Mcm5 Represses Endodermal Migration through Cxcr4a-itgb1b Cascade Instead of Cell Cycle Control
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish Maintenance
2.2. Plasmid Construction
2.3. Generation of the Tg(Hsp70l:mcm5-T2A-mCherry) Transgenic Line
2.4. MO and mRNA Injection
2.5. Real-Time qRT-PCR and Primers
2.6. Whole-Mount In Situ Hybridization
2.7. Immunostaining
2.8. Cell Apoptosis Staining
2.9. Cell Cycle Analysis
2.10. Statistical Analysis
3. Results
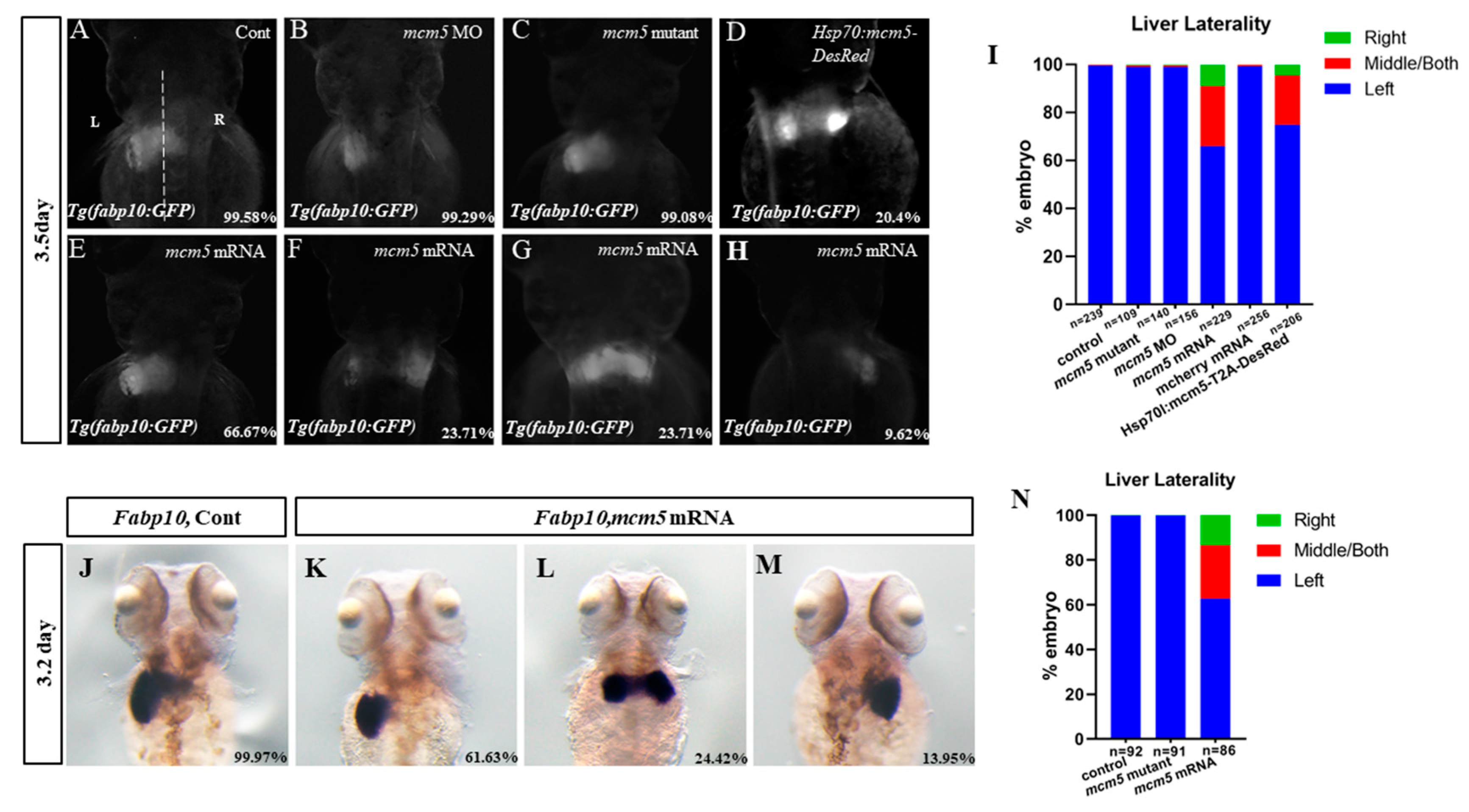
3.1. Mcm5 Overexpression Gives Rise to Liver Bifida
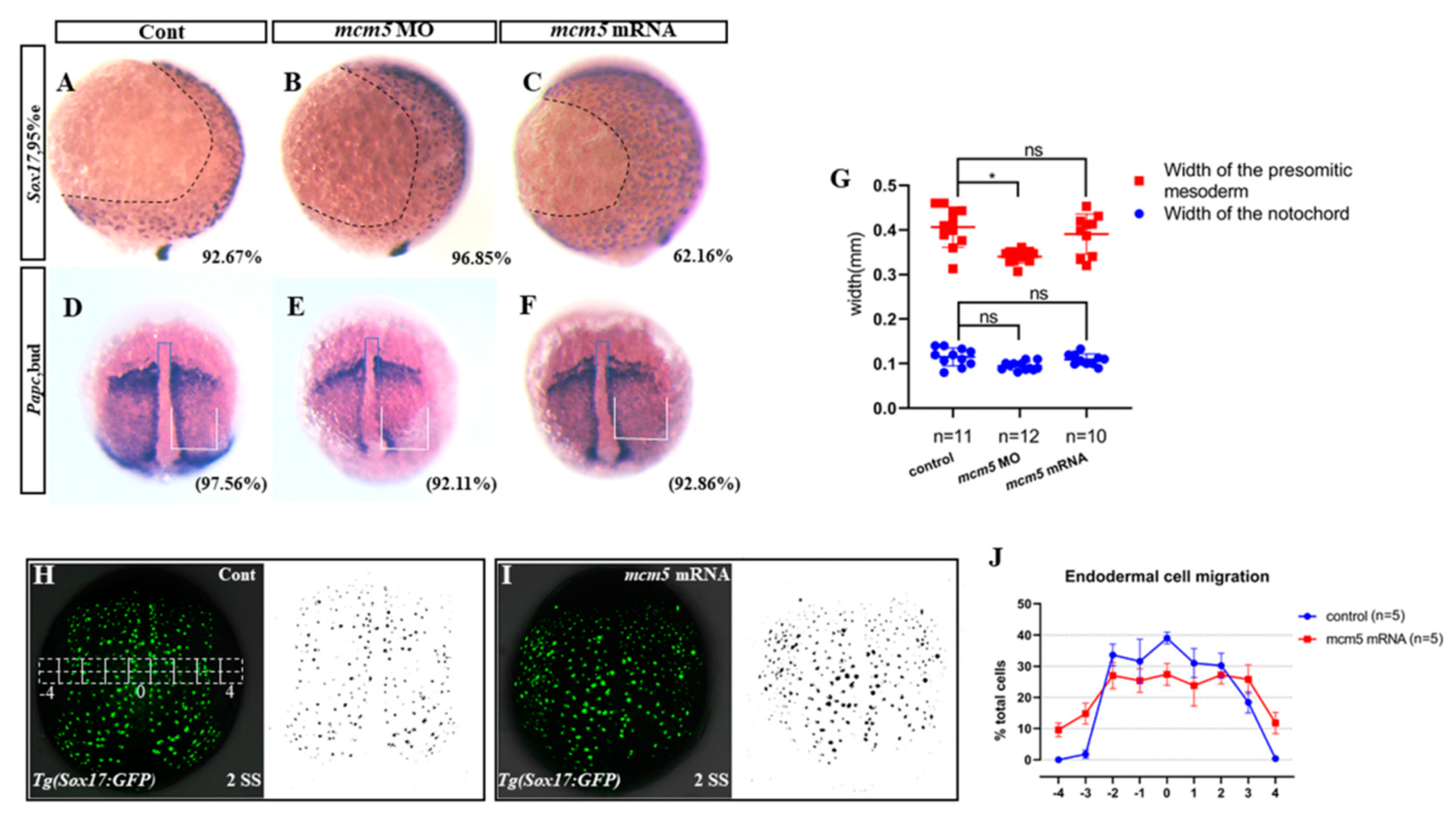
3.2. MCM5 Represses Endodermal Migration during Gastrulation
3.3. Overexpression of mcm5 did Not Result in Cell Cycle Progress Defect
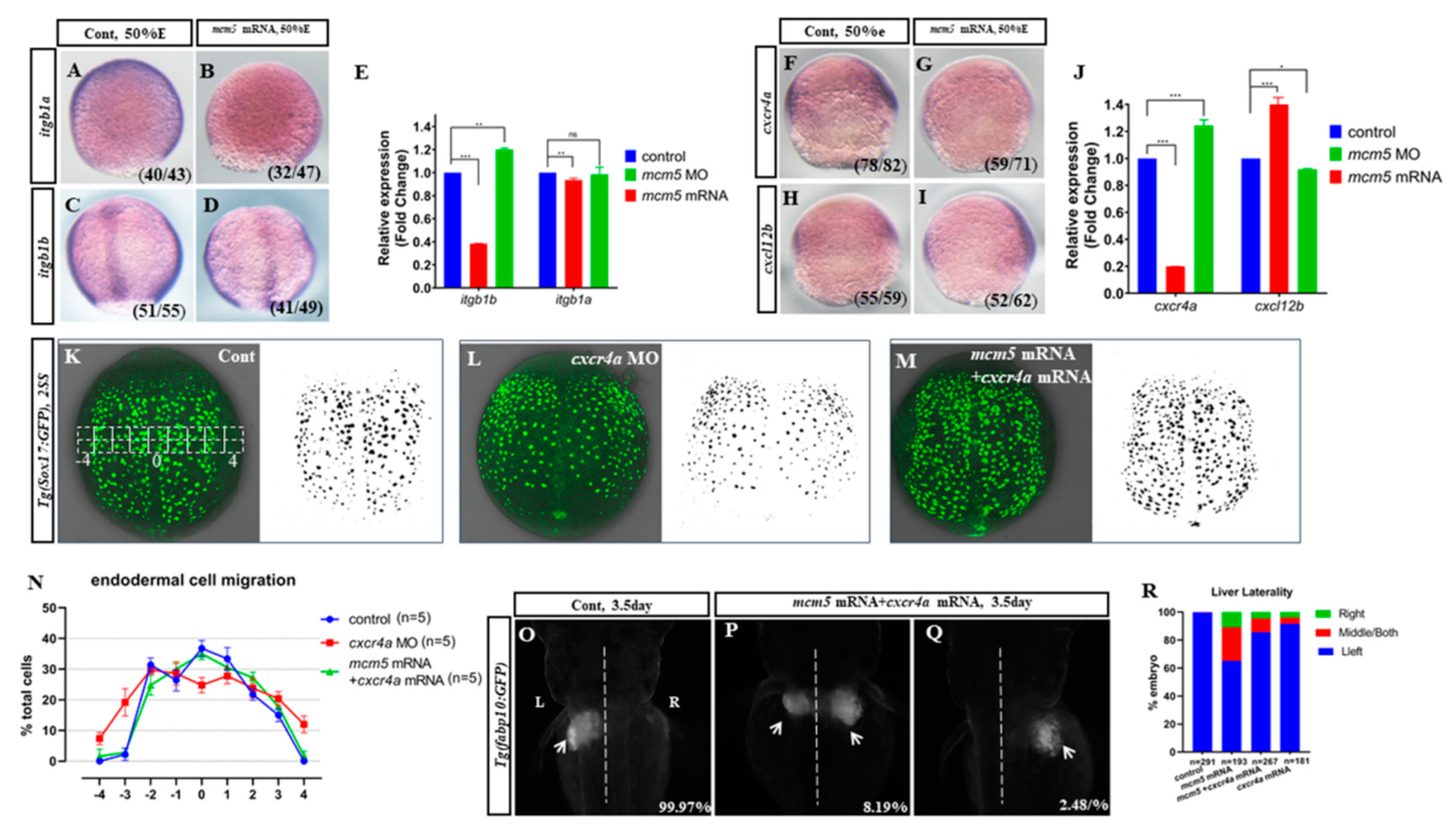
3.4. Cxcr4a-itgb1b Cascade Mediates MCM5 to Regulate Endodermal Migration
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costa, A.; Onesti, S. The MCM complex: (Just) A replicative helicase? Biochem. Soc. Trans. 2008, 36 Pt 1, 136–140. [Google Scholar] [CrossRef] [Green Version]
- Shima, N.; Alcaraz, A.; Liachko, I.; Buske, T.R.; Andrews, C.A.; Munroe, R.J.; Hartford, S.A.; Tye, B.K.; Schimenti, J.C. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 2007, 39, 93–98. [Google Scholar] [CrossRef]
- Kunnev, D.; Rusiniak, M.E.; Kudla, A.; Freeland, A.; Cady, G.K.; Pruitt, S.C. DNA damage response and tumorigenesis in Mcm2-deficient mice. Oncogene 2010, 29, 3630–3638. [Google Scholar] [CrossRef] [Green Version]
- Edwards, M.C.; Tutter, A.V.; Cvetic, C.; Gilbert, C.H.; Prokhorova, T.A.; Walter, J.C. MCM2–7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 2002, 277, 33049–33057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortez, D.; Glick, G.; Elledge, S.J. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA 2004, 101, 10078–10083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibarra, A.; Schwob, E.; Mendez, J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc. Natl. Acad. Sci. USA 2008, 105, 8956–8961. [Google Scholar] [CrossRef] [Green Version]
- Macheret, M.; Halazonetis, T.D. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 2018, 555, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Coster, G.; Frigola, J.; Beuron, F.; Morris, E.P.; Diffley, J.F. Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol. Cell 2014, 55, 666–677. [Google Scholar] [CrossRef] [Green Version]
- Ryu, S.; Holzschuh, J.; Erhardt, S.; Ettl, A.K.; Driever, W. Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc. Natl. Acad. Sci. USA 2005, 102, 18467–18472. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Huang, S.; Zhao, H.; Cao, K.; Gan, J.; Yang, C.; Xu, Z.; Li, S.; Su, B. Zebrafish Minichromosome Maintenance Protein 5 Gene Regulates the Development and Migration of Facial Motor Neurons via Fibroblast Growth Factor Signaling. Dev. Neurosci. 2021, 43, 84–94. [Google Scholar] [CrossRef]
- Nair, S.; Schilling, T.F. Chemokine signaling controls endodermal migration during zebrafish gastrulation. Science 2008, 322, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Lin, F. S1pr2/Galpha13 signaling controls myocardial migration by regulating endoderm convergence. Development 2013, 140, 789–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Zhu, C.; Ning, G.; Yang, L.; Cao, Y.; Huang, S.; Wang, Q. Chemokine signaling links cell-cycle progression and cilia formation for left-right symmetry breaking. PLoS Biol. 2019, 17, e3000203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.; Gang, K.; Yang, Q.; Liu, X.; Tang, X.; Lu, H.; He, J.; Luo, L. The CCCH-type zinc finger transcription factor Zc3h8 represses NF-kappaB-mediated inflammation in digestive organs in zebrafish. J. Biol. Chem. 2018, 293, 11971–11983. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, S.; Ma, J.; Li, C.; Zhang, Y.; Luo, L. NF-kappaB and Snail1a coordinate the cell cycle with gastrulation. J. Cell Biol. 2009, 184, 805–815. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Guo, Z.; Zhang, Y.; Liu, M.; Chen, B.; Cao, K.; Wu, Y.; Yang, M.; Yin, W.; Zhao, H.; et al. Aplnra/b Sequentially Regulate Organ Left-Right Patterning via Distinct Mechanisms. Int. J. Biol. Sci. 2019, 15, 1225–1239. [Google Scholar] [CrossRef]
- Cheng, W.; Guo, L.; Zhang, Z.; Soo, H.M.; Wen, C.; Wu, W.; Peng, J. HNF factors form a network to regulate liver-enriched genes in zebrafish. Dev. Biol. 2006, 294, 482–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, S.; Ahmad, N.; Rebagliati, M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development 2003, 130, 2303–2316. [Google Scholar] [CrossRef] [Green Version]
- Dalton, S. Linking the Cell Cycle to Cell Fate Decisions. Trends Cell Biol. 2015, 25, 592–600. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Michowski, W.; Inuzuka, H.; Shimizu, K.; Nihira, N.T.; Chick, J.M.; Li, N.; Geng, Y.; Meng, A.Y.; Ordureau, A.; et al. G1 cyclins link proliferation, pluripotency and differentiation of embryonic stem cells. Nat. Cell Biol. 2017, 19, 177–188. [Google Scholar] [CrossRef]
- Yi, X.; Yu, J.; Ma, C.; Li, L.; Luo, L.; Li, H.; Ruan, H.; Huang, H. Yap1/Taz are essential for the liver development in zebrafish. Biochem. Biophys. Res. Commun. 2018, 503, 131–137. [Google Scholar] [CrossRef]
- Mizoguchi, T.; Verkade, H.; Heath, J.K.; Kuroiwa, A.; Kikuchi, Y. Sdf1/Cxcr4 signaling controls the dorsal migration of endodermal cells during zebrafish gastrulation. Development 2008, 135, 2521–2529. [Google Scholar] [CrossRef] [Green Version]
- Parker, M.W.; Botchan, M.R.; Berger, J.M. Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 107–144. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Liu, Y.; Kong, D. Mechanism of chromosomal DNA replication initiation and replication fork stabilization in eukaryotes. Sci. China Life Sci. 2014, 57, 482–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, N.E.; Schwacha, A. The Mcm2–7 replicative helicase: A promising chemotherapeutic target. Biomed Res. Int. 2014, 2014, 549719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, S.; Diaz, M.; Flach, J.; Rodriguez-Acebes, S.; Lopez-Contreras, A.J.; Martinez, D.; Canamero, M.; Fernandez-Capetillo, O.; Isern, J.; Passegue, E.; et al. Replication stress caused by low MCM expression limits fetal erythropoiesis and hematopoietic stem cell functionality. Nat. Commun. 2015, 6, 8548. [Google Scholar] [CrossRef] [PubMed]
- Bagley, B.N.; Keane, T.M.; Maklakova, V.I.; Marshall, J.G.; Lester, R.A.; Cancel, M.M.; Paulsen, A.R.; Bendzick, L.E.; Been, R.A.; Kogan, S.C.; et al. A dominantly acting murine allele of Mcm4 causes chromosomal abnormalities and promotes tumorigenesis. PLoS Genet. 2012, 8, e1003034. [Google Scholar] [CrossRef]
- McNairn, A.J.; Chuang, C.H.; Bloom, J.C.; Wallace, M.D.; Schimenti, J.C. Female-biased embryonic death from inflammation induced by genomic instability. Nature 2019, 567, 105–108. [Google Scholar] [CrossRef]
- Ma, D.; Wang, L.; Wang, S.; Gao, Y.; Wei, Y.; Liu, F. Foxn1 maintains thymic epithelial cells to support T-cell development via mcm2 in zebrafish. Proc. Natl. Acad. Sci. USA 2012, 109, 21040–21045. [Google Scholar] [CrossRef] [Green Version]
- Casar Tena, T.; Maerz, L.D.; Szafranski, K.; Groth, M.; Blatte, T.J.; Donow, C.; Matysik, S.; Walther, P.; Jeggo, P.A.; Burkhalter, M.D.; et al. Resting cells rely on the DNA helicase component MCM2 to build cilia. Nucleic Acids Res. 2019, 47, 134–151. [Google Scholar] [CrossRef] [PubMed]
- DaFonseca, C.J.; Shu, F.; Zhang, J.J. Identification of two residues in MCM5 critical for the assembly of MCM complexes and Stat1-mediated transcription activation in response to IFN-gamma. Proc. Natl. Acad. Sci. USA 2001, 98, 3034–3039. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Xia, J.; Liu, M.; Chen, B.; Yang, M.; Yu, X.; Ou, Y.; Li, S.; Liu, X.; Feng, Y.; et al. Mcm5 Represses Endodermal Migration through Cxcr4a-itgb1b Cascade Instead of Cell Cycle Control. Biomolecules 2022, 12, 286. https://doi.org/10.3390/biom12020286
Zhang Y, Xia J, Liu M, Chen B, Yang M, Yu X, Ou Y, Li S, Liu X, Feng Y, et al. Mcm5 Represses Endodermal Migration through Cxcr4a-itgb1b Cascade Instead of Cell Cycle Control. Biomolecules. 2022; 12(2):286. https://doi.org/10.3390/biom12020286
Chicago/Turabian StyleZhang, Yu, Jiamin Xia, Min Liu, Bingyu Chen, Min Yang, Xiaoping Yu, Yu Ou, Shurong Li, Xindong Liu, Yi Feng, and et al. 2022. "Mcm5 Represses Endodermal Migration through Cxcr4a-itgb1b Cascade Instead of Cell Cycle Control" Biomolecules 12, no. 2: 286. https://doi.org/10.3390/biom12020286
APA StyleZhang, Y., Xia, J., Liu, M., Chen, B., Yang, M., Yu, X., Ou, Y., Li, S., Liu, X., Feng, Y., Su, B., & Huang, S. (2022). Mcm5 Represses Endodermal Migration through Cxcr4a-itgb1b Cascade Instead of Cell Cycle Control. Biomolecules, 12(2), 286. https://doi.org/10.3390/biom12020286





