cell. Biol. 1991, 11, 4045–4052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Görner, W.; Durchschlag, E.; Martinez-Pastor, M.T.; Estruch, F.; Ammerer, G.; Hamilton, B.; Ruis, H.; Schüller, C. Nuclear Localization of the C2H2 Zinc Finger Protein Msn2p Is Regulated by Stress and Protein Kinase A Activity. Genes Dev. 1998, 12, 586–597. [Google Scholar] [CrossRef]
- Lee, P.; Paik, S.-M.; Shin, C.-S.; Huh, W.-K.; Hahn, J.-S. Regulation of Yeast Yak1 Kinase by PKA and Autophosphorylation-Dependent 14-3-3 Binding. Mol. Microbiol. 2011, 79, 633–646. [Google Scholar] [CrossRef]
- Lee, P.; Cho, B.-R.; Joo, H.-S.; Hahn, J.-S. Yeast Yak1 Kinase, a Bridge between PKA and Stress-Responsive Transcription Factors, Hsf1 and Msn2/Msn4. Mol. Microbiol. 2008, 70, 882–895. [Google Scholar] [CrossRef]
- Malcher, M.; Schladebeck, S.; Mösch, H.-U. The Yak1 Protein Kinase Lies at the Center of a Regulatory Cascade Affecting Adhesive Growth and Stress Resistance in Saccharomyces cerevisiae. Genetics 2011, 187, 717–730. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.; Kim, M.S.; Paik, S.-M.; Choi, S.-H.; Cho, B.-R.; Hahn, J.-S. Rim15-Dependent Activation of Hsf1 and Msn2/4 Transcription Factors by Direct Phosphorylation in Saccharomyces cerevisiae. FEBS Lett. 2013, 587, 3648–3655. [Google Scholar] [CrossRef] [Green Version]
- Cameroni, E.; Hulo, N.; Roosen, J.; Winderickx, J.; De Virgilio, C. The Novel Yeast PAS Kinase Rim 15 Orchestrates G0-Associated Antioxidant Defense Mechanisms. Cell Cycle 2004, 3, 462–468. [Google Scholar] [CrossRef] [Green Version]
- Toda, T.; Cameron, S.; Sass, P.; Wigler, M. SCH9, a Gene of Saccharomyces cerevisiae That Encodes a Protein Distinct from, but Functionally and Structurally Related to, CAMP-Dependent Protein Kinase Catalytic Subunits. Genes Dev. 1988, 2, 517–527. [Google Scholar] [CrossRef] [Green Version]
- Schmelzle, T.; Beck, T.; Martin, D.E.; Hall, M.N. Activation of the RAS/Cyclic AMP Pathway Suppresses a TOR Deficiency in Yeast. Mol. Cell. Biol. 2004, 24, 338–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurita-Martinez, S.A.; Cardenas, M.E. Tor and Cyclic AMP-Protein Kinase A: Two Parallel Pathways Regulating Expression of Genes Required for Cell Growth. Eukaryot. Cell 2005, 4, 63–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araki, T.; Uesono, Y.; Oguchi, T.; Toh-E, A. LAS24/KOG1, a Component of the TOR Complex 1 (TORC1), Is Needed for Resistance to Local Anesthetic Tetracaine and Normal Distribution of Actin Cytoskeleton in Yeast. Genes Genet. Syst. 2005, 80, 325–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramachandran, V.; Herman, P.K. Antagonistic Interactions between the CAMP-Dependent Protein Kinase and Tor Signaling Pathways Modulate Cell Growth in Saccharomyces cerevisiae. Genetics 2011, 187, 441–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crauwels, M.; Donaton, M.C.; Pernambuco, M.B.; Winderickx, J.; de Winde, J.H.; Thevelein, J.M. The Sch9 Protein Kinase in the Yeast Saccharomyces cerevisiae Controls CAPK Activity and Is Required for Nitrogen Activation of the Fermentable-Growth-Medium-Induced (FGM) Pathway. Microbiology 1997, 143, 2627–2637. [Google Scholar] [CrossRef]
- Rudra, D.; Warner, J.R. What Better Measure than Ribosome Synthesis? Genes Dev. 2004, 18, 2431–2436. [Google Scholar] [CrossRef] [Green Version]
- Warner, J.R. The Economics of Ribosome Biosynthesis in Yeast. Trends Biochem. Sci. 1999, 24, 437–440. [Google Scholar] [CrossRef]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef]
- Jorgensen, P.; Rupes, I.; Sharom, J.R.; Schneper, L.; Broach, J.R.; Tyers, M. A Dynamic Transcriptional Network Communicates Growth Potential to Ribosome Synthesis and Critical Cell Size. Genes Dev. 2004, 18, 2491–2505. [Google Scholar] [CrossRef] [Green Version]
- Slattery, M.G.; Heideman, W. Coordinated Regulation of Growth Genes in Saccharomyces cerevisiae. Cell Cycle 2007, 6, 1210–1219. [Google Scholar] [CrossRef]
- Wang, Y.; Pierce, M.; Schneper, L.; Güldal, C.G.; Zhang, X.; Tavazoie, S.; Broach, J.R. Ras and Gpa2 Mediate One Branch of a Redundant Glucose Signaling Pathway in Yeast. PLoS Biol. 2004, 2, e128. [Google Scholar] [CrossRef] [PubMed]
- Zaman, S.; Lippman, S.I.; Schneper, L.; Slonim, N.; Broach, J.R. Glucose Regulates Transcription in Yeast through a Network of Signaling Pathways. Mol. Syst. Biol. 2009, 5, 245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunkel, J.; Luo, X.; Capaldi, A.P. Integrated TORC1 and PKA Signaling Control the Temporal Activation of Glucose-Induced Gene Expression in Yeast. Nat. Commun. 2019, 10, 3558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosio, M.C.; Fermi, B.; Spagnoli, G.; Levati, E.; Rubbi, L.; Ferrari, R.; Pellegrini, M.; Dieci, G. Abf1 and Other General Regulatory Factors Control Ribosome Biogenesis Gene Expression in Budding Yeast. Nucleic Acids Res. 2017, 45, 4493–4506. [Google Scholar] [CrossRef] [Green Version]
- Fingerman, I.; Nagaraj, V.; Norris, D.; Vershon, A.K. Sfp1 Plays a Key Role in Yeast Ribosome Biogenesis. Eukaryot. Cell 2003, 2, 1061–1068. [Google Scholar] [CrossRef] [Green Version]
- Dequard-Chablat, M.; Riva, M.; Carles, C.; Sentenac, A. RPC19, the Gene for a Subunit Common to Yeast RNA Polymerases A (I) and C (III). J. Biol. Chem. 1991, 266, 15300–15307. [Google Scholar] [CrossRef]
- Hughes, J.D.; Estep, P.W.; Tavazoie, S.; Church, G.M. Computational Identification of Cis-Regulatory Elements Associated with Groups of Functionally Related Genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000, 296, 1205–1214. [Google Scholar] [CrossRef]
- Liko, D.; Slattery, M.G.; Heideman, W. Stb3 Binds to Ribosomal RNA Processing Element Motifs That Control Transcriptional Responses to Growth in Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 26623–26628. [Google Scholar] [CrossRef] [Green Version]
- Humphrey, E.L.; Shamji, A.F.; Bernstein, B.E.; Schreiber, S.L. Rpd3p Relocation Mediates a Transcriptional Response to Rapamycin in Yeast. Chem. Biol. 2004, 11, 295–299. [Google Scholar] [CrossRef] [Green Version]
- Kurdistani, S.K.; Robyr, D.; Tavazoie, S.; Grunstein, M. Genome-Wide Binding Map of the Histone Deacetylase Rpd3 in Yeast. Nat. Genet. 2002, 31, 248–254. [Google Scholar] [CrossRef]
- Rohde, J.R.; Cardenas, M.E. The Tor Pathway Regulates Gene Expression by Linking Nutrient Sensing to Histone Acetylation. Mol. Cell. Biol. 2003, 23, 629–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lippman, S.I.; Broach, J.R. Protein Kinase A and TORC1 Activate Genes for Ribosomal Biogenesis by Inactivating Repressors Encoded by Dot6 and Its Homolog Tod6. Proc. Natl. Acad. Sci. USA 2009, 106, 19928–19933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes Hallett, J.E.; Luo, X.; Capaldi, A.P. State Transitions in the TORC1 Signaling Pathway and Information Processing in Saccharomyces cerevisiae. Genetics 2014, 198, 773–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, A.; Bodenmiller, B.; Uotila, A.; Stahl, M.; Wanka, S.; Gerrits, B.; Aebersold, R.; Loewith, R. Characterization of the Rapamycin-Sensitive Phosphoproteome Reveals That Sch9 Is a Central Coordinator of Protein Synthesis. Genes Dev. 2009, 23, 1929–1943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deminoff, S.J.; Howard, S.C.; Hester, A.; Warner, S.; Herman, P.K. Using Substrate-Binding Variants of the CAMP-Dependent Protein Kinase to Identify Novel Targets and a Kinase Domain Important for Substrate Interactions in Saccharomyces cerevisiae. Genetics 2006, 173, 1909–1917. [Google Scholar] [CrossRef] [Green Version]
- Kasten, M.M.; Stillman, D.J. Identification of the Saccharomyces cerevisiae Genes STB1-STB5 Encoding Sin3p Binding Proteins. Mol. Gen. Genet. MGG 1997, 256, 376–386. [Google Scholar] [CrossRef]
- Budovskaya, Y.V.; Stephan, J.S.; Deminoff, S.J.; Herman, P.K. An Evolutionary Proteomics Approach Identifies Substrates of the CAMP-Dependent Protein Kinase. Proc. Natl. Acad. Sci. USA 2005, 102, 13933–13938. [Google Scholar] [CrossRef] [Green Version]
- Liko, D.; Conway, M.K.; Grunwald, D.S.; Heideman, W. Stb3 Plays a Role in the Glucose-Induced Transition from Quiescence to Growth in Saccharomyces cerevisiae. Genetics 2010, 185, 797–810. [Google Scholar] [CrossRef] [Green Version]
- Powers, T.; Walter, P. Regulation of Ribosome Biogenesis by the Rapamycin-Sensitive TOR-Signaling Pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 1999, 10, 987–1000. [Google Scholar] [CrossRef] [Green Version]
- Hall, D.B.; Wade, J.T.; Struhl, K. An HMG Protein, Hmo1, Associates with Promoters of Many Ribosomal Protein Genes and throughout the RRNA Gene Locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006, 26, 3672–3679. [Google Scholar] [CrossRef] [Green Version]
- Schawalder, S.B.; Kabani, M.; Howald, I.; Choudhury, U.; Werner, M.; Shore, D. Growth-Regulated Recruitment of the Essential Yeast Ribosomal Protein Gene Activator Ifh1. Nature 2004, 432, 1058–1061. [Google Scholar] [CrossRef] [PubMed]
- Rudra, D.; Zhao, Y.; Warner, J.R. Central Role of Ifh1p-Fhl1p Interaction in the Synthesis of Yeast Ribosomal Proteins. EMBO J. 2005, 24, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Wade, J.T.; Hall, D.B.; Struhl, K. The Transcription Factor Ifh1 Is a Key Regulator of Yeast Ribosomal Protein Genes. Nature 2004, 432, 1054–1058. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.E.; Soulard, A.; Hall, M.N. TOR Regulates Ribosomal Protein Gene Expression via PKA and the Forkhead Transcription Factor FHL1. Cell 2004, 119, 969–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lempiäinen, H.; Uotila, A.; Urban, J.; Dohnal, I.; Ammerer, G.; Loewith, R.; Shore, D. Sfp1 Interaction with TORC1 and Mrs6 Reveals Feedback Regulation on TOR Signaling. Mol. Cell 2009, 33, 704–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, C.; Struhl, K. Protein Kinase A Mediates Growth-Regulated Expression of Yeast Ribosomal Protein Genes by Modulating RAP1 Transcriptional Activity. Mol. Cell. Biol. 1994, 14, 1920–1928. [Google Scholar] [CrossRef] [Green Version]
- Bosio, M.C.; Negri, R.; Dieci, G. Promoter Architectures in the Yeast Ribosomal Expression Program. Transcription 2011, 2, 71–77. [Google Scholar] [CrossRef] [Green Version]
- De Sanctis, V.; La Terra, S.; Bianchi, A.; Shore, D.; Burderi, L.; Di Mauro, E.; Negri, R. In Vivo Topography of Rap1p-DNA Complex at Saccharomyces cerevisiae TEF2 UAS(RPG) during Transcriptional Regulation. J. Mol. Biol. 2002, 318, 333–349. [Google Scholar] [CrossRef]
- Durocher, D.; Jackson, S.P. The FHA Domain. FEBS Lett. 2002, 513, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; McIntosh, K.B.; Rudra, D.; Schawalder, S.; Shore, D.; Warner, J.R. Fine-Structure Analysis of Ribosomal Protein Gene Transcription. Mol. Cell. Biol. 2006, 26, 4853–4862. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.S.; Hahn, J.-S. Role of CK2-Dependent Phosphorylation of Ifh1 and Crf1 in Transcriptional Regulation of Ribosomal Protein Genes in Saccharomyces cerevisiae. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2016, 1859, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Rudra, D.; Mallick, J.; Zhao, Y.; Warner, J.R. Potential Interface between Ribosomal Protein Production and Pre-RRNA Processing. Mol. Cell. Biol. 2007, 27, 4815–4824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorgensen, P.; Nishikawa, J.L.; Breitkreutz, B.-J.; Tyers, M. Systematic Identification of Pathways That Couple Cell Growth and Division in Yeast. Science 2002, 297, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Albert, B.; Kos-Braun, I.C.; Henras, A.K.; Dez, C.; Rueda, M.P.; Zhang, X.; Gadal, O.; Kos, M.; Shore, D. A Ribosome Assembly Stress Response Regulates Transcription to Maintain Proteome Homeostasis. eLife 2019, 8, e45002. [Google Scholar] [CrossRef]
- Marion, R.M.; Regev, A.; Segal, E.; Barash, Y.; Koller, D.; Friedman, N.; O’Shea, E.K. Sfp1 Is a Stress- and Nutrient-Sensitive Regulator of Ribosomal Protein Gene Expression. Proc. Natl. Acad. Sci. USA 2004, 101, 14315–14322. [Google Scholar] [CrossRef] [Green Version]
- Pluta, K.; Lefebvre, O.; Martin, N.C.; Smagowicz, W.J.; Stanford, D.R.; Ellis, S.R.; Hopper, A.K.; Sentenac, A.; Boguta, M. Maf1p, a Negative Effector of RNA Polymerase III in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 5031–5040. [Google Scholar] [CrossRef] [Green Version]
- Upadhya, R.; Lee, J.; Willis, I.M. Maf1 Is an Essential Mediator of Diverse Signals That Repress RNA Polymerase III Transcription. Mol. Cell 2002, 10, 1489–1494. [Google Scholar] [CrossRef]
- Moir, R.D.; Lee, J.; Haeusler, R.A.; Desai, N.; Engelke, D.R.; Willis, I.M. Protein Kinase A Regulates RNA Polymerase III Transcription through the Nuclear Localization of Maf1. Proc. Natl. Acad. Sci. USA 2006, 103, 15044–15049. [Google Scholar] [CrossRef] [Green Version]
- Roberts, D.N.; Wilson, B.; Huff, J.T.; Stewart, A.J.; Cairns, B.R. Dephosphorylation and Genome-Wide Association of Maf1 with Pol III-Transcribed Genes during Repression. Mol. Cell 2006, 22, 633–644. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Moir, R.D.; Willis, I.M. Regulation of RNA Polymerase III Transcription Involves SCH9-Dependent and SCH9-Independent Branches of the Target of Rapamycin (TOR) Pathway. J. Biol. Chem. 2009, 284, 12604–12608. [Google Scholar] [CrossRef] [Green Version]
- Oficjalska-Pham, D.; Harismendy, O.; Smagowicz, W.J.; Gonzalez de Peredo, A.; Boguta, M.; Sentenac, A.; Lefebvre, O. General Repression of RNA Polymerase III Transcription Is Triggered by Protein Phosphatase Type 2A-Mediated Dephosphorylation of Maf1. Mol. Cell 2006, 22, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Noda, T.; Ohsumi, Y. Tor, a Phosphatidylinositol Kinase Homologue, Controls Autophagy in Yeast. J. Biol. Chem. 1998, 273, 3963–3966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-Mediated Induction of Autophagy via an Apg1 Protein Kinase Complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budovskaya, Y.V.; Stephan, J.S.; Reggiori, F.; Klionsky, D.J.; Herman, P.K. The Ras/CAMP-Dependent Protein Kinase Signaling Pathway Regulates an Early Step of the Autophagy Process in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 20663–20671. [Google Scholar] [CrossRef] [Green Version]
- Yorimitsu, T.; Zaman, S.; Broach, J.R.; Klionsky, D.J. Protein Kinase A and Sch9 Cooperatively Regulate Induction of Autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 4180–4189. [Google Scholar] [CrossRef] [Green Version]
- Stephan, J.S.; Yeh, Y.-Y.; Ramachandran, V.; Deminoff, S.J.; Herman, P.K. The Tor and PKA Signaling Pathways Independently Target the Atg1/Atg13 Protein Kinase Complex to Control Autophagy. Proc. Natl. Acad. Sci. USA 2009, 106, 17049–17054. [Google Scholar] [CrossRef] [Green Version]
- Funakoshi, T.; Matsuura, A.; Noda, T.; Ohsumi, Y. Analyses of APG13 Gene Involved in Autophagy in Yeast, Saccharomyces cerevisiae. Gene 1997, 192, 207–213. [Google Scholar] [CrossRef]
- Kabeya, Y.; Noda, N.N.; Fujioka, Y.; Suzuki, K.; Inagaki, F.; Ohsumi, Y. Characterization of the Atg17-Atg29-Atg31 Complex Specifically Required for Starvation-Induced Autophagy in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2009, 389, 612–615. [Google Scholar] [CrossRef]
- Fujioka, Y.; Suzuki, S.W.; Yamamoto, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Akada, R.; Inagaki, F.; Ohsumi, Y.; Noda, N.N. Structural Basis of Starvation-Induced Assembly of the Autophagy Initiation Complex. Nat. Struct. Mol. Biol. 2014, 21, 513–521. [Google Scholar] [CrossRef]
- Suzuki, K.; Kirisako, T.; Kamada, Y.; Mizushima, N.; Noda, T.; Ohsumi, Y. The Pre-Autophagosomal Structure Organized by Concerted Functions of APG Genes Is Essential for Autophagosome Formation. EMBO J. 2001, 20, 5971–5981. [Google Scholar] [CrossRef]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and MTOR Regulate Autophagy through Direct Phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamada, Y.; Yoshino, K.; Kondo, C.; Kawamata, T.; Oshiro, N.; Yonezawa, K.; Ohsumi, Y. Tor Directly Controls the Atg1 Kinase Complex to Regulate Autophagy. Mol. Cell. Biol. 2010, 30, 1049–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Raucci, S.; Jaquenoud, M.; Hatakeyama, R.; Stumpe, M.; Rohr, R.; Reggiori, F.; De Virgilio, C.; Dengjel, J. Multilayered Control of Protein Turnover by TORC1 and Atg1. Cell Rep. 2019, 28, 3486–3496.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farré, J.-C.; Subramani, S. Mechanistic Insights into Selective Autophagy Pathways: Lessons from Yeast. Nat. Rev. Mol. Cell Biol. 2016, 17, 537–552. [Google Scholar] [CrossRef]
- Vidan, S.; Mitchell, A.P. Stimulation of Yeast Meiotic Gene Expression by the Glucose-Repressible Protein Kinase Rim15p. Mol. Cell. Biol. 1997, 17, 2688–2697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, V.; Moradas-Ferreira, P. Oxidative Stress and Signal Transduction in Saccharomyces cerevisiae: Insights into Ageing, Apoptosis and Diseases. Mol. Aspects Med. 2001, 22, 217–246. [Google Scholar] [CrossRef]
- Pedruzzi, I.; Bürckert, N.; Egger, P.; De Virgilio, C. Saccharomyces cerevisiae Ras/CAMP Pathway Controls Post-Diauxic Shift Element-Dependent Transcription through the Zinc Finger Protein Gis1. EMBO J. 2000, 19, 2569–2579. [Google Scholar] [CrossRef]
- Talarek, N.; Cameroni, E.; Jaquenoud, M.; Luo, X.; Bontron, S.; Lippman, S.; Devgan, G.; Snyder, M.; Broach, J.R.; De Virgilio, C. Initiation of the TORC1-Regulated G0 Program Requires Igo1/2, Which License Specific MRNAs to Evade Degradation via the 5’-3’ MRNA Decay Pathway. Mol. Cell 2010, 38, 345–355. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Talarek, N.; De Virgilio, C. Initiation of the Yeast G0 Program Requires Igo1 and Igo2, Which Antagonize Activation of Decapping of Specific Nutrient-Regulated MRNAs. RNA Biol. 2011, 8, 14–17. [Google Scholar] [CrossRef] [Green Version]
- Juanes, M.A.; Khoueiry, R.; Kupka, T.; Castro, A.; Mudrak, I.; Ogris, E.; Lorca, T.; Piatti, S. Budding Yeast Greatwall and Endosulfines Control Activity and Spatial Regulation of PP2A(Cdc55) for Timely Mitotic Progression. PLoS Genet. 2013, 9, e1003575. [Google Scholar] [CrossRef] [Green Version]
- Talarek, N.; Gueydon, E.; Schwob, E. Homeostatic Control of START through Negative Feedback between Cln3-Cdk1 and Rim15/Greatwall Kinase in Budding Yeast. eLife 2017, 6, e26233. [Google Scholar] [CrossRef] [PubMed]
- Barbet, N.C.; Schneider, U.; Helliwell, S.B.; Stansfield, I.; Tuite, M.F.; Hall, M.N. TOR Controls Translation Initiation and Early G1 Progression in Yeast. Mol. Biol. Cell 1996, 7, 25–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanke, V.; Cameroni, E.; Uotila, A.; Piccolis, M.; Urban, J.; Loewith, R.; De Virgilio, C. Caffeine Extends Yeast Lifespan by Targeting TORC1. Mol. Microbiol. 2008, 69, 277–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedruzzi, I.; Dubouloz, F.; Cameroni, E.; Wanke, V.; Roosen, J.; Winderickx, J.; De Virgilio, C. TOR and PKA Signaling Pathways Converge on the Protein Kinase Rim15 to Control Entry into G0. Mol. Cell 2003, 12, 1607–1613. [Google Scholar] [CrossRef]
- Ogawa, N.; DeRisi, J.; Brown, P.O. New Components of a System for Phosphate Accumulation and Polyphosphate Metabolism in Saccharomyces cerevisiae Revealed by Genomic Expression Analysis. Mol. Biol. Cell 2000, 11, 4309–4321. [Google Scholar] [CrossRef] [Green Version]
- Persson, B.L.; Lagerstedt, J.O.; Pratt, J.R.; Pattison-Granberg, J.; Lundh, K.; Shokrollahzadeh, S.; Lundh, F. Regulation of Phosphate Acquisition in Saccharomyces cerevisiae. Curr. Genet. 2003, 43, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Wanke, V.; Pedruzzi, I.; Cameroni, E.; Dubouloz, F.; De Virgilio, C. Regulation of G0 Entry by the Pho80-Pho85 Cyclin-CDK Complex. EMBO J. 2005, 24, 4271–4278. [Google Scholar] [CrossRef] [Green Version]
- Taylor, B.L.; Zhulin, I.B. PAS Domains: Internal Sensors of Oxygen, Redox Potential, and Light. Microbiol. Mol. Biol. Rev. MMBR 1999, 63, 479–506. [Google Scholar] [CrossRef] [Green Version]
- Griffioen, G.; Anghileri, P.; Imre, E.; Baroni, M.D.; Ruis, H. Nutritional Control of Nucleocytoplasmic Localization of CAMP-Dependent Protein Kinase Catalytic and Regulatory Subunits in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 1449–1456. [Google Scholar] [CrossRef] [Green Version]
- Griffioen, G.; Thevelein, J.M. Molecular Mechanisms Controlling the Localisation of Protein Kinase A. Curr. Genet. 2002, 41, 199–207. [Google Scholar] [CrossRef]
- Estruch, F.; Carlson, M. Two Homologous Zinc Finger Genes Identified by Multicopy Suppression in a SNF1 Protein Kinase Mutant of Saccharomyces cerevisiae. Mol. Cell. Biol. 1993, 13, 3872–3881. [Google Scholar] [CrossRef] [Green Version]
- Treger, J.M.; Schmitt, A.P.; Simon, J.R.; McEntee, K. Transcriptional Factor Mutations Reveal Regulatory Complexities of Heat Shock and Newly Identified Stress Genes in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 26875–26879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Pastor, M.T.; Marchler, G.; Schüller, C.; Marchler-Bauer, A.; Ruis, H.; Estruch, F. The Saccharomyces cerevisiae Zinc Finger Proteins Msn2p and Msn4p Are Required for Transcriptional Induction through the Stress Response Element (STRE). EMBO J. 1996, 15, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.P.; McEntee, K. Msn2p, a Zinc Finger DNA-Binding Protein, Is the Transcriptional Activator of the Multistress Response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1996, 93, 5777–5782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Görner, W.; Durchschlag, E.; Wolf, J.; Brown, E.L.; Ammerer, G.; Ruis, H.; Schüller, C. Acute Glucose Starvation Activates the Nuclear Localization Signal of a Stress-Specific Yeast Transcription Factor. EMBO J. 2002, 21, 135–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Düvel, K.; Santhanam, A.; Garrett, S.; Schneper, L.; Broach, J.R. Multiple Roles of Tap42 in Mediating Rapamycin-Induced Transcriptional Changes in Yeast. Mol. Cell 2003, 11, 1467–1478. [Google Scholar] [CrossRef]
- Santhanam, A.; Hartley, A.; Düvel, K.; Broach, J.R.; Garrett, S. PP2A Phosphatase Activity Is Required for Stress and Tor Kinase Regulation of Yeast Stress Response Factor Msn2p. Eukaryot. Cell 2004, 3, 1261–1271. [Google Scholar] [CrossRef] [Green Version]
- Reiter, W.; Klopf, E.; De Wever, V.; Anrather, D.; Petryshyn, A.; Roetzer, A.; Niederacher, G.; Roitinger, E.; Dohnal, I.; Görner, W.; et al. Yeast Protein Phosphatase 2A-Cdc55 Regulates the Transcriptional Response to Hyperosmolarity Stress by Regulating Msn2 and Msn4 Chromatin Recruitment. Mol. Cell. Biol. 2013, 33, 1057–1072. [Google Scholar] [CrossRef] [Green Version]
- De Wever, V.; Reiter, W.; Ballarini, A.; Ammerer, G.; Brocard, C. A Dual Role for PP1 in Shaping the Msn2-Dependent Transcriptional Response to Glucose Starvation. EMBO J. 2005, 24, 4115–4123. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.-Y.; Lin, Y.-Y.; Sheu, J.-C.; Wu, J.-T.; Lee, F.-J.; Chen, Y.; Lin, M.-I.; Chiang, F.-T.; Tai, T.-Y.; Berger, S.L.; et al. Acetylation of Yeast AMPK Controls Intrinsic Aging Independently of Caloric Restriction. Cell 2011, 146, 969–979. [Google Scholar] [CrossRef] [Green Version]
- Kemp, B.E.; Graves, D.J.; Benjamini, E.; Krebs, E.G. Role of Multiple Basic Residues in Determining the Substrate Specificity of Cyclic AMP-Dependent Protein Kinase. J. Biol. Chem. 1977, 252, 4888–4894. [Google Scholar] [CrossRef]
- Mok, J.; Kim, P.M.; Lam, H.Y.K.; Piccirillo, S.; Zhou, X.; Jeschke, G.R.; Sheridan, D.L.; Parker, S.A.; Desai, V.; Jwa, M.; et al. Deciphering Protein Kinase Specificity through Large-Scale Analysis of Yeast Phosphorylation Site Motifs. Sci. Signal. 2010, 3, ra12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plank, M.; Perepelkina, M.; Müller, M.; Vaga, S.; Zou, X.; Bourgoint, C.; Berti, M.; Saarbach, J.; Haesendonckx, S.; Winssinger, N.; et al. Chemical Genetics of AGC-Kinases Reveals Shared Targets of Ypk1, Protein Kinase A and Sch9. Mol. Cell. Proteom. MCP 2020, 19, 655–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crespo, J.L.; Daicho, K.; Ushimaru, T.; Hall, M.N. The GATA Transcription Factors GLN3 and GAT1 Link TOR to Salt Stress in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 34441–34444. [Google Scholar] [CrossRef] [Green Version]
- Veisova, D.; Macakova, E.; Rezabkova, L.; Sulc, M.; Vacha, P.; Sychrova, H.; Obsil, T.; Obsilova, V. Role of Individual Phosphorylation Sites for the 14-3-3-Protein-Dependent Activation of Yeast Neutral Trehalase Nth1. Biochem. J. 2012, 443, 663–670. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Sreenivas, A.; Ostrander, D.B.; Carman, G.M. Phosphorylation of Saccharomyces cerevisiae Choline Kinase on Ser30 and Ser85 by Protein Kinase A Regulates Phosphatidylcholine Synthesis by the CDP-Choline Pathway. J. Biol. Chem. 2002, 277, 34978–34986. [Google Scholar] [CrossRef] [Green Version]
- Cherry, J.R.; Johnson, T.R.; Dollard, C.; Shuster, J.R.; Denis, C.L. Cyclic AMP-Dependent Protein Kinase Phosphorylates and Inactivates the Yeast Transcriptional Activator ADR1. Cell 1989, 56, 409–419. [Google Scholar] [CrossRef]
- Chang, Y.-W.; Howard, S.C.; Herman, P.K. The Ras/PKA Signaling Pathway Directly Targets the Srb9 Protein, a Component of the General RNA Polymerase II Transcription Apparatus. Mol. Cell 2004, 15, 107–116. [Google Scholar] [CrossRef]
- Takeda, E.; Jin, N.; Itakura, E.; Kira, S.; Kamada, Y.; Weisman, L.S.; Noda, T.; Matsuura, A. Vacuole-Mediated Selective Regulation of TORC1-Sch9 Signaling Following Oxidative Stress. Mol. Biol. Cell 2018, 29, 510–522. [Google Scholar] [CrossRef] [Green Version]
- Huh, W.-K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global Analysis of Protein Localization in Budding Yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Zhang, Z.; Cottignie, I.; Van Zeebroeck, G.; Thevelein, J.M. Nutrient Transceptors Physically Interact with the Yeast S6/Protein Kinase B Homolog, Sch9, a TOR Kinase Target. Biochem. J. 2021, 478, 357–375. [Google Scholar] [CrossRef] [PubMed]
- Heitman, J.; Movva, N.; Hall, M. Targets for Cell Cycle Arrest by the Immunosuppressant Rapamycin in Yeast. Science 1991, 253, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Moriya, H.; Shimizu-Yoshida, Y.; Omori, A.; Iwashita, S.; Katoh, M.; Sakai, A. Yak1p, a DYRK Family Kinase, Translocates to the Nucleus and Phosphorylates Yeast Pop2p in Response to a Glucose Signal. Genes Dev. 2001, 15, 1217–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.; Shen, Y.; Gao, W.; Dong, J. Role of Sch9 in Regulating Ras-CAMP Signal Pathway in Saccharomyces cerevisiae. FEBS Lett. 2011, 585, 3026–3032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tudisca, V.; Recouvreux, V.; Moreno, S.; Boy-Marcotte, E.; Jacquet, M.; Portela, P. Differential Localization to Cytoplasm, Nucleus or P-Bodies of Yeast PKA Subunits under Different Growth Conditions. Eur. J. Cell Biol. 2010, 89, 339–348. [Google Scholar] [CrossRef]
- Stracka, D.; Jozefczuk, S.; Rudroff, F.; Sauer, U.; Hall, M.N. Nitrogen Source Activates TOR (Target of Rapamycin) Complex 1 via Glutamine and Independently of Gtr/Rag Proteins. J. Biol. Chem. 2014, 289, 25010–25020. [Google Scholar] [CrossRef] [Green Version]
- Shamji, A.F.; Kuruvilla, F.G.; Schreiber, S.L. Partitioning the Transcriptional Program Induced by Rapamycin among the Effectors of the Tor Proteins. Curr. Biol. 2000, 10, 1574–1581. [Google Scholar] [CrossRef] [Green Version]
- Komeili, A.; Wedaman, K.P.; O’Shea, E.K.; Powers, T. Mechanism of Metabolic Control. Target of Rapamycin Signaling Links Nitrogen Quality to the Activity of the Rtg1 and Rtg3 Transcription Factors. J. Cell Biol. 2000, 151, 863–878. [Google Scholar] [CrossRef] [Green Version]
- Hirimburegama, K.; Durnez, P.; Keleman, J.; Oris, E.; Vergauwen, R.; Mergelsberg, H.; Thevelein, J.M. Nutrient-Induced Activation of Trehalase in Nutrient-Starved Cells of the Yeast Saccharomyces cerevisiae: CAMP Is Not Involved as Second Messenger. J. Gen. Microbiol. 1992, 138, 2035–2043. [Google Scholar] [CrossRef] [Green Version]
- Durnez, P.; Pernambuco, M.B.; Oris, E.; Argüelles, J.C.; Mergelsberg, H.; Thevelein, J.M. Activation of Trehalase during Growth Induction by Nitrogen Sources in the Yeast Saccharomyces cerevisiae Depends on the Free Catalytic Subunits of CAMP-Dependent Protein Kinase, but Not on Functional Ras Proteins. Yeast 1994, 10, 1049–1064. [Google Scholar] [CrossRef]






| Observed Phenotype | Reference | Interaction Type * |
|---|---|---|
| bcy1Δ rescues growth defect of sch9Δ | Toda, 1988 [70] | TOR + PKA |
| SCH9-overexpression rescues inviability of tpk1Δ tpk2Δ tpk3Δ, ras1Δ ras2Δ and cyr1Δ | Toda, 1988 [70] | PKA + TOR |
| SCH9 rescues temperature-sensitivity of cdc25-ts | Toda, 1988 [70] | PKA + TOR |
| ras2V19-, CDC25- or TPK1-overexpression increase rapamycin resistance of gat1Δ gln3Δ | Schmelzle, 2004 [71] | TOR + PKA |
| bcy1Δ increases rapamycin resistance of gat1Δ gln3Δ | Schmelzle, 2004 [71] | TOR + PKA |
| ira2Δ, bcy1Δ and rasV19 mutations increase rapamycin resistance | Zurita-Martinez, 2005 [72] | TOR + PKA |
| ras2Δ, tpk1Δ, tpk2Δ and tpk3Δ increase rapamycin sensitivity | Zurita-Martinez, 2005 [72] | TOR + PKA |
| tpk1Δ tpk2Δ tpk3Δ yak1Δ and tpk1Δ tpk2Δ tpk3Δ msn2Δ msn4Δ increase rapamycin sensitivity | Zurita-Martinez, 2005 [72] | TOR AND PKA |
| BCY1 and PDE2 overexpression rescue temperature sensitivity of kog1-ts | Araki, 2005 [73] | TOR -PKA |
| ras1Δ ras2-23 mutations increase rapamycin resistance (1.5–2.5 ng/mL) | Ramachandran, 2011 [74] | TOR -PKA |
| bcy1Δ increases rapamycin sensitivity (1.5 ng/mL) | Ramachandran, 2011 [74] | TOR -PKA |
| Expression of rasV19 causes rapamycin sensitivity (3 ng/mL) | Ramachandran, 2011 [74] | TOR -PKA |
| Overexpression of PDE2 causes rapamycin resistance (3 ng/mL) and rescues temperature-sensitivity of tor2-ts | Ramachandran, 2011 [74] | TOR -PKA |
| rasV19 mutant shows synthetic growth defect with tor1Δ and tor1Δ tor2-ts and with tor2-ts at non-permissive temperature | Ramachandran, 2011 [74] | TOR -PKA |
| Rapamycin treatment increases phosphorylation of PKA targets Srb9, Rim15 (after 2 h) and Cki1 (~2–3 h) | Ramachandran, 2011 [74] | TOR -| PKA |
| sch9Δ has increased basal trehalase activity during growth on glycerol, but magnitude of increase after glucose addition is decreased | Crauwels, 1997 [75] | TOR -| PKA; TOR -> PKA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plank, M. Interaction of TOR and PKA Signaling in S. cerevisiae. Biomolecules 2022, 12, 210. https://doi.org/10.3390/biom12020210
Plank M. Interaction of TOR and PKA Signaling in S. cerevisiae. Biomolecules. 2022; 12(2):210. https://doi.org/10.3390/biom12020210
Chicago/Turabian StylePlank, Michael. 2022. "Interaction of TOR and PKA Signaling in S. cerevisiae" Biomolecules 12, no. 2: 210. https://doi.org/10.3390/biom12020210






