The Novel Protease Activities of JMJD5–JMJD6–JMJD7 and Arginine Methylation Activities of Arginine Methyltransferases Are Likely Coupled
Abstract
:1. Introduction
2. Arginine Methylation and Phase Separation for Ribonucleoproteins (RNPs)
3. Arginine Methylation of Histone Subunits and Transcription Activation
4. Arginine Methylation and Transcription Repression
5. The Ubiquitous Arginine Methylation and Potential Final Destination
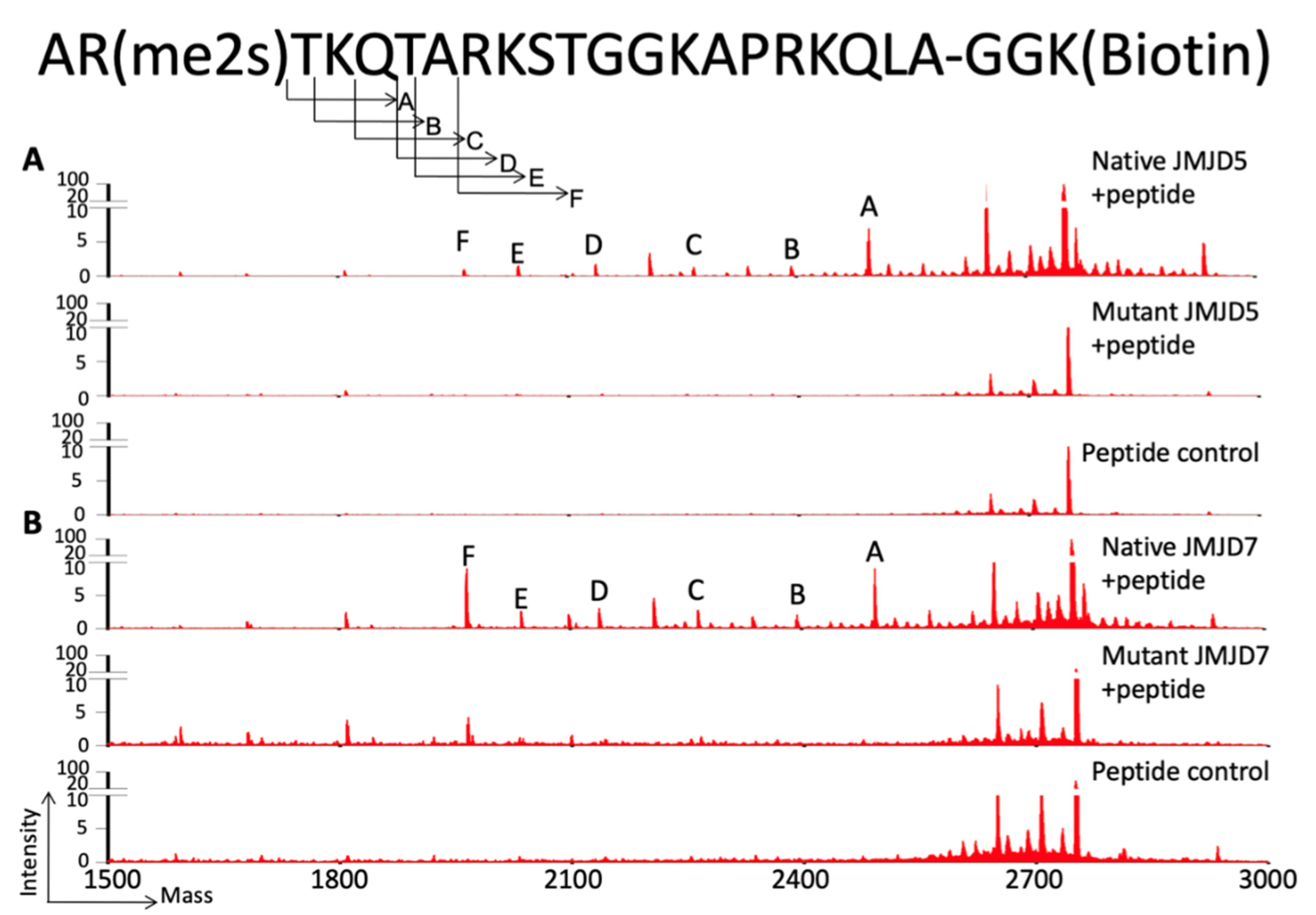
6. The Novel Protease Activities of JMJD5 Arginine-Methylated Histone Tails Coupled with CDK9 to Release Paused Pol II
7. JMJD6 Cleaves MePCE to Disrupt the 7SK snRNP Complex to Release p-TEFb
8. The Protease Activity of JMJD7 on Histone Tails and Beyond
9. Cancers Coupled with Upregulations of JMJD5/JMJD6/JMJD7 and PRMTs
10. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paik, W.K.; Kim, S. Enzymatic methylation of protein fractions from calf thymus nuclei. Biochem. Biophys. Res. Commun. 1967, 29, 14–20. [Google Scholar] [CrossRef]
- Hwang, J.W.; Cho, Y.; Bae, G.U.; Kim, S.N.; Kim, Y.K. Protein arginine methyltransferases: Promising targets for cancer therapy. Exp. Mol. Med. 2021, 53, 788–808. [Google Scholar] [CrossRef] [PubMed]
- Rakow, S.; Pullamsetti, S.S.; Bauer, U.M.; Bouchard, C. Assaying epigenome functions of PRMTs and their substrates. Methods 2020, 175, 53–65. [Google Scholar] [CrossRef]
- Guccione, E.; Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 642–657. [Google Scholar] [CrossRef]
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gayatri, S.; Bedford, M.T. Readers of histone methylarginine marks. Biochim. Biophys. Acta 2014, 1839, 702–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofweber, M.; Dormann, D. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 2019, 294, 7137–7150. [Google Scholar] [CrossRef] [Green Version]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef] [Green Version]
- Lafontaine, D.L.J.; Riback, J.A.; Bascetin, R.; Brangwynne, C.P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2021, 22, 165–182. [Google Scholar] [CrossRef]
- Lee, S.; Liu, H.; Hill, R.; Chen, C.; Hong, X.; Crawford, F.; Kingsley, M.; Zhang, Q.; Liu, X.; Chen, Z.; et al. JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes. Elife 2020, 9, e53930. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, T.; Price, D.H. RNA polymerase II elongation control. Annu. Rev. Biochem. 2012, 81, 119–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [Green Version]
- Bachand, F. Protein arginine methyltransferases: From unicellular eukaryotes to humans. Eukaryot. Cell 2007, 6, 889–898. [Google Scholar] [CrossRef] [Green Version]
- Larsen, S.C.; Sylvestersen, K.B.; Mund, A.; Lyon, D.; Mullari, M.; Madsen, M.V.; Daniel, J.A.; Jensen, L.J.; Nielsen, M.L. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 2016, 9, rs9. [Google Scholar] [CrossRef] [PubMed]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef] [Green Version]
- Gan, B.; Chen, S.; Liu, H.; Min, J.; Liu, K. Structure and function of eTudor domain containing TDRD proteins. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 119–132. [Google Scholar] [CrossRef]
- Chang, B.; Chen, Y.; Zhao, Y.; Bruick, R.K. JMJD6 is a histone arginine demethylase. Science 2007, 318, 444–447. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Ma, Q.; Wong, K.; Li, W.; Ohgi, K.; Zhang, J.; Aggarwal, A.; Rosenfeld, M.G. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 2013, 155, 1581–1595. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Ali, S.; Duan, X.; Liu, S.; Du, J.; Liu, C.; Dai, H.; Zhou, M.; Zhou, L.; Yang, L.; et al. JMJD1B Demethylates H4R3me2s and H3K9me2 to Facilitate Gene Expression for Development of Hematopoietic Stem and Progenitor Cells. Cell Rep. 2018, 23, 389–403. [Google Scholar] [CrossRef] [Green Version]
- Walport, L.J.; Hopkinson, R.J.; Chowdhury, R.; Schiller, R.; Ge, W.; Kawamura, A.; Schofield, C.J. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 2016, 7, 11974. [Google Scholar] [CrossRef] [PubMed]
- Bonnici, J.; Tumber, A.; Kawamura, A.; Schofield, C.J. Inhibitors of both the N-methyl lysyl- and arginyl-demethylase activities of the JmjC oxygenases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20170071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Wang, C.; Lee, S.; Deng, Y.; Wither, M.; Oh, S.; Ning, F.; Dege, C.; Zhang, Q.; Liu, X.; et al. Clipping of arginine-methylated histone tails by JMJD5 and JMJD7. Proc. Natl. Acad. Sci. USA 2017, 114, E7717–E7726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Wang, C.; Lee, S.; Ning, F.; Wang, Y.; Zhang, Q.; Chen, Z.; Zang, J.; Nix, J.; Dai, S.; et al. Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7. Sci. Rep. 2018, 8, 3275. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ramachandran, S.; Fong, N.; Phang, T.; Lee, S.; Parsa, P.; Liu, X.; Harmacek, L.; Danhorn, T.; Song, T.; et al. JMJD5 couples with CDK9 to release the paused RNA polymerase II. Proc. Natl. Acad. Sci. USA 2020, 117, 19888–19895. [Google Scholar] [CrossRef]
- Liu, H.; Lee, S.; Zhang, Q.; Chen, Z.; Zhang, G. The potential underlying mechanism of the leukemia caused by MLL-fusion and potential treatments. Mol. Carcinog. 2020, 59, 839–851. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Thandapani, P.; O’Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol. Cell 2013, 50, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Choi, J.M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699.e16. [Google Scholar] [CrossRef] [Green Version]
- Ryan, V.H.; Dignon, G.L.; Zerze, G.H.; Chabata, C.V.; Silva, R.; Conicella, A.E.; Amaya, J.; Burke, K.A.; Mittal, J.; Fawzi, N.L. Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol. Cell 2018, 69, 465–479.e7. [Google Scholar] [CrossRef]
- Qamar, S.; Wang, G.; Randle, S.J.; Ruggeri, F.S.; Varela, J.A.; Lin, J.Q.; Phillips, E.C.; Miyashita, A.; Williams, D.; Strohl, F.; et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-pi Interactions. Cell 2018, 173, 720–734.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef] [Green Version]
- Hofweber, M.; Hutten, S.; Bourgeois, B.; Spreitzer, E.; Niedner-Boblenz, A.; Schifferer, M.; Ruepp, M.D.; Simons, M.; Niessing, D.; Madl, T.; et al. Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell 2018, 173, 706–719.e13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogaert, E.; Boeynaems, S.; Kato, M.; Guo, L.; Caulfield, T.R.; Steyaert, J.; Scheveneels, W.; Wilmans, N.; Haeck, W.; Hersmus, N.; et al. Molecular Dissection of FUS Points at Synergistic Effect of Low-Complexity Domains in Toxicity. Cell Rep. 2018, 24, 529–537.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolzhanskaya, N.; Merz, G.; Aletta, J.M.; Denman, R.B. Methylation regulates the intracellular protein-protein and protein-RNA interactions of FMRP. J. Cell Sci. 2006, 119, 1933–1946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, W.C.; Gayatri, S.; Reineke, L.C.; Sbardella, G.; Bedford, M.T.; Lloyd, R.E. Arginine Demethylation of G3BP1 Promotes Stress Granule Assembly. J. Biol. Chem. 2016, 291, 22671–22685. [Google Scholar] [CrossRef] [Green Version]
- Tsai, W.C.; Reineke, L.C.; Jain, A.; Jung, S.Y.; Lloyd, R.E. Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1. J. Biol. Chem. 2017, 292, 18886–18896. [Google Scholar] [CrossRef] [Green Version]
- Arribas-Layton, M.; Dennis, J.; Bennett, E.J.; Damgaard, C.K.; Lykke-Andersen, J. The C-Terminal RGG Domain of Human Lsm4 Promotes Processing Body Formation Stimulated by Arginine Dimethylation. Mol. Cell. Biol. 2016, 36, 2226–2235. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, K.; Nakayama, H.; Yoshimura, M.; Masuda, A.; Dohmae, N.; Matsumoto, S.; Tsujimoto, M. PRMT1 is required for RAP55 to localize to processing bodies. RNA Biol. 2012, 9, 610–623. [Google Scholar] [CrossRef] [Green Version]
- Poornima, G.; Shah, S.; Vignesh, V.; Parker, R.; Rajyaguru, P.I. Arginine methylation promotes translation repression activity of eIF4G-binding protein, Scd6. Nucleic Acids Res. 2016, 44, 9358–9368. [Google Scholar] [CrossRef] [Green Version]
- Rajyaguru, P.; She, M.; Parker, R. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell 2012, 45, 244–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wall, M.L.; Lewis, S.M. Methylarginines within the RGG-Motif Region of hnRNP A1 Affect Its IRES Trans-Acting Factor Activity and Are Required for hnRNP A1 Stress Granule Localization and Formation. J. Mol. Biol. 2017, 429, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Kaehler, C.; Guenther, A.; Uhlich, A.; Krobitsch, S. PRMT1-mediated arginine methylation controls ATXN2L localization. Exp. Cell Res. 2015, 334, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Stetler, A.; Winograd, C.; Sayegh, J.; Cheever, A.; Patton, E.; Zhang, X.; Clarke, S.; Ceman, S. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum. Mol. Genet. 2006, 15, 87–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roovers, E.F.; Kaaij, L.J.T.; Redl, S.; Bronkhorst, A.W.; Wiebrands, K.; de Jesus Domingues, A.M.; Huang, H.Y.; Han, C.T.; Riemer, S.; Dosch, R.; et al. Tdrd6a Regulates the Aggregation of Buc into Functional Subcellular Compartments that Drive Germ Cell Specification. Dev. Cell 2018, 46, 285–301.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friesen, W.J.; Massenet, S.; Paushkin, S.; Wyce, A.; Dreyfuss, G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 2001, 7, 1111–1117. [Google Scholar] [CrossRef]
- Yamazaki, T.; Chen, S.; Yu, Y.; Yan, B.; Haertlein, T.C.; Carrasco, M.A.; Tapia, J.C.; Zhai, B.; Das, R.; Lalancette-Hebert, M.; et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012, 2, 799–806. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Ling, S.C.; Qiu, J.; Albuquerque, C.P.; Zhou, Y.; Tokunaga, S.; Li, H.; Qiu, H.; Bui, A.; Yeo, G.W.; et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat. Commun. 2015, 6, 6171. [Google Scholar] [CrossRef]
- Boke, E.; Ruer, M.; Wuhr, M.; Coughlin, M.; Lemaitre, R.; Gygi, S.P.; Alberti, S.; Drechsel, D.; Hyman, A.A.; Mitchison, T.J. Amyloid-like Self-Assembly of a Cellular Compartment. Cell 2016, 166, 637–650. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Ilin, S.; Wang, W.; Duncan, E.M.; Wysocka, J.; Allis, C.D.; Patel, D.J. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006, 442, 91–95. [Google Scholar] [CrossRef]
- Liu, K.; Chen, C.; Guo, Y.; Lam, R.; Bian, C.; Xu, C.; Zhao, D.Y.; Jin, J.; MacKenzie, F.; Pawson, T.; et al. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc. Natl. Acad. Sci. USA 2010, 107, 18398–18403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courchaine, E.M.; Barentine, A.E.S.; Straube, K.; Lee, D.R.; Bewersdorf, J.; Neugebauer, K.M. DMA-tudor interaction modules control the specificity of in vivo condensates. Cell 2021, 184, 3612–3625.e17. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Huang, S.M.; Stallcup, M.R. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 2000, 275, 40810–40816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, S.S.; Chen, D.; Lee, Y.H.; Stallcup, M.R. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem. 2001, 276, 1089–1098. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Ma, H.; Hong, H.; Koh, S.S.; Huang, S.M.; Schurter, B.T.; Aswad, D.W.; Stallcup, M.R. Regulation of transcription by a protein methyltransferase. Science 1999, 284, 2174–2177. [Google Scholar] [CrossRef]
- Strahl, B.D.; Briggs, S.D.; Brame, C.J.; Caldwell, J.A.; Koh, S.S.; Ma, H.; Cook, R.G.; Shabanowitz, J.; Hunt, D.F.; Stallcup, M.R.; et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 2001, 11, 996–1000. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Huang, Z.Q.; Xia, L.; Feng, Q.; Erdjument-Bromage, H.; Strahl, B.D.; Briggs, S.D.; Allis, C.D.; Wong, J.; Tempst, P.; et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 2001, 293, 853–857. [Google Scholar] [CrossRef]
- Ma, H.; Baumann, C.T.; Li, H.; Strahl, B.D.; Rice, R.; Jelinek, M.A.; Aswad, D.W.; Allis, C.D.; Hager, G.L.; Stallcup, M.R. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr. Biol. 2001, 11, 1981–1985. [Google Scholar] [CrossRef] [Green Version]
- Schurter, B.T.; Koh, S.S.; Chen, D.; Bunick, G.J.; Harp, J.M.; Hanson, B.L.; Henschen-Edman, A.; Mackay, D.R.; Stallcup, M.R.; Aswad, D.W. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 2001, 40, 5747–5756. [Google Scholar] [CrossRef]
- Tang, J.; Kao, P.N.; Herschman, H.R. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J. Biol. Chem. 2000, 275, 19866–19876. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Chen, T.; Hebert, J.; Li, E.; Richard, S. A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol. Cell. Biol. 2009, 29, 2982–2996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawlak, M.R.; Scherer, C.A.; Chen, J.; Roshon, M.J.; Ruley, H.E. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol. Cell. Biol. 2000, 20, 4859–4869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, N.; Lee, J.; Kim, J.; Shen, J.; Hu, M.C.; Aldaz, C.M.; Bedford, M.T. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. USA 2003, 100, 6464–6468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, D.; Vemulapalli, V.; Lu, Y.; Shen, J.; Aoyagi, S.; Fry, C.J.; Yang, Y.; Foulds, C.E.; Stossi, F.; Trevino, L.S.; et al. CARM1 methylates MED12 to regulate its RNA-binding ability. Life Sci. Alliance 2018, 1, e201800117. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Li, Q.; Yang, C.; Huo, D.; Wang, X.; Ai, C.; Kong, Y.; Sun, X.; Wang, W.; Zhou, Y.; et al. PRMT2 links histone H3R8 asymmetric dimethylation to oncogenic activation and tumorigenesis of glioblastoma. Nat. Commun. 2018, 9, 4552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouchard, C.; Sahu, P.; Meixner, M.; Notzold, R.R.; Rust, M.B.; Kremmer, E.; Feederle, R.; Hart-Smith, G.; Finkernagel, F.; Bartkuhn, M.; et al. Genomic Location of PRMT6-Dependent H3R2 Methylation Is Linked to the Transcriptional Outcome of Associated Genes. Cell Rep. 2018, 24, 3339–3352. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.E.; Wang, C.; Xu, S.; Cho, Y.W.; Wang, L.; Feng, X.; Baldridge, A.; Sartorelli, V.; Zhuang, L.; Peng, W.; et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2013, 2, e01503. [Google Scholar] [CrossRef]
- Wang, C.; Lee, J.E.; Lai, B.; Macfarlan, T.S.; Xu, S.; Zhuang, L.; Liu, C.; Peng, W.; Ge, K. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc. Natl. Acad. Sci. USA 2016, 113, 11871–11876. [Google Scholar] [CrossRef] [Green Version]
- Lai, B.; Lee, J.E.; Jang, Y.; Wang, L.; Peng, W.; Ge, K. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res. 2017, 45, 6388–6403. [Google Scholar] [CrossRef]
- Yan, J.; Chen, S.A.; Local, A.; Liu, T.; Qiu, Y.; Dorighi, K.M.; Preissl, S.; Rivera, C.M.; Wang, C.; Ye, Z.; et al. Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res. 2018, 28, 387. [Google Scholar] [CrossRef] [Green Version]
- Hu, D.; Gao, X.; Morgan, M.A.; Herz, H.M.; Smith, E.R.; Shilatifard, A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol. 2013, 33, 4745–4754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahl, J.A.; Jung, I.; Aanes, H.; Greggains, G.D.; Manaf, A.; Lerdrup, M.; Li, G.; Kuan, S.; Li, B.; Lee, A.Y.; et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016, 537, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, C.; Liu, W.; Li, J.; Li, C.; Kou, X.; Chen, J.; Zhao, Y.; Gao, H.; Wang, H.; et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 2016, 537, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zheng, H.; Huang, B.; Li, W.; Xiang, Y.; Peng, X.; Ming, J.; Wu, X.; Zhang, Y.; Xu, Q.; et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 2016, 537, 553–557. [Google Scholar] [CrossRef]
- Xia, W.; Xu, J.; Yu, G.; Yao, G.; Xu, K.; Ma, X.; Zhang, N.; Liu, B.; Li, T.; Lin, Z.; et al. Resetting histone modifications during human parental-to-zygotic transition. Science 2019, 365, 353–360. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, B.; Mao, F.; Xu, J.; Miao, H.; Zou, Z.; Phuc Khoa, L.T.; Jang, Y.; Cai, S.; Witkin, M.; et al. HOXA9 Reprograms the Enhancer Landscape to Promote Leukemogenesis. Cancer Cell 2018, 34, 643–658.e5. [Google Scholar] [CrossRef] [Green Version]
- Dhar, S.S.; Lee, S.H.; Kan, P.Y.; Voigt, P.; Ma, L.; Shi, X.; Reinberg, D.; Lee, M.G. Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4. Genes Dev. 2012, 26, 2749–2762. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Qin, S.; Chen, T.Y.; Lei, M.; Dhar, S.S.; Ho, J.C.; Dong, A.; Loppnau, P.; Li, Y.; Lee, M.G.; et al. Structural insights into trans-histone regulation of H3K4 methylation by unique histone H4 binding of MLL3/4. Nat. Commun. 2019, 10, 36. [Google Scholar] [CrossRef]
- Zhang, Y.; Jang, Y.; Lee, J.E.; Ahn, J.; Xu, L.; Holden, M.R.; Cornett, E.M.; Krajewski, K.; Klein, B.J.; Wang, S.P.; et al. Selective binding of the PHD6 finger of MLL4 to histone H4K16ac links MLL4 and MOF. Nat. Commun. 2019, 10, 2314. [Google Scholar] [CrossRef]
- Cheung, N.; Fung, T.K.; Zeisig, B.B.; Holmes, K.; Rane, J.K.; Mowen, K.A.; Finn, M.G.; Lenhard, B.; Chan, L.C.; So, C.W. Targeting Aberrant Epigenetic Networks Mediated by PRMT1 and KDM4C in Acute Myeloid Leukemia. Cancer Cell 2016, 29, 32–48. [Google Scholar] [CrossRef] [Green Version]
- Whetstine, J.R.; Nottke, A.; Lan, F.; Huarte, M.; Smolikov, S.; Chen, Z.; Spooner, E.; Li, E.; Zhang, G.; Colaiacovo, M.; et al. Reversal of Histone Lysine Trimethylation by the JMJD2 Family of Histone Demethylases. Cell 2006, 125, 467–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Zang, J.; Whetstine, J.; Hong, X.; Davrazou, F.; Kutateladze, T.G.; Simpson, M.; Mao, Q.; Pan, C.H.; Dai, S.; et al. Structural Insights into Histone Demethylation by JMJD2 Family Members. Cell 2006, 125, 691–702. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zang, J.; Kappler, J.; Hong, X.; Crawford, F.; Wang, Q.; Lan, F.; Jiang, C.; Whetstine, J.; Dai, S.; et al. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc. Natl. Acad. Sci. USA 2007, 104, 10818–10823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klose, R.J.; Yamane, K.; Bae, Y.; Zhang, D.; Erdjument-Bromage, H.; Tempst, P.; Wong, J.; Zhang, Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 2006, 442, 312–316. [Google Scholar] [CrossRef]
- Guccione, E.; Bassi, C.; Casadio, F.; Martinato, F.; Cesaroni, M.; Schuchlautz, H.; Luscher, B.; Amati, B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 2007, 449, 933–937. [Google Scholar] [CrossRef]
- Migliori, V.; Muller, J.; Phalke, S.; Low, D.; Bezzi, M.; Mok, W.C.; Sahu, S.K.; Gunaratne, J.; Capasso, P.; Bassi, C.; et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol. 2012, 19, 136–144. [Google Scholar] [CrossRef]
- Girardot, M.; Hirasawa, R.; Kacem, S.; Fritsch, L.; Pontis, J.; Kota, S.K.; Filipponi, D.; Fabbrizio, E.; Sardet, C.; Lohmann, F.; et al. PRMT5-mediated histone H4 arginine-3 symmetrical dimethylation marks chromatin at G + C-rich regions of the mouse genome. Nucleic Acids Res. 2014, 42, 235–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, W.; Kim, J.; Roeder, R.G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 2004, 117, 735–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daujat, S.; Bauer, U.M.; Shah, V.; Turner, B.; Berger, S.; Kouzarides, T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 2002, 12, 2090–2097. [Google Scholar] [CrossRef] [Green Version]
- Pollack, B.P.; Kotenko, S.V.; He, W.; Izotova, L.S.; Barnoski, B.L.; Pestka, S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 1999, 274, 31531–31542. [Google Scholar] [CrossRef] [Green Version]
- Scaglione, A.; Patzig, J.; Liang, J.; Frawley, R.; Bok, J.; Mela, A.; Yattah, C.; Zhang, J.; Teo, S.X.; Zhou, T.; et al. PRMT5-mediated regulation of developmental myelination. Nat. Commun. 2018, 9, 2840. [Google Scholar] [CrossRef]
- Pal, S.; Vishwanath, S.N.; Erdjument-Bromage, H.; Tempst, P.; Sif, S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 2004, 24, 9630–9645. [Google Scholar] [CrossRef] [Green Version]
- Waldmann, T.; Izzo, A.; Kamieniarz, K.; Richter, F.; Vogler, C.; Sarg, B.; Lindner, H.; Young, N.L.; Mittler, G.; Garcia, B.A.; et al. Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenet. Chromatin 2011, 4, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; An, W.; Cao, R.; Xia, L.; Erdjument-Bromage, H.; Chatton, B.; Tempst, P.; Roeder, R.G.; Zhang, Y. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 2003, 12, 475–487. [Google Scholar] [CrossRef]
- Ayyanathan, K.; Lechner, M.S.; Bell, P.; Maul, G.G.; Schultz, D.C.; Yamada, Y.; Tanaka, K.; Torigoe, K.; Rauscher, F.J., 3rd. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes Dev. 2003, 17, 1855–1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritsch, L.; Robin, P.; Mathieu, J.R.; Souidi, M.; Hinaux, H.; Rougeulle, C.; Harel-Bellan, A.; Ameyar-Zazoua, M.; Ait-Si-Ali, S. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol. Cell 2010, 37, 46–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, M.A.; Bannister, A.J.; Gottgens, B.; Foster, S.D.; Bartke, T.; Green, A.R.; Kouzarides, T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature 2009, 461, 819–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Nott, T.J.; Jin, J.; Pawson, T. Deciphering arginine methylation: Tudor tells the tale. Nat. Rev. Mol. Cell Biol. 2011, 12, 629–642. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Adams, M.K.; Banks, C.A.S.; Thornton, J.L.; Kempf, C.G.; Zhang, Y.; Miah, S.; Hao, Y.; Sardiu, M.E.; Killer, M.; Hattem, G.L.; et al. Differential Complex Formation via Paralogs in the Human Sin3 Protein Interaction Network. Mol. Cell. Proteom. 2020, 19, 1468–1484. [Google Scholar] [CrossRef]
- Chen, M.J.; Dixon, J.E.; Manning, G. Genomics and evolution of protein phosphatases. Sci. Signal. 2017, 10, eaag1796. [Google Scholar] [CrossRef]
- Wilkinson, K.D. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997, 11, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Zang, J.; White, J.; Wang, C.; Pan, C.H.; Zhao, R.; Murphy, R.C.; Dai, S.; Henson, P.; Kappler, J.W.; et al. Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 14568–14572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Rizzo, P.A.; Krishnan, S.; Trievel, R.C. Crystal structure and functional analysis of JMJD5 indicate an alternate specificity and function. Mol. Cell. Biol. 2012, 32, 4044–4052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsia, D.A.; Tepper, C.G.; Pochampalli, M.R.; Hsia, E.Y.; Izumiya, C.; Huerta, S.B.; Wright, M.E.; Chen, H.W.; Kung, H.J.; Izumiya, Y. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc. Natl. Acad. Sci. USA 2010, 107, 9671–9676. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, S.E.; Islam, M.S.; Gannon, J.M.; Markolovic, S.; Hopkinson, R.J.; Ge, W.; Schofield, C.J.; Chowdhury, R. JMJD5 is a human arginyl C-3 hydroxylase. Nat. Commun. 2018, 9, 1180. [Google Scholar] [CrossRef]
- Oh, S.; Janknecht, R. Histone demethylase JMJD5 is essential for embryonic development. Biochem. Biophys. Res. Commun. 2012, 420, 61–65. [Google Scholar] [CrossRef]
- Ishimura, A.; Minehata, K.; Terashima, M.; Kondoh, G.; Hara, T.; Suzuki, T. Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression. Development 2012, 139, 749–759. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.; Shin, S.; Janknecht, R. The small members of the JMJD protein family: Enzymatic jewels or jinxes? Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 406–418. [Google Scholar] [CrossRef]
- Shen, J.; Xiang, X.; Chen, L.; Wang, H.; Wu, L.; Sun, Y.; Ma, L.; Gu, X.; Liu, H.; Wang, L.; et al. JMJD5 cleaves monomethylated histone H3 N-tail under DNA damaging stress. EMBO Rep. 2017, 18, 2131–2143. [Google Scholar] [CrossRef]
- Meinhart, A.; Cramer, P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 2004, 430, 223–226. [Google Scholar] [CrossRef]
- Noble, C.G.; Hollingworth, D.; Martin, S.R.; Ennis-Adeniran, V.; Smerdon, S.J.; Kelly, G.; Taylor, I.A.; Ramos, A. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat. Struct. Mol. Biol. 2005, 12, 144–151. [Google Scholar] [CrossRef]
- Lunde, B.M.; Reichow, S.L.; Kim, M.; Suh, H.; Leeper, T.C.; Yang, F.; Mutschler, H.; Buratowski, S.; Meinhart, A.; Varani, G. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2010, 17, 1195–1201. [Google Scholar] [CrossRef]
- Becker, R.; Loll, B.; Meinhart, A. Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2008, 283, 22659–22669. [Google Scholar] [CrossRef] [Green Version]
- Ni, Z.; Xu, C.; Guo, X.; Hunter, G.O.; Kuznetsova, O.V.; Tempel, W.; Marcon, E.; Zhong, G.; Guo, H.; Kuo, W.W.; et al. RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation. Nat. Struct. Mol. Biol. 2014, 21, 686–695. [Google Scholar] [CrossRef] [Green Version]
- Glover-Cutter, K.; Kim, S.; Espinosa, J.; Bentley, D.L. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008, 15, 71–78. [Google Scholar] [CrossRef] [Green Version]
- Chapman, R.D.; Heidemann, M.; Albert, T.K.; Mailhammer, R.; Flatley, A.; Meisterernst, M.; Kremmer, E.; Eick, D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 2007, 318, 1780–1782. [Google Scholar] [CrossRef]
- Schuller, R.; Forne, I.; Straub, T.; Schreieck, A.; Texier, Y.; Shah, N.; Decker, T.M.; Cramer, P.; Imhof, A.; Eick, D. Heptad-Specific Phosphorylation of RNA Polymerase II CTD. Mol. Cell 2016, 61, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, J.C.; Ebmeier, C.C.; Podell, E.R.; Heimiller, J.; Taatjes, D.J.; Cech, T.R. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012, 26, 2690–2695. [Google Scholar] [CrossRef] [Green Version]
- Kwon, I.; Kato, M.; Xiang, S.; Wu, L.; Theodoropoulos, P.; Mirzaei, H.; Han, T.; Xie, S.; Corden, J.L.; McKnight, S.L. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 2013, 155, 1049–1060. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Halanski, M.A.; Radonovich, M.F.; Kashanchi, F.; Peng, J.; Price, D.H.; Brady, J.N. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 2000, 20, 5077–5086. [Google Scholar] [CrossRef] [Green Version]
- Marshall, N.F.; Peng, J.; Xie, Z.; Price, D.H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 1996, 271, 27176–27183. [Google Scholar] [CrossRef] [Green Version]
- Hsin, J.P.; Manley, J.L. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012, 26, 2119–2137. [Google Scholar] [CrossRef] [Green Version]
- Ni, Z.; Schwartz, B.E.; Werner, J.; Suarez, J.R.; Lis, J.T. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 2004, 13, 55–65. [Google Scholar] [CrossRef]
- Chen, F.X.; Smith, E.R.; Shilatifard, A. Born to run: Control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2018, 19, 464–478. [Google Scholar] [CrossRef]
- Chun, Y.; Joo, Y.J.; Suh, H.; Batot, G.; Hill, C.P.; Formosa, T.; Buratowski, S. Selective kinase inhibition shows that Bur1 (Cdk9) phosphorylates the Rpb1 linker in vivo. Mol. Cell. Biol. 2019, 39, e00602-18. [Google Scholar] [CrossRef] [Green Version]
- Keogh, M.C.; Podolny, V.; Buratowski, S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 2003, 23, 7005–7018. [Google Scholar] [CrossRef] [Green Version]
- Mavrich, T.N.; Jiang, C.; Ioshikhes, I.P.; Li, X.; Venters, B.J.; Zanton, S.J.; Tomsho, L.P.; Qi, J.; Glaser, R.L.; Schuster, S.C.; et al. Nucleosome organization in the Drosophila genome. Nature 2008, 453, 358–362. [Google Scholar] [CrossRef]
- Schones, D.E.; Cui, K.; Cuddapah, S.; Roh, T.Y.; Barski, A.; Wang, Z.; Wei, G.; Zhao, K. Dynamic regulation of nucleosome positioning in the human genome. Cell 2008, 132, 887–898. [Google Scholar] [CrossRef] [Green Version]
- Weber, C.M.; Ramachandran, S.; Henikoff, S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell 2014, 53, 819–830. [Google Scholar] [CrossRef] [Green Version]
- Voong, L.N.; Xi, L.; Sebeson, A.C.; Xiong, B.; Wang, J.P.; Wang, X. Insights into Nucleosome Organization in Mouse Embryonic Stem Cells through Chemical Mapping. Cell 2016, 167, 1555–1570.e15. [Google Scholar] [CrossRef] [Green Version]
- Vangimalla, S.S.; Ganesan, M.; Kharbanda, K.K.; Osna, N.A. Bifunctional Enzyme JMJD6 Contributes to Multiple Disease Pathogenesis: New Twist on the Old Story. Biomolecules 2017, 7, 41. [Google Scholar] [CrossRef]
- Fadok, V.A.; Bratton, D.L.; Rose, D.M.; Pearson, A.; Ezekewitz, R.A.; Henson, P.M. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 2000, 405, 85–90. [Google Scholar] [CrossRef]
- Cikala, M.; Alexandrova, O.; David, C.N.; Proschel, M.; Stiening, B.; Cramer, P.; Bottger, A. The phosphatidylserine receptor from Hydra is a nuclear protein with potential Fe(II) dependent oxygenase activity. BMC Cell Biol. 2004, 5, 26. [Google Scholar] [CrossRef] [Green Version]
- Cui, P.; Qin, B.; Liu, N.; Pan, G.; Pei, D. Nuclear localization of the phosphatidylserine receptor protein via multiple nuclear localization signals. Exp. Cell Res. 2004, 293, 154–163. [Google Scholar] [CrossRef]
- Bose, J.; Gruber, A.D.; Helming, L.; Schiebe, S.; Wegener, I.; Hafner, M.; Beales, M.; Kontgen, F.; Lengeling, A. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 2004, 3, 15. [Google Scholar] [CrossRef] [Green Version]
- Webby, C.J.; Wolf, A.; Gromak, N.; Dreger, M.; Kramer, H.; Kessler, B.; Nielsen, M.L.; Schmitz, C.; Butler, D.S.; Yates, J.R., 3rd; et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 2009, 325, 90–93. [Google Scholar] [CrossRef]
- Han, G.; Li, J.; Wang, Y.; Li, X.; Mao, H.; Liu, Y.; Chen, C.D. The hydroxylation activity of Jmjd6 is required for its homo-oligomerization. J. Cell. Biochem. 2012, 113, 1663–1670. [Google Scholar] [CrossRef]
- Neumann, B.; Coakley, S.; Giordano-Santini, R.; Linton, C.; Lee, E.S.; Nakagawa, A.; Xue, D.; Hilliard, M.A. EFF-1-mediated regenerative axonal fusion requires components of the apoptotic pathway. Nature 2015, 517, 219–222. [Google Scholar] [CrossRef]
- Yang, H.; Chen, Y.Z.; Zhang, Y.; Wang, X.; Zhao, X.; Godfroy, J.I., 3rd; Liang, Q.; Zhang, M.; Zhang, T.; Yuan, Q.; et al. A lysine-rich motif in the phosphatidylserine receptor PSR-1 mediates recognition and removal of apoptotic cells. Nat. Commun. 2015, 6, 5717. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Kim, S.J.; Jeong, D.G.; Lee, S.M.; Ryu, S.E. Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau. J. Biol. Chem. 2003, 278, 7558–7563. [Google Scholar] [CrossRef] [Green Version]
- Li, M.O.; Sarkisian, M.R.; Mehal, W.Z.; Rakic, P.; Flavell, R.A. Phosphatidylserine receptor is required for clearance of apoptotic cells. Science 2003, 302, 1560–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, M.K.; Mochizuki, K.; Zhou, M.; Jeong, H.S.; Brady, J.N.; Ozato, K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 2005, 19, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yik, J.H.; Chen, R.; He, N.; Jang, M.K.; Ozato, K.; Zhou, Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 2005, 19, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Jeronimo, C.; Forget, D.; Bouchard, A.; Li, Q.; Chua, G.; Poitras, C.; Therien, C.; Bergeron, D.; Bourassa, S.; Greenblatt, J.; et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 2007, 27, 262–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xhemalce, B.; Robson, S.C.; Kouzarides, T. Human RNA methyltransferase BCDIN3D regulates microRNA processing. Cell 2012, 151, 278–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.; Yang, Z.; Chen, R.; Zhou, Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010, 38, 360–369. [Google Scholar] [CrossRef] [Green Version]
- Quaresma, A.J.C.; Bugai, A.; Barboric, M. Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb. Nucleic Acids Res. 2016, 44, 7527–7539. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Morlock, H.; Hanes, S.D. The Bin3 RNA methyltransferase is required for repression of caudal translation in the Drosophila embryo. Dev. Biol. 2011, 352, 104–115. [Google Scholar] [CrossRef] [Green Version]
- Peterlin, B.M.; Brogie, J.E.; Price, D.H. 7SK snRNA: A noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA 2012, 3, 92–103. [Google Scholar] [CrossRef] [Green Version]
- Rahman, S.; Sowa, M.E.; Ottinger, M.; Smith, J.A.; Shi, Y.; Harper, J.W.; Howley, P.M. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 2011, 31, 2641–2652. [Google Scholar] [CrossRef] [Green Version]
- Konuma, T.; Yu, D.; Zhao, C.; Ju, Y.; Sharma, R.; Ren, C.; Zhang, Q.; Zhou, M.M.; Zeng, L. Structural Mechanism of the Oxygenase JMJD6 Recognition by the Extraterminal (ET) Domain of BRD4. Sci. Rep. 2017, 7, 16272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luck, K.; Kim, D.K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Gudmundsdottir, B.; Gudmundsson, K.O.; Klarmann, K.D.; Singh, S.K.; Sun, L.; Singh, S.; Du, Y.; Coppola, V.; Stockwin, L.; Nguyen, N.; et al. POGZ Is Required for Silencing Mouse Embryonic beta-like Hemoglobin and Human Fetal Hemoglobin Expression. Cell Rep. 2018, 23, 3236–3248. [Google Scholar] [CrossRef] [PubMed]
- Suliman-Lavie, R.; Title, B.; Cohen, Y.; Hamada, N.; Tal, M.; Tal, N.; Monderer-Rothkoff, G.; Gudmundsdottir, B.; Gudmundsson, K.O.; Keller, J.R.; et al. Pogz deficiency leads to transcription dysregulation and impaired cerebellar activity underlying autism-like behavior in mice. Nat. Commun. 2020, 11, 5836. [Google Scholar] [CrossRef]
- Liu, Y.; Arai, A.; Kim, T.; Kim, S.; Park, N.H.; Kim, R.H. Histone Demethylase Jmjd7 Negatively Regulates Differentiation of Osteoclast. Chin. J. Dent. Res. 2018, 21, 113–118. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, Y.; Li, J.; Chang, I.; Wang, C.Y. A novel read-through transcript JMJD7-PLA2G4B regulates head and neck squamous cell carcinoma cell proliferation and survival. Oncotarget 2017, 8, 1972–1982. [Google Scholar] [CrossRef] [Green Version]
- Markolovic, S.; Zhuang, Q.; Wilkins, S.E.; Eaton, C.D.; Abboud, M.I.; Katz, M.J.; McNeil, H.E.; Lesniak, R.K.; Hall, C.; Struwe, W.B.; et al. The Jumonji-C oxygenase JMJD7 catalyzes (3S)-lysyl hydroxylation of TRAFAC GTPases. Nat. Chem. Biol. 2018, 14, 688–695. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, C.; Huang, X.; Shen, Y.; Shen, J.; Ying, K. Differential proteome profiling of pleural effusions from lung cancer and benign inflammatory disease patients. Biochim. Biophys. Acta 2012, 1824, 692–700. [Google Scholar] [CrossRef]
- Zhang, R.; Huang, Q.; Li, Y.; Song, Y.; Li, Y. JMJD5 is a potential oncogene for colon carcinogenesis. Int. J. Clin. Exp. Pathol. 2015, 8, 6482–6489. [Google Scholar]
- Wang, H.J.; Pochampalli, M.; Wang, L.Y.; Zou, J.X.; Li, P.S.; Hsu, S.C.; Wang, B.J.; Huang, S.H.; Yang, P.; Yang, J.C.; et al. KDM8/JMJD5 as a dual coactivator of AR and PKM2 integrates AR/EZH2 network and tumor metabolism in CRPC. Oncogene 2019, 38, 17–32. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Chen, S.; Yang, Y.; Ma, X.; Shao, B.; Yang, S.; Wei, Y.; Wei, X. Jumonji domain-containing protein 6 protein and its role in cancer. Cell Prolif. 2020, 53, e12747. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Tie, Y.; Fang, Z.; Wu, X.; Yi, T.; Huang, S.; Liang, X.; Qian, Y.; Wang, X.; Pi, R.; et al. Jumonji domain-containing 6 (JMJD6) identified as a potential therapeutic target in ovarian cancer. Signal Transduct. Target. Ther. 2019, 4, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, M.; Sun, Y.; Xi, Z.; Milazzo, G.; Poulos, R.C.; Bartenhagen, C.; Bell, J.L.; Mayoh, C.; Ho, N.; Tee, A.E.; et al. JMJD6 is a tumorigenic factor and therapeutic target in neuroblastoma. Nat. Commun. 2019, 10, 3319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, T.E.; Liau, B.B.; Wallace, L.C.; Morton, A.R.; Xie, Q.; Dixit, D.; Factor, D.C.; Kim, L.J.Y.; Morrow, J.J.; Wu, Q.; et al. Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature 2017, 547, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Kwok, J.; O’Shea, M.; Hume, D.A.; Lengeling, A. Jmjd6, a JmjC Dioxygenase with Many Interaction Partners and Pleiotropic Functions. Front. Genet. 2017, 8, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poulard, C.; Rambaud, J.; Lavergne, E.; Jacquemetton, J.; Renoir, J.M.; Tredan, O.; Chabaud, S.; Treilleux, I.; Corbo, L.; Le Romancer, M. Role of JMJD6 in Breast Tumourigenesis. PLoS ONE 2015, 10, e0126181. [Google Scholar] [CrossRef]
- Wang, F.; He, L.; Huangyang, P.; Liang, J.; Si, W.; Yan, R.; Han, X.; Liu, S.; Gui, B.; Li, W.; et al. JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol. 2014, 12, e1001819. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.F.; Miller, L.D.; Chan, X.B.; Black, M.A.; Pang, B.; Ong, C.W.; Salto-Tellez, M.; Liu, E.T.; Desai, K.V. JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer. Breast Cancer Res. 2012, 14, R85. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Yang, Y.; Zhang, X. MiR-770 inhibits tumorigenesis and EMT by targeting JMJD6 and regulating WNT/beta-catenin pathway in non-small cell lung cancer. Life Sci. 2017, 188, 163–171. [Google Scholar] [CrossRef]
- Zhou, D.X.; Zhou, D.; Zhan, S.Q.; Wang, P.; Qin, K.; Gan, W.; Lin, X.F. Inhibition of JMJD6 expression reduces the proliferation, migration and invasion of neuroglioma stem cells. Neoplasma 2017, 64, 700–708. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Lu, X.; Huang, J.; He, H.; Chen, L.; Liu, Y.; Wang, H.; Xu, Y.; Xing, S.; Ruan, X.; et al. Epigenome screening highlights that JMJD6 confers an epigenetic vulnerability and mediates sunitinib sensitivity in renal cell carcinoma. Clin. Transl. Med. 2021, 11, e328. [Google Scholar] [CrossRef]
- Paschalis, A.; Welti, J.; Neeb, A.J.; Yuan, W.; Figueiredo, I.; Pereira, R.; Ferreira, A.; Riisnaes, R.; Rodrigues, D.N.; Jimenez-Vacas, J.M.; et al. JMJD6 Is a Druggable Oxygenase That Regulates AR-V7 Expression in Prostate Cancer. Cancer Res. 2021, 81, 1087–1100. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhang, R.; Liu, Y.; Fang, Z.; Zhang, H.; Fan, Y.; Yang, S.; Xiang, R. Discovery of a new class of JMJD6 inhibitors and structure-activity relationship study. Bioorg. Med. Chem. Lett. 2021, 44, 128109. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Thinnes, C.C.; Holt-Martyn, J.P.; Chowdhury, R.; McDonough, M.A.; Schofield, C.J. Inhibition of JMJD6 by 2-Oxoglutarate Mimics. ChemMedChem 2022, 17, e202100398. [Google Scholar] [CrossRef] [PubMed]
- Saran, A.R.; Kalinowska, D.; Oh, S.; Janknecht, R.; DiTacchio, L. JMJD5 links CRY1 function and proteasomal degradation. PLoS Biol. 2018, 16, e2006145. [Google Scholar] [CrossRef] [Green Version]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Wei, P.; Zhang, Q.; Chen, Z.; Liu, J.; Zhang, G. The Novel Protease Activities of JMJD5–JMJD6–JMJD7 and Arginine Methylation Activities of Arginine Methyltransferases Are Likely Coupled. Biomolecules 2022, 12, 347. https://doi.org/10.3390/biom12030347
Liu H, Wei P, Zhang Q, Chen Z, Liu J, Zhang G. The Novel Protease Activities of JMJD5–JMJD6–JMJD7 and Arginine Methylation Activities of Arginine Methyltransferases Are Likely Coupled. Biomolecules. 2022; 12(3):347. https://doi.org/10.3390/biom12030347
Chicago/Turabian StyleLiu, Haolin, Pengcheng Wei, Qianqian Zhang, Zhongzhou Chen, Junfeng Liu, and Gongyi Zhang. 2022. "The Novel Protease Activities of JMJD5–JMJD6–JMJD7 and Arginine Methylation Activities of Arginine Methyltransferases Are Likely Coupled" Biomolecules 12, no. 3: 347. https://doi.org/10.3390/biom12030347
APA StyleLiu, H., Wei, P., Zhang, Q., Chen, Z., Liu, J., & Zhang, G. (2022). The Novel Protease Activities of JMJD5–JMJD6–JMJD7 and Arginine Methylation Activities of Arginine Methyltransferases Are Likely Coupled. Biomolecules, 12(3), 347. https://doi.org/10.3390/biom12030347






