The Role of Epigenetic Modifications in Human Cancers and the Use of Natural Compounds as Epidrugs: Mechanistic Pathways and Pharmacodynamic Actions
Abstract
1. Introduction
2. Overview on Cancer
3. Epigenetic Modifications
4. Implication of Epigenetic Perturbations in Cancer Diseases
4.1. DNA Methylation and Cancer
4.2. mRNA Methylation and Cancer
4.3. Histone Modification and Cancer
4.3.1. Histone Acetylation and Cancer
4.3.2. Histone Methylation and Cancer
4.4. Chromatin Remodeling
4.5. RNAs Modification and Cancer
4.5.1. MicroRNAs and Cancer
4.5.2. Long Non-Coding RNAs and Cancer
5. Natural Compounds as Epidrugs against Cancer
5.1. Flavonoids Targeting Epigenetic Pathways in Cancer
5.1.1. Anthocyanidins
5.1.2. Anthocyanins
5.1.3. Cyanidin
5.1.4. Pelargonidin
5.1.5. Delphinidin
| Molecule (Origin) | Used Model | Key Results and Conclusion (⇒) | Refs. |
|---|---|---|---|
| Anthocyanidins | |||
| Anthocyanins (Raspberry) | Colorectal T116, Caco2, and SW480 cells |
| [134] |
| Cyanidin (Not reported) | MCF7 breast cancer cells |
| [137] |
| Delphinidin (Purchased) | Mouse epidermal JB6 P+ cell line Human hepatocellular HepG2-C8 cell line |
| [143] |
| Pelargonidin (Purchased) | Mouse skin epidermal JB6 (JB6 P+) cells HepG2-C8 cells |
| [140] |
| Pelargonidin (Not reported) |
inhibitor for cancer treatment | [139] | |
| Biflavonoids | |||
| Amentoflavone (Selaginella tamariscina) | Glioma cell line: U87, LV229, U251, LN18 and U373 |
| [144] |
| Flavans | |||
| Kazinol Q (Purchased) | LNCaP prostate and MCF-7 and MDAMB-231 breast cancer |
| [145] |
| Flavanols | |||
| Catechin, Epicatechin, (-)-epigallocatechin-3-O-gallate (Purchased) | Human breast cancer cell lines (MCF-7 and MDA-MB-231) |
enzymes through increased formation of SAH (a potent noncompetitive inhibitor of DNMTs) | [146] |
| (-)-Epigallocatechin-3-gallate (Not reported) | Female SKH-1 hairless mice |
| [147] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Urinary bladder transitional cell carcinoma (T24 cells), prostate adenocarcinoma (PC3 cells), and colorectal adenocarcinoma (HT29 cells) |
| [148] |
| (-)-Epigallocatechin-3-gallate (Not reported) | Esophageal squamous cell carcinoma cell lines, KYSE 510, 150 and 450 |
| [149] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human breast cancer cell lines (MCF-7 and MDA-MB-231) |
| [146] |
| (-)-Epigallocatechin-3-gallate (Not reported) | Wild-type and APC 1309 knock-out mice |
| [150] |
| (-)-Epigallocatechin-3-gallate (Purchased) | TK6, Jurkat, and KG-1 leukemia cell models |
and RG108 caused concentration-dependent demethylation of genomic DNA, whereas EGCG failed to induce significant effects | [2] |
| (-)-Epigallocatechin-3-gallate (Purchased) | MCF-7 breast cancer and HL60 leukemia cells |
| [151] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human oral squamous cell carcinoma cell lines HSC3, HSC4, SCC9, SCC25 and human cervical cancer cell line HeLa |
| [152] |
| (-)-Epigallocatechin-3-gallate (Purchased) | SCC-13, A431 and HaCaT skin cancer cells |
regulatory mechanisms | [153] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human lung carcinoma cell lines H1703, H460 and A549 and colorectal cancer cell line HCT116 |
| [154] |
| (-)-Epigallocatechin-3-gallate (Not reported) | - |
| [155] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Breast cancer MCF-7 and MDA-MB-231 cells and normal control MCF10A cells |
| [156] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human skin cancer A431 and squamous cell carcinoma (SCC) 13 |
| [157] |
| (-)-Epigallocatechin-3-gallate (Purchased) | HCT116, HEK293 and MRC5 cells |
cells by EGCG could be caused by the cancer-specific induction of reactive oxygen species (ROS) and epigenetic modulation of expression of apoptosis-related genes, such as hTERT | [158] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human T lymphocyte leukemic Jurkat cells |
| [159] |
| (-)-Epigallocatechin-3-gallate (Purchased) | SKOV3-ip1 and SKOV3TR-ip2 cells Ovarian cancer cell lines |
treatment can inhibit ovarian cancer cells by creating DNA damage through decreasing hTERT and Bcl-2 expression. | [160] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human colon cancer cell lines HT-29 and HCT 116 |
other DNMT and HDAC inhibitors, could be beneficial to treat colon cancer | [161] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human breast carcinoma cell lines MCF7 and MDA MB 231 |
| [162] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human colon adenocarcinoma cell line HT29 cells |
| [163] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human Burkitt’s Lymphoma CA46 cells |
| [164] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human established PDA cell lines BxPc-3 and MIA-PaCa2 and human hTERT-HPNE immortalized pancreatic duct cells CRL-1097 |
| [165] |
| (-)-Epigallocatechin-3-gallate (Not reported) | Breast cancer cell lines MCF-7 and MDA-MB-231 |
| [166] |
| (-)-Epigallocatechin-3-gallate (Purchased) | RKO (CRL-2577), HCT-116 (CCL-247) and HT-29 (HTB-38) colorectal cancer cells |
| [167] |
| (-)-Epigallocatechin-3-gallate (Purchased) | CAL-27 human oral squamous cell carcinoma cell line |
| [168] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human cervical carcinoma cell line HeLa |
| [169] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human breast cancer cell lines (MCF-7, MDA-MB 231) and non-tumorigenic MCF-10A breast epithelial cells |
| [170] |
| (-)-Epigallocatechin-3-gallate (Purchased) | A549 and DDP-resistant A549/DDP human lung adenocarcinoma cell |
candidate genes | [171] |
| (-)-Epigallocatechin-3-gallate (Not reported) | Human APL NB4 and HL-60 cells |
| [172] |
| (-)-Epigallocatechin-3-gallate (Purchased) | ERα (+) MCF-7 and ERα (-) MDA-MB-157, MDA-MB-231, and HCC1806 breast cancer |
proliferation of breast cancer cells | [173] |
| (-)-Epigallocatechin-3-gallate (Purchased) | MCF7 and MDA-MB-231 breast cancer cells |
promote cancer cell death and reactivate DNA methylation-silenced tumor suppressor genes in breast cancer cells with different invasive potential | [174] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human cervical cancer cell line Hela |
eugenol–amarogentin, demonstrates a better chemotherapeutic effect on the Hela cell line, which might be due to the epigenetic modification, and in particular DNA hypomethylation through downregulation of DNMT1 | [175] |
| (-)-Epigallocatechin-3-gallate (Not reported) | Myeloid leukemia NB4 and K562 cells |
| [176] |
| (-)-Epigallocatechin-3-gallate (Not reported) | Human prostate cancer DUPRO and LNCaP cells |
| [177] |
| (-)-Epigallocatechin-3-gallate (Purchased) | Human breast cancer cell lines MCF-7 (cat. no. HTB-22) and MDA-MB 231 |
| [178] |
| (-)-Epigallocatechin-3-gallate (Not reported) | Human microvascular endothelial cells (HMEC-1) and human umbilical vein endothelial cells (HUVECs) |
| [179] |
| (-)-Epigallocatechin-3-gallate (Purchased) | ERα (+) MCF-7 and ERα (–) MDA-MB-157, MDA-MB- 231, and HCC1806 breast cancer cells |
| [180] |
| Flavanones | |||
| Hesperetin (Not reported) | NCI-60 cell line |
and MLH1 may be effective in circumventing chemoresistance in breast cancer | [181] |
| Hesperidin (Purchased) | Human prostate carcinoma cells DU145 |
| [182] |
| Hesperidin (Not reported) | HL60 human leukaemia cancer Diethylnitrosamine-induced hepatocarcinogen |
therapy purposes | [183] |
| Naringenin (Not reported) | Mesangial cell (MMCs) |
| [184] |
| Naringenin (Purchased) | Human colon adenocarcinoma (CaCo-2) |
| [185] |
| Flavanonols | |||
| Taxifolin (Purchased) | Human hepatoma HepG2 cells |
epigenetic pathway | [186] |
| Flavones | |||
| 3,6-dihydroxyflavone (3,6-DHF) (Purchased) | Human breast epithelial MCF-10A cells |
| [187] |
| 3,6-dihydroxyflavone (3,6-DHF) (Purchased) | MCF-10A and MDA-MB-231 cells Mammary gland and tumor samples (in vivo) |
hypermethylation, and consequently upregulates miR-34a in breast carcinogenesis | [188] |
| Apigenin (Purchased) | Human prostate cancer LNCaP and DU145 cells and transformed human prostate epithelial RWPE-1 cells |
| [189] |
| Apigenin (Purchased) | Human prostate cancer cell lines 22Rv1 and PC-3 |
| [190] |
| Apigenin (Purchased) | Skin epidermal JB6 P+ cells |
| [191] |
| Apigenin (Purchased) | MDA-MB-231 human breast cancer cells |
| [192] |
| Baicalein (Purchased) | MCF7 breast cancer cells |
| [137] |
| Casticin (Purchased) | MGC803 gastric cancer cells |
of gastric cancer MGC803 cells by upregulating RECK gene expression and reducing intracellular methylation levels | [193] |
| Casticin (Not reported) | MHCC97H, SK-Hep-1, and L02 hepatic cancer cells |
| [194] |
| Chrysin (Purchased) | Human prostate cancer LNCaP and DU145 cells and transformed human prostate epithelial RWPE-1 cells |
| [189] |
| Diosmin (Purchased) | Human breast cancer cells MCF-7, MDA-MB-231 and SK-BR-3 |
| [195] |
| Diosmin (Purchased) | Human prostate carcinoma cells (DU145) |
chromosomal damage in DU145 prostate cancer cells, which, in turn, may provoke apoptotic cell death and may have implications for diosmin-based anticancer therapy | [182] |
| Isovitexin (Purchased) | Human OS U2OS and MG63 cell lines |
| [196] |
| Luteolin (Purchased) | Human lung cancer cells LNM35 |
| [197] |
| Luteolin (Not reported) | Human breast cancer BT474, MCF-7 and MDA-MB-231 cells |
| [198] |
| Luteolin (Purchased) | Human HT-29 colon cancer and SNU-407 cells |
| [199] |
| Luteolin (Purchased) | Human colon cancer cell line BE |
and DNMT1 using luteolin could be an interesting way to prevent and/or treat colorectal cancers | [200] |
| Luteolin (Not reported) | KB cells and Hnscc cancer xenograft mouse |
| [201] |
| Luteolin (Purchased) | Human colorectal cancer HCT116 cells |
| [202] |
| Luteolin (Purchased) | Human prostate cancer LNCaP and DU145 cells and transformed human prostate epithelial RWPE-1 cells |
| [189] |
| Naringin (Purchased) | Human prostate carcinoma cells (DU145) |
| [182] |
| Pectolinarigenin (Purchased) | 143B, HOS and MG63 osteosarcoma |
| [203] |
| Flavonolignans | |||
| Silibinin (Purchased) | Human prostate cancer cell lines DU145 and PC3 |
| [204] |
| Silibinin (Purchased) | Human bladder cell lines RT4 and T24 |
| [205] |
| Silibinin (Purchased) | SW480 and SW620 cells colon adenocarcinoma |
activity in colorectal SW480 and metastatic SW620 cells, and exerted synergistic effects with HDAC inhibitors on cancer cell death | [206] |
| Silibinin (Not reported) | Human NSCLC H1299 cells |
| [207] |
| Flavonols | |||
| Fisetin (Purchased) | Human breast cancer cell lines (MCF-7 and MDA-MB-231) |
| [146] |
| Galangin (Not reported) | MCF7 breast cancer cells |
| [137] |
| Galangin (Purchased) | Human neuroblastoma SH-SY5Y cells |
mechanism of polyphenols against alzheimer’s disease through an epigenic modifications of AD-related genes | [208] |
| Kaempferol (Not reported) | Human-derived cell lines HepG2, Hep3B |
activity | [209] |
| Kaempferol (Purchased) | Human GC cell lines (AGS, SNU-216, NCI-N87, SNU-638, and MKN-74) |
| [210] |
| Kaempferol (Purchased) | T24 and 5637 bladder cancer cell lines |
| [211] |
| Morin (Purchased) | Human ovarian cancer cell lines, A2780 and SKOV-3 |
| [212] |
| Myricetin (Purchased) | Human breast cancer cell lines (MCF-7 and MDA-MB-231) |
enzymes through increased formation of SAH (a potent noncompetitive inhibitor of DNMTs) | [146] |
| Myricetin (Purchased) | MCF7 breast cancer cells |
| [137] |
| Quercetin (Purchased) | Human breast cancer cell lines (MCF-7 and MDA-MB-231) |
enzymes through increased formation of SAH (a potent noncompetitive inhibitor of DNMTs) | [146] |
| Quercetin (Not reported) | Human colon cancer cell line RKO cells |
gene promoter | [213] |
| Quercetin (Not reported) | Human breast cancer cell lines MCF-7 and MDA-MB-231 |
| [214] |
| Quercetin (Purchased) | Human osteosarcoma 143B cell line |
217-KRAS axis | [215] |
| Quercetin (Purchased) | Human Pancreatic ductal adenocarcinoma PDA cell lines BxPc-3 and MIA-PaCa2 and human hTERT-HPNE immortalized pancreatic duct cells CRL-1097 |
| [165] |
| Quercetin (Purchased) | Human cervical cancer cells HeLa cells |
| [216] |
| Quercetin (Purchased) | MDA-MB-231, MDA-MB-468, T47D, BT-20, and MCF-7 breast cancer cells |
| [217] |
| Quercetin (Purchased) | Human leukemia HL-60 cells |
| [218] |
| Quercetin (Purchased) | SNU719 cells, a gastric carcinoma cell line |
| [219] |
| Quercetin (Purchased) | Male ICR mice |
| [220] |
| Quercetin (Purchased) | Male Syrian hamsters |
| [221] |
| Quercetin (Purchased) | Mouse intestinal epithelial cell line (MODE-K) |
| [222] |
| Quercetin (Not reported) | Human prostate cancer PC3 and LNCaP cell lines |
| [223] |
| Quercetin (Purchased) | Human esophageal 9706 cancer cell |
and histone acetylation, acting as histone deacetylase inhibitor mediated via epigenetic-NF-κB cascade signaling | [224] |
| Isoflavones | |||
| Biochanin A (Not reported) | Esophageal KYSE 510 and KYSE 150 cell lines |
| [225] |
| Biochanin A (Purchased) | Daphnia magna Straus (clone K6) | ⇒ The isoflavones biochanin A do not induce an effect on overall D. magna DNA methylation at exposure concentrations for which effects on reproduction were observed | [226] |
| Daidzein (Purchased) | Breast cancer cell lines, MCF-7 and MDA-MB 231 |
| [227] |
| Daidzein (Purchased) | Prostate cell lines (PC-3, DU-145, LNCaP |
such as the promoter CpG island demethylation of tumor suppressor genes, might be related to the protective effect of daidzein on prostate cancer | [228] |
| Daidzein (Not reported) | Esophageal KYSE 510 and KYSE 150 cell lines |
| [225] |
| Equol (Purchased) | Breast cancer cell lines, MCF-7 and MDA-MB 231 |
| [227] |
| Genistein (Not reported) | Esophageal KYSE 510 and KYSE 150 cell lines |
may contribute to its chemopreventive activity | [225] |
| Genistein (Purchased) | breast cancer cell lines, MCF-7 and MDA-MB 231 |
| [227] |
| Genistein (Purchased) | Prostate cell lines (PC-3, DU-145, LNCaP |
such as the promoter CpG island demethylation of tumor suppressor genes, might be related to the protective effect of genistein on prostate cancer | [228] |
5.1.6. Biflavonoids
5.1.7. Amentoflavone
5.1.8. Flavans
5.1.9. Kazinol Q
5.1.10. Flavanols
5.1.11. Catechin, Epicatechin, and (-)-epigallocatechin-3-O-gallate
5.1.12. (-)-epigallocatechin-3-O-gallate
5.1.13. Flavanones
5.1.14. Hesperetin
5.1.15. Hesperidin
5.1.16. Naringenin
5.1.17. Flavanonols
5.1.18. Taxifolin
5.1.19. Flavones
5.1.20. 3,6-dihydroxyflavone (3,6-DHF)
5.1.21. Apigenin
5.1.22. Baicalein
5.1.23. Casticin
5.1.24. Chrysin
5.1.25. Diosmin
5.1.26. Isovitexin
5.1.27. Luteolin
5.1.28. Naringin
5.1.29. Pectolinarigenin
5.1.30. Flavonolignans
5.1.31. Silibinin
5.1.32. Flavonols
5.1.33. Fisetin
5.1.34. Galangin
5.1.35. Kaempferol
5.1.36. Morin
5.1.37. Myricetin
5.1.38. Quercetin
5.1.39. Isoflavones
5.1.40. Biochanin A
5.1.41. Daidzein
5.1.42. Equol
5.1.43. Genistein
5.2. Phenolic Acids Targeting Epigenetic Pathways in Cancer
5.2.1. Caffeic Acid
5.2.2. Gallic Acid
5.2.3. Other Phenolic Acids
| Bioactive | Origin | Key Results | Refs. |
|---|---|---|---|
| Caffeic acid | Purchased | Inhibited the M.SssI DNMT-mediated DNA methylation (IC50 = 3.0 μM) Inhibited the human DNMT1-mediated DNA methylation (IC50 = 2.3 μM) Inhibited the DNA methylation predominantly through a non-competitive mechanism, largely due to the increased formation of S-adenosyl-L-homocysteine Inhibited the methylation of the promoter region of the RARβ gene | [281] |
| Purchased | Moderately inhibited the HDAC activity (IC50 = 2.54 mM) | [282] | |
| Purchased | Attenuated the CSCs-like properties by the microRNA-148a (miR-148a)-mediated inhibition of transforming growth factor beta (TGFβ)-SMAD2 signaling pathway both in vitro and in vivo Enhanced the expression of miR-148a by inducing DNA methylation | [283] | |
| Not reported | Decreased the viability in MCF-7 and MDA-MB-231 cells, after 24 h of administration Decreased the cell growth in time depending on time in both cell lines No methylation effect on caspase-8 in MDA-MB-231 cell lines, but stimulation was carried out via the intrinsic pathway | [280] | |
| Gallic acid | Purchased | Arrested the cytotoxicity in EAhy926 and HBEC-5i cells, dose-dependently, induced by the combination Reduced the formation of microparticles with anti-apoptotic effects Reversed the DNMT1 depletions at the protein level Inhibited the proteasome activities | [285] |
| Purchased | Changed the methylome of lung cancer and pre-malignant oral cell lines Reduced both nuclear and cytoplasmic DNMT1 and DNMT3B within 1 week Exhibited a strong cytotoxicity against the lung cancer cell line H1299 Reactivated the growth arrest and DNA damage-inducible 45 (GADD45) signaling pathway Induced an anti-tumor pathway epigenetically reactivated by DNA demethylation in lung cancer cell | [284] | |
| Rosmarinic acid | Purchased | Inhibited the DNMT activity (up to 88% inhibition) No effect on the methylation pattern or the expression of RASSF1A, GSTP1 or HIN-1 in MCF7 cells No effect on the global methylation of histone H3 Increased GSTP1 transcript level No effect on the expression of GSTP1 in MCF7 cells Increased DNMT1 transcript level Reduced the DNMT1 protein level in MCF7 cells | [137] |
| Protocatechuic acid | Purchased | Inhibited the DNMT activity No effect on the methylation pattern or the expression of RASSF1A, GSTP1 or HIN-1 in MCF7 cells No effect on the global methylation of histone H3 No effect on DNMT1 transcription or on DNMT1 protein level | [137] |
| Sinapic acid | Purchased | Inhibited the DNMT activity No effect on the methylation pattern or the expression of RASSF1A, GSTP1 or HIN-1 in MCF7 cells No effect on the global methylation of histone H3 No effect on DNMT1 transcription or on DNMT1 protein level | [137] |
| Syringic acid | Purchased | Inhibited the DNMT activity No effect on the methylation pattern or the expression of RASSF1A, GSTP1 or HIN-1 in MCF7 cells No effect on the global methylation of histone H3 No effect on DNMT1 transcription or on DNMT1 protein level | [137] |
5.3. Stilbenes and Ketones as Targeting Epigenetic Pathways in Cancer
5.3.1. Resveratrol
5.3.2. Pterostilbene
5.3.3. Calebin-A
5.3.4. Curcumin
| Bioactive | Origin | Key Results | Refs. |
|---|---|---|---|
| Resveratrol (RSV) | Not reported | Activated Sirt1 and inhibited the expression of Survivin, which caused a more profound inhibitory effect on Brca1 mutant cancer cells than on Brca1-wild-type cancer cells both in vitro and in vivo | [349] |
| Purchased | Inhibited the DNMT activity No effect on the methylation pattern or the expression of RASSF1A, GSTP1 or HIN-1 in MCF7 cells No effect on the global methylation of histone H3 | [137] | |
| Purchased | Modulated the recruitment of the AhR and ERα to the BRCA-1 promoter Antagonized the TCDD (Tetrachlorodibenzo-p-dioxin)-induced histone modifications at the BRCA-1 gene Attenuated the epigenetic changes (reduced association at the BRCA-1 gene of AcH4 and AcH3K9; increased association of mMH3K9, DNMT1, and MBD2; and accumulation of DNA strand breaks) | [289] | |
| Purchased | Methylated, partially, the RARbeta2 promoter in the tested fragment in MCF-7 cells Improved the action of 2CdA (2-chloro-2′-deoxyadenosine) and F-ara-A (9-beta-D-arabinosyl-2- fluoroadenine) on RARbeta2 methylation and/or expression | [303] | |
| Purchased | Decreased the levels of several oncogenic microRNAs targeting genes encoding Dicer1, a cytoplasmic RNase III producing mature microRNAs from their immediate precursors, tumor-suppressor factors such as PDCD4 or PTEN, as well as key effectors of the TGFβ signaling pathway Increased the levels of miR-663, a tumor-suppressor microRNA targeting TGFβ1 transcripts Decreased the transcriptional activity of SMADs, the main effectors of the canonical TGFβ pathway | [301] | |
| Not reported | RSV + tamoxifen significantly reduced tumor cell viability more than either drug alone RSV + tamoxifen effectively blocked breast cancer growth Reduced the STAT3 acetylation, restored ERα expression and sensitized TNBC (triple-negative breast cancer) cells to antiestrogen therapy Inhibited the tumor growth in mice bearing A2058 human melanoma tumors, accompanied by disruption of STAT3- DNMT1 complex formation and demethylation of several tumor-suppressor gene promoters | [312] | |
| Purchased | Reduced the PTEN promoter methylation in MCF-7 cells Downregulated the DNMT and upregulated the p21 Improved the inhibitory effects of 2CdA and F-ara-A on PTEN methylation in MCF-7 cells | [292] | |
| Purchased | Upregulated the gene encoding the histone H2B ubiquitin ligase RNF20 (ring finger protein 20), a chromatin modifying enzyme and putative tumor suppressor, in MDA-MB-231 cells Increased all histone markers tested (H3K4me3, H3K9K14Ac, H4K16Ac, and H4tetraAc) at most analysed regions in MDA-MB-231 cells Increased only H3K4me3 at all regions examined in MCF-7 cells Inhibited MDA-MB-231 breast cancer cell growth in a time- and dose-dependent manner Increased p21 expression in a dose-dependent manner in MDA-MB-231 cells Increased RNF20 expression during a 72-h period Increased the p21 mRNA Increased the H3K4me3 in the promoter and 5’ coding region of p21 | [302] | |
| Purchased | Decreased the transcript levels of all the DNMTs investigated (DNMT1, DNMT3a, and DNMT3b) Decreased the protein levels of DNMT1, HDAC1, and MeCP2 | [162] | |
| Not reported | Decreased the DNMT3b in tumor tissue Increased the DNMT3b in normal tissue Increased miR21, −129, −204, and −489 >twofold in tumor and decreased the same miRs in normal tissue 10–50% Influenced the tumor vs. normal tissue DNMT3b and miRNA expression | [293] | |
| Purchased | RSV + PTS inhibited the growth of TNBCs RSV + PTS downregulated the SIRT1 (a type III HDAC) Res + PTS decreased the γ-H2AX and telomerase expression, which resulted in significant growth inhibition, apoptosis, and cell cycle arrest in HCC1806 and MDA-MB-157 breast cancer cells RSV + PTS downregulated the DNMT enzymes with no significant effects on DNMT enzyme expression in MCF10A control cells | [314] | |
| Purchased | Induced the glioma cell apoptosis through activation of tristetraprolin (TTP) Increased the TTP expression in U87MG human glioma cells TTP induced by RSV destabilized the urokinase plasminogen activator and urokinase plasminogen activator receptor mRNAs by binding to the ARE regions containing the 3′ untranslated regions of their mRNAs TTP induced by RSV suppressed cell growth and induced apoptosis in the human glioma cells | [304] | |
| Purchased | Induced an anti-myofibroblast activity in three fBMF lines Inhibited the expression of fibrogenic genes at the mRNA and protein levels in a dose- and time-dependent manner Downregulated the ZEB1 in fBMFs, which was mediated by epigenetic mechanisms, such as the upregulated expression of miR-200c and the enhancer of zeste homolog 2 (EZH2), as well as the trimethylated lysine 27 of histone H3 (H3K27me3) Increased the binding of H3K27me3 to the ZEB1 promoter | [305] | |
| Purchased | Caused significant reactivation of PAX1 expression in Caski cells Induced a striking correlation between PAX1 reactivation and Ubiquitin-like with PHD and RING finger domains 1 (UHRF1) downregulation in HeLa, SiHa, and Caski cell lines Reactivation of PAX1 was due to its effect on HDAC mediated through downregulation of UHRF1 which can regulate both DNA methylation and histone acetylation | [307] | |
| Purchased | Decreased gene promoter hypermethylation Increased DNA hypomethylation Restored the hypomethylated and hypermethylated status of key tumor suppressor genes and oncogenes, respectively | [308] | |
| Purchased | Inhibited the cell growth in a dose- and time dependent manner in both cell lines Decreased DNA methylation within promoters of potential tumor suppressor genes Increased DNA methylation within regulatory regions of potential oncogenes Increased the methylation levels (in breast cancer cells) of CpG sites within enhancers of MAML2 and GLI2 toward the levels present in normal cells Decreased the methylation of CpG sites in SEMA3A enhancer and in the CpG island of GLI2 Downregulated the MAML2 in both cancer cell lines Decreased the expression of the tested genes in MCF10CA1h cells Altered DNA methylation patterns in breast cancer cells Reduced invasive properties of breast cancer cells and their ability to anchorage-independent growth | [309] | |
| Not reported | Decreased the gene expression APOBEC3B and DNMT1 Increased the expression of TET1 in both of cell lines | [294] | |
| Not reported | Inhibited the total DNMT activity and DNMT3a protein expression Regulated the tumor suppressor gene SFRP5 expression Regulated the Wnt/β-catenin signaling pathway | [295] | |
| Not reported | Suppressed the cell migration through epigenetic regulation Recruited HDAC1 and DNMT3a to the promoter region of the focal adhesion kinase (FAK), a key factor involved in cell adhesion Enhanced the promoter methylation Attenuated the protein expression | [350] | |
| Purchased | RSV + GSPs synergistically decreased cell viability and cell proliferation in both cell lines RSV + GSPs synergistically induced apoptosis in MDA-MB-231 cells by upregulating Bax expression and downregulating Bcl-2 expression RSV + GSPs synergistically reduced DNMT and HDAC activities in MDA-MB-231 and MCF-7 cells | [351] | |
| Purchased | Inhibited the proliferation of cancer cell lines Induced the senescence along with increase in SA-β-gal activity and regulation of senescence-associated molecular markers p38MAPK, p27, p21, Rb and p-Rb protein Induced a mitochondrial dysfunction with reduction in mitochondrial membrane potential Downregulation of MT-ND1, MT-ND6 and ATPase8 in transcript level and downregulation of PGC-1α in protein level Decreased DNMT1 and increased DLC1 expression Induced cancer cellular senescence through DLC1 in a ROS-dependent manner Increased ROS production to induce DNA damage with p-CHK1 upregulation and result in cancer cellular senescence | [311] | |
| Purchased | Decreased the cell proliferation and induced the DNA damage in all cell lines Reduced the number of colonies in all cell lines Increased the rates of apoptosis in the TP53 wild type cells and downregulation of AKT, mTOR, and SRC Modulated the DNMT1 gene Induced a cell cycle arrest at S phase with PLK1 downregulation in the TP53 mutated cells | [299] | |
| Purchased | Inhibited the cell growth in a dose- and time-dependent manner in both breast cancer cell lines Attenuated the invasive capacity and anchorage independent growth by 7 μm Activated the tumor suppressor gene SEMA3A epigenetically in breast cancer cells Decreased the DNMT3A occupancy within SEMA3A promoter Increased the nuclear factor 1C occupancy at SEMA3A promoter Increased the SALL3 expression by 1.5-fold upon 9-day treatment of MCF10CA1a cells with 7 μm PTS | [298] | |
| Purchased | Increased the expression of BRCA1, p53, and p21 Decreased the expression of protein arginine methyltransferase 5 (PRMT5) and enhancer of Zeste homolog 2 (EZH2) in both breast cancer cells Decreased the lysine deacetylase (KDAC) activity and expression of KDAC1-3 Increased the expression of lysine acetyltransferase KAT2A/3B Increased the global level of H3K9ac and H3K27ac marks Reduced the enrichment of repressive histone marks (H4R3me2s and H3K27me3) Increased the abundance of activating histone marks (H3K9/27ac) within the proximal promoter region of BRCA1, p53, and p21 | [290] | |
| Purchased | Increased the ZFP36 (Zinc finger protein 36) expression and decreased the mRNA levels of ZFP36 target genes in A549 lung cancer cells Suppressed the expression of DNA (cytosine-5)-methyltransferase 1 and induced the demethylation of the ZFP36 promoter | [310] | |
| Not reported | RSV-induced ATP2A3 upregulation correlated with about 50% of reduced HDAC activity and reduced nuclear HDAC2 expression and occupancy on ATP2A3 promoter, increasing the global acetylation of histone H3 and the enrichment of histone mark H3K27Ac on the proximal promoter of the ATP2A3 gene in MDA-MB-231 cells Boosted the HAT activity Reduced the DNMT activity Reduced the expression of Methyl- DNA binding proteins MeCP2 and MBD2 | [296] | |
| Purchased | RSV + clofarabine inhibited the cell growth and induced caspase-3-dependent apoptosis RSV + clofarabine downregulated the DNMT1 and upregulated the CDKN1A, with a concomitant enhanced decrease in DNMT1 protein level RSV + clofarabine induced a concurrent methylation-mediated RARB and PTEN reactivation | [300] | |
| Purchased | Increased the CRABP2 expression and RA sensitivity of THJ-11T and UW228-2 cells Partially (3/5) demethylated five CpG methylation sites at the CRABP2 promoter region (of both cell lines) Reduced DNMT1, DNMT3A, and DNMT3B in UW228-2 cells and reduced DNMT1 and DNMT3A in THJ-11T cells in a time-related fashion | [297] | |
| Curcumin (Cur) | Curcuma longa | Inhibited the p300/CREB-binding protein (CBP) HAT activity Inhibited the p300-mediated acetylation of p53 in vivo Repressed specifically the p300/CBP HAT activity-dependent transcriptional activation from chromatin but not a DNA template Inhibited the acetylation of HIV-Tat protein in vitro by p300 as well as proliferation of the virus | [321] |
| Purchased | Inhibited the histone acetylation in the absence or presence of trichostatin A (the specific HDAC inhibitor) No effect on the in vitro activity of HDAC Inhibited the HAT activity both in vivo and in vitro Diminished or enhanced the ROS generation at low or high concentrations, respectively Induced the histone hypoacetylation in vivo | [320] | |
| Purchased | Inhibited the proliferation of B-NHL cell line Raji cells with a 36-h IC50 value of 24.1 ± 2.0 μmol/L Induced the Raji cell apoptosis Downregulated the expression levels of HDAC1, HDAC3, and HDAC8 proteins Upregulated the Ac-histone H4 protein expression | [316] | |
| Purchased | Inhibited the proliferation potency on Raji cells in vitro (IC50 = 25 μmol/L) Decreased the amounts of p300, HDAC1, and HDAC3 in a dose-dependent manner Reversed the protection degradation of HDAC1 and p300 by MG-132 Prevented the degradation of IκBα Inhibited the nuclear translocation of the NF-κB (nuclear factor kappa B)/p65 subunit and the expression of Notch 1 | [317] | |
| Not reported | Altered the miRNA expression in human pancreatic cells, upregulating miRNA-22 and downregulating miRNA-199a The upregulation of miRNA-22 expression suppressed expression of target genes SP1 transcription factor (SP1) and estrogen receptor 1 (ESR1), while inhibiting miRNA-22 with antisense enhanced SP1 and ESR1 expression | [297] | |
| Purchased | Inhibited the HDAC activity (IC50 = 115 μM) | [282] | |
| Not reported | Covalently blocked the catalytic thiolate of C1226 of DNMT1 to exert the inhibitory effect Inhibited the activity of M. SssI (IC50 =3 0 nM) Induced a global DNA hypomethylation in a leukemia cell line | [328] | |
| Purchased | Downregulated the expression of EZH2 (enhancer of zeste homolog 2) in a dose- and time- dependent manner Decreased the proliferation in the MDA-MB-435 cell Accumulated the cells in the G1 phase of the cell cycle Stimulated three major members of the mitogen-activated protein kinase (MAPK) pathway: c-Jun NH2-terminal kinase (JNK), extracellular signal-regulated kinase (ERK), and p38 kinase | [338] | |
| Not reported | A small (15–20%) decrease in methylation No detectable DNA demethylation in three human cancer cell lines | [155] | |
| Not reported | Inhibited the PDE1–5 (phosphodiesterase) activities (IC50 ≅ 10−5 M) Decreased the PDE1 and PDE4 activities Increased the intracellular cGMP levels in a dose-dependent manner Inhibited cell proliferation and cell cycle progression by accumulating cells in the S- and G2/M-phases with enhanced expressions of cyclin- dependent kinase inhibitors Decreased the expressions of PDE1A, cyclin A, and the epigenetic integrator UHRF1 and DNMT1 | [331] | |
| Purchased | Reversed the methylation status of the first 5 CpGs in the promoter region of the Nrf2 gene Reduced the anti-mecyt antibody binding to the first 5 CpGs of the Nrf2 promoter | [339] | |
| Purchased | Induced the apoptosis and cell cycle arrest at the G2/M phase in medulloblastoma cells Reduced the HDAC4 expression and activity and increased tubulin acetylation Reduced tumor growth and significantly increased survival in the Smo/Smo transgenic medulloblastoma mouse model | [318] | |
| Not reported | Disturbed the Sp1/NF-κB complex on DNMT1, DNMT3a, and DNMT3b promoter thereby decreasing the transactivation activity of these complexes and downregulating their mRNA and protein expression levels in AML cells in vitro and in vivo Induced a cell cycle arrest in the S-phase Increased SubG1 cell fraction | [352] | |
| Purchased | Reduced miR-21 promoter activity and expression in a dose-dependent manner by inhibiting AP-1 binding to the promoter Induced the expression of the tumor suppressor Pdcd4 (programmed cell death protein 4), which is a target of miR-21 Arrested the Rko and HCT116 cells in the G2/M phase with increasing concentrations Inhibited the tumor growth, invasion and in vivo metastasis in the chicken-embryo-metastasis assay Inhibited the miR-21 expression in primary tumors generated in vivo by Rko and HCT116 cells | [325] | |
| Not reported | Induced a tumor- suppressive miRNA, miR-203, in bladder cancer Induced a DNA hypermethylation of the miR-203 promoter Induced a hypomethylation of the miR-203 promoter and upregulated the miR-203 expression Induced a downregulation of miR-203 target genes Akt2 and Src | [326] | |
| Purchased | Demethylated the first 14 CpG sites of the CpG island of the Neurog1 gene Restored the expression of this cancer-related CpG-methylation epigenome marker gene Possessed limited effects on the expression of epigenetic modifying proteins MBD2, MeCP2, DNMT1, and DNMT3a Decreased the MeCP2 binding to the promoter of Neurog1 Induced different effects on the protein expression of HDACs Increased the expression of HDAC1, 4, 5, and 8 but decreased the HDAC3 Decreased the total HDAC activity Decreased the enrichment of H3K27me3 at the Neurog1 promoter region as well as at the global level | [326] | |
| Purchased | Enhanced the mRNA and protein levels of ras-association domain family protein 1A (RASSF1A), 1 hypermethylation-silenced TSG, and decreased its promoter methylation in breast cancer cells Decreased the DNA methylation activity of nuclear extract Downregulated the mRNA and protein levels of DNMT1 in MCF-7 cells | [327] | |
| Purchased | Inhibited the cell viability and proliferation in CRC cell lines Induced the demethylation of specific CpG loci in CRC cells No induction of global DNA methylation changes Validated the methylation changes at several randomly selected loci including KM-HN-1, PTPRO, WT1, and GATA4 Supported the DNA methylation alterations by corresponding changes in gene expression at both up- and downregulated genes in various CRC cell lines | [330] | |
| Purchased | Decreased the transcript levels of all the DNMTs investigated (DNMT1, DNMT3a, and DNMT3b) Decreased the protein levels of DNMT1, HDAC1, and MeCP2 | [162] | |
| Purchased | Downregulated the DNMT1 expression in AML cells in vitro and ex vivo Inhibited the expression of p65 and Sp1 and their association with the DNMT1 promoter Reactivated the epigenetically silenced p15INK4B Increased the proportion of cells in SubG1 phase of the cell cycle, induced the caspase cleavage, and inhibited the AML cell growth in vitro Inhibited the AML cell growth and downregulated the DNMT1 expression in vivo | [332] | |
| Purchased | Decreased the cell number and viability Increased the apoptotic events and superoxide level Induced toxic effects when even as low as 1 μM concentration Induced a decrease in AgNOR protein pools, which may be mediated by global DNA hypermethylation | [353] | |
| Purchased | Inhibited the cell proliferation and increased the apoptosis rate through the upregulation of PTEN associated with a decreased DNA methylation level Reduced the DNMT3b in vivo and in vitro Increased the MiR-29b in activated HSCs Targeted the DNMT3b | [333] | |
| Purchased | Suppressed anchorage-independent growth of HT29 cells Decreased the methylation of the DLEC1 promoter in HT29 cells Increased the transcription of DLEC1 in HT29 cells Altered the expression of epigenetic modifying enzymes in HT29 cells | [340] | |
| Purchased | Increased the expression of RARβ at the mRNA and protein levels in tested cancer cells Decreased the RARβ promoter methylation in lung cancer A549 and H460 cells Downregulated the mRNA levels of DNMT3b Exhibited protective effect against weight loss, because of tumor burden, in a lung cancer xenograft node mice model Repressed the tumor growth Increased the RARβ mRNA and decreased the DNMT3b mRNA in vitro | [334] | |
| Purchased | Cur dependent overexpression of miR-29a and miR-185 can downregulate the expression of DNMT1, 3A, and 3B, and subsequently overexpresses MEG3 Induced the DNA hypomethylation and the reexpression of silenced tumor suppressor genes in hepatocellular cancer | [335] | |
| Purchased | Caused significant reactivation of PAX1 expression in HeLa and SiHa cells Induced a striking correlation between PAX1 reactivation and Ubiquitin-like with PHD and RING finger domains 1 (UHRF1) downregulation in HeLa, SiHa, and Caski cell lines Reactivation of PAX1 was due to its effect on HDAC mediated through downregulation of UHRF1 which can regulate both DNA methylation and histone acetylation | [307] | |
| Not reported | Induced the DLC1 expression by demethylation of DLC1 promoter in MDA-MB- 361 cells Inhibited the proliferation of MDA-MB-361 cells via DLC1 Inhibited the DNMT1 expression Inhibited the Sp1 expression to regulate DNMT1 and DLC1 Inhibited the breast cancer growth in vivo | [354] | |
| Not reported | Cur + quercetin inhibited DNMT, causing global hypomethylation, restoring AR (Androgen Receptor) mRNA and protein levels and causing apoptosis via mitochondrial depolarization of PC3 and DU145 Increased the AR mRNA levels 1.5–2.0 fold in PC3 and DU145 cells Cur + quercetin increased the AR mRNA levels by approximately 3-fold in both the cell lines Induced the apoptosis in a significant number of cells Cur + quercetin induced apoptosis through mitochondrial depolarization in 34% PC3 cells and 28% DU145 cells in 48 h Cur + quercetin induced the expression of AR in androgen refractory PC3 and DU145-gene reporter assay Cur + quercetin increased the AR protein levels in PC3 and DU145 cells and remained unchanged after transfection of these cells with scrambled ARsiRNA | [223] | |
| Purchased | Increased the expression of TCF21 (transcription factor 21) Downregulated the expression of DNMT1 | [355] | |
| Not reported | Reversed the liver injury in vivo and in vitro, possibly through downregulation of DNMT1, α-SMA, and Col1α1 and by demethylation of the key genes | [356] | |
| Purchased | Reduced the viability of the MCF-7 cells in a dose-dependent manner (IC50 = 20 μM) Inhibited the growth with increased amount of cell deaths at higher concentration of 20 and 30 μM Induced the DNA damage in MCF-7 cells Reversed the hypermethylation Reactivated the glutathione S-transferase pi 1 (GSTP1) protein expression in MCF-7 cells at 10 μM Induced a complete reversal of GSTP1 promoter hypermethylation and caused a re-expression of GSTP1 | [341] | |
| Not reported | Restored the miR-143 and miR-145 expression in prostate cancer cell lines Reduced the methylation of CpG dinucleotides in miR-143 promoter Reduced the expression of DNMT1 and DNMT3B Restored the miR-143 and miR-145 expression via hypomethylation Enhanced the radiation-induced cancer cell growth inhibition and apoptosis Reduced the radiation-induced autophagy in PC3 and DU145 cells | [295] | |
| Purchased | Cur + decitabine inhibited the cancer cell colony formation and migration, the total DNMT activity, and DNMT3a protein expression Cur + decitabine regulated the tumor suppressor gene SFRP5 expression through Wnt/β-catenin signaling pathway | [345] | |
| Purchased | Inhibited the apoptotic effects, and reduced the expression of DNMT1 The relative expression of DNMT1 gene was 0.7 to 0.3 | [337] | |
| Purchased | Affected the proliferation of all four cancer cell lines in time- and dose-dependent manner after 24 h Increased the relative expression of p21 mRNA as well as p21 protein in all four cell lines after 24 h Altered the DNMT expression and induced global DNA hypomethylation Reduced the methylation specific amplification with an increase in corresponding unmethylation specific amplification Increased the KLF4 and p53 expression Increased the p21 proximal promoter occupancy by KLF4 | [290] | |
| Purchased | Suppressed the proliferation of myeloma cells Induced the apoptosis in myeloma cells Inhibited the expression of mTOR (mammalian target of rapamycin) in NCI-H929 cells Induced a hypermethylation of the mTOR promoter region Upregulated the expression of DNMT3 | [306] | |
| Purchased | Cur + quercetin enhanced the BRCA1 (breast cancer type 1 susceptibility protein) expression levels in TNBC (triple-negative breast cancer) Cur + quercetin inhibited the cell survival of TNBC cell lines Cur + quercetin enhanced the BRCA1 expression Cur + quercetin induced the histone acetylation of BRCA1 promoter Cur + quercetin inhibited the migratory ability of TNBC cell lines and regulated genes involved in tumor migration Cur + quercetin downregulated the T47D cells in BRCA1 | [217] | |
| Purchased | Cur (20 μM) + DAC (5 μM) inhibited the cancer cell colony formation, the migration through EMT (epithelial–mesenchymal transition) process regulation, the total DNMT activity, especially in DNMT3a protein expression Cur (20 μM) + DAC (5 μM) regulated the tumor suppressor gene SFRP5 expression involved in the Wnt/β-catenin signaling pathway | [346] | |
| Purchased | Suppressed the proliferation of HCC-38, UACC-3199, and T47D cell lines Increased the mRNA and protein levels of BRCA1 in HCC-38 and UACC-3199 cells by reducing promoter methylation Cur + DAC induced the demethylation of the -379 CpG site in the BRCA1 promoter in the HCC-38 cells Downregulated the expression of DNMT1 and upregulated the TET1 and DNMT3 in HCC-38 cells Downregulated the miR-29b in HCC-38 cells Decreased the mRNA and protein levels of SNCG (proto-oncogene γ synuclein) in T47D and HCC-38 cells by inducing promoter methylation Downregulated the expression of DNMT1 and TET1 and upregulated the DNMT3 in T47D cells Upregulated the miR-29b in T47D cells | [347] | |
| Purchased | The IC50 value for MCF-7 were 15.73 μM The IC50 value for PA-1 were 12.89 μM Cur (IC25) + 6-TG-CNPs (6-thioguanine encapsulated chitosan nanoparticles) (IC25) on PA-1 and MCF-7 showed % cell viability of 43.67 ± 0.02 and 49.77 ± 0.05, respectively Possessed in vitro cytotoxicity potential in terms of % cell viability, early apoptosis, G2/M phase arrest, and DNA demethylation activity on PA-1 cells | [348] | |
| Purchased | Indicated a dose- and time-dependent significant antiproliferative effects (IC50~5 μM) Indicated significant apoptotic effects in all different periods Increased significantly the ERα gene expression quantity | [322] | |
| Purchased | Inhibited the proliferation, colony formation, and migration of hGCCs in a dose- and time-dependent fashion Increased ROS levels and triggered mitochondrial damage, DNA damage, and apoptosis of hGCCs Cur-induced DNA demethylation of hGCCs was mediated by the damaged DNA repair-p53-p21/GADD45A-cyclin/CDK-Rb/E2F-DNMT1 axis | [323] | |
| Calebin-A | Not reported | Inhibited the proliferation of MPNST and primary neurofibroma cells in a dose-dependent manner. Increased the population in the G0/G1 phase but decreased in G2/M phase Reduced the expression of phosphorylated-AKT, -ERK1/2, survivin, hTERT, and acetyl H3 proteins Reduced the promoter DNA copies of survivin (BRIC5) and hTERT genes Reduced the enzyme activity of HAT, but not that of HDAC | [315] |
| Pterostilbene (PTS) | Purchased | RSV + PTS inhibited the growth of TNBCs RSV + PTS downregulated the SIRT1 (a type III HDAC) RSV + PTS decreased the γ-H2AX and telomerase expression, which resulted in significant growth inhibition, apoptosis, and cell cycle arrest in HCC1806 and MDA-MB-157 breast cancer cells RSV + PTS downregulated the DNMT enzymes with no significant effects on DNMT enzyme expression in MCF10A control cells | [314] |
| Purchased | Inhibited the cell growth in a dose- and time dependent manner in both cell lines Increased the methylation at CpG#2 and/or CpG#1 in both cancer cells Downregulated the MAML2 in both cancer cell lines Decreased the expression of the tested genes in MCF10CA1h cells Altered DNA methylation patterns in breast cancer cells Reduced invasive properties of breast cancer cells and their ability to anchorage-independent growth | [309] | |
| Purchased | Inhibited the cell growth in a dose- and time-dependent manner in both breast cancer cell lines Attenuated the invasive capacity and anchorage independent growth by 15 μm Identifyed 364 hypomethylated CpG sites at day 4 of RSV treatment compared to 990 hypomethylated CpG sites at day 9 of treatment Activated the tumor suppressor gene SEMA3A epigenetically in breast cancer cells Decreased the DNMT3A occupancy within SEMA3A promoter Increased the nuclear factor 1C occupancy at SEMA3A promoter Increased the SALL3 expression by 2.5-fold upon 9-day treatment of MCF10CA1a cells with 15 μm RSV | [298] |
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2-HG | 2-HydroxyGlutarate |
| 5-caC | 5-Carboxylcytosine |
| 5-fC | 5-Formylcytosine |
| 5-hmC | 5-HydroxymethylCytosine |
| 5-mC | 5-MethylCytosine |
| ACF | ATP-utilizing Chromatin Assembly and Remodeling Factor |
| ALKBH5 | α-Ketoglutarate-dependent dioxygenase alkB Homolog 5 |
| AML | Acute Myeloid Leukemia |
| APC | Adenomatous Polyposis Coli |
| Arg | Arginine |
| ARID1A | AT-Rich Interaction Domain 1A |
| ATG | Autophagy Related |
| BAF | BRG1/BRM Associated Factor |
| Bcl-2 | B-Cell Lymphoma 2 |
| BET | Bromodomain and Extraterminal Domain |
| BNIP3 | BCL2 Interacting Protein 3 |
| BPTF | Bromodomain PHD Finger Transcription Factor |
| BRCA | Breast Cancer |
| Brg1 | Brahma-Related Gene 1 |
| CA9 | Carbonic Anhydrase 9 |
| CCND2 | Cyclin D2 |
| CCT4 | Chaperonin-Containing tailless complex polypeptide, subunit 4 |
| CDC37 | Cell Division Cycle 37 |
| CDH1 | Cadherin 1 |
| CDH3 | Cadherin 3 |
| CDKN2A | Cyclin Dependent Kinase Inhibitor 2A |
| CERF | CECR2-Containing Remodeling Factor |
| CGIs | CpG Islands |
| CHD | Chromodomain-Helicase-DNA-binding protein |
| CHRAC | Chromatin Accessibility Complex |
| CML | Chronic Myeloid Leukemia |
| COX2 | Cyclooxygenase 2 |
| CRC | Colorectal Cancer |
| CTHRC1 | Collagen Triple Helix Repeat Containing 1 |
| DAMP | Damage-associated molecular pattern |
| DAPK | Death associated protein kinase 1 |
| DGGR8 | DiGeorge syndrome critical region gene 8 |
| DLBCL | Diffuse Large B-Cell Lymphoma |
| DMRs | Differentially Methylated Regions |
| DNA | Deoxyribonucleic Acid |
| DNMT | DNA MethylTransferases |
| DOT1L | Disruptor of Telomeric silencing 1-Like |
| EGFR | Epidermal Growth Factor Receptor |
| EMT | Epithelial-Mesenchymal Transition |
| ER | Estrogen Receptor |
| ERCC1 | Excision Repair Cross-Complementation Group 1 |
| ERK | Extracellular signal-Regulated Kinase |
| ERV | Endogenous Retro-Viruses |
| ESCC | Esophageal Squamous Cell Carcinoma |
| EZH2 | Enhancer of Zeste Homolog 2 |
| FHIT | Fragile Histidine Triad diadenosine triphosphatase |
| FTO | Fat-mass and obesity associated protein |
| GDHO | Global DNA Hypomethylation |
| GNAT | GCN5-related N-acetyl transferases |
| GSTP1 | Glutathione S-Transferase P1 |
| H | Histone |
| HAT | Histone Acetyl Transferase |
| HATi | HAT inhibitor |
| HCAs | Heterocyclic Amines |
| HCC | Human hepatocellular Cancer |
| HDAC | Histone Deacetylases |
| HDM | Histone Demethylases |
| HIF | Hypoxia-Inducible Factor |
| HK2 | Hexokinase 2 Enzyme |
| hMLH1 | Human MutL Homolog 1 |
| HMMR | Hyualuronan-Mediated Motility Receptor |
| hMSH2 | Human MutS Homolog 2 |
| HMT | Histone Methyltransferases |
| HNRNP | Heterogeneous Nuclear Ribo-Nucleo-Proteins |
| HNSCC | Head and Neck Squamous Cell Carcinoma |
| hOGG1 | Human 8-Oxoguanine DNA N-Glycosylase 1 |
| HOTAIR | Hox Transcript Antisense RNA |
| HOXA5 | Homeobox A5 |
| hOXC | Human homeobox C |
| HRAS | Harvey Rat Sarcoma viral oncogene homolog |
| HSP90 | Heat Shock Protein 90 |
| HSPGs | Heparan Sulfate ProteoGlycans |
| hTERT | Human Telomerase Reverse Transcriptase |
| IDH1 | Isocitrate Dehydrogenase NADP+1 |
| IGF2BPs | Insulin-like Growth Factor 2 mRNA-Binding Proteins |
| IL10 | Interleukin 10 |
| IL11 | Interleukin-11 |
| INO80 | Inositol requiring 80 |
| ISWI | Imitation switch |
| K | Lysines |
| KDM | Lysine-Specific Demethylase |
| KG | Ketoglutarate |
| KMT | Lysine-specific Methyltransferase |
| let-7a | Lethal-7a |
| lincRNA | Large Intergenic Non-Coding RNA |
| LINES | Long interspersed transposable elements |
| lncRNA | Long non coding RNA |
| LSD | Lysine-Specific Demethylases |
| Lys | Lysine |
| m6A: N6 | Methyladenosine |
| MALAT1 | Metastasis-Associated Lung Adenocarcinoma Transcript 1 |
| MAPK | Mitogen-Activated Protein Kinase |
| MBD | Methyl Binding Protein |
| MBPs | Methyl-CpG Binding Proteins |
| MCRS1 | Microspherule protein 1 |
| MDR1 | Multi-Drug Resistance 1 |
| MECP | Methyl CpG binding Protein 2 |
| MEF2 | Myocyte Enhancer Factor 2 |
| METTL | Methyltransferase-Like Protein |
| MGMT | O6-Methylguanine-DNA Methyltransferase |
| miRNA | Micro RNA |
| mRNAs | Messenger RNAs |
| MTA | Metastasis-Associated Gene |
| MTOR | Mammalian Target of Rapamycin |
| MyoD | Myogenic Activator |
| NCR | Natural Cytotoxicity Receptor |
| ncRNAs | Non-coding RNAs |
| NF-κB | Nuclear Factor Kappa light chain enhancer of activated B cells |
| NK | Natural Killer |
| NoRC | Nucleolar-Remodeling Complex |
| NSCLC | Non-Small-Cell Lung Cancer |
| NuRD | Nucleosome-Remodeling Deacetylase |
| NURF | Nucleolar-Remodeling Factor |
| PAX2 | Paired box 2 |
| PDCD4 | Programmed Cell Death 4 |
| PI3K | Phosphoinositide 3-Kinase |
| piRNA | PIWI-interacting RNA |
| PRC2 | Polycomb Repressive Complex 2 |
| PTEN | Phosphatase and Tensin Homolog |
| PTM | Post-Transcriptional Modifications |
| R | Arginine |
| RB gene | Retinoblastoma gene |
| RB1 | RB transcriptional corepressor 1 |
| RBBP | Retinoblastoma Binding Protein |
| RBM15 | RNA-Binding Motif protein 15 |
| RBM15B | RNA-Binding Motif protein 15B |
| RISC | RNA Induced Silencing Complex |
| RNA | Ribonucleic Acid |
| ROD1 | Regulator of Differentiation 1 |
| ROS | Reactive Oxygen Species |
| rRNAs | Ribosomal RNAs |
| RSF | Remodeling and Spacing Factor |
| RUVBL1 | RuvB Like AAA ATPase 1 |
| RUVBL2 | RuvB Like AAA ATPase 2 |
| S | Serine |
| SAM | S-Adenosyl-Methionine |
| Ser | Serine |
| SERPINE2 | Serpin family E member 2 |
| SINES | Short Interspersed Transposable Elements |
| SIRT | Sirtuin |
| SNF2H | Sucrose Non-Fermenting 2 Homologue |
| SNF2L | Sucrose Non-Fermenting 2 Like |
| sno-RNA | Small Nucleolar RNA |
| STAT3 | Signal Transducer and Activator of Transcription 3 |
| SWI/SNF | Switching defective/sucrose nonfermenting |
| TDG | Thymine-DNA glycosylase |
| TE | Transposable elements |
| TET | Ten eleven translocation |
| TET2 | TET methylcytosine dioxygenase 2 |
| TGF-β1 | Transformting growth factor beta 1 |
| Thr | Threonine |
| TLS | Translocated in liposarcoma |
| TOP2A | Topoisomerase II α |
| tRNAs | Transfer RNAs |
| TSS | Transcription Start Site |
| T-UCR | Transcribed Ultraconcerved Regions |
| TUG1 | Taurine-Upregulated Gene 1 |
| Tyr: T | Tyrosine |
| Tyr | Tyrosine |
| UHRF1 | Ubiquitin like with PHD and Ring Finger domains 1 |
| UTR | Untranslated region |
| VEGF | Vascular Endothelial Growth Factor |
| VHL | Von Hippel-Lindau syndrome |
| VIRMA | Vir-like m6A Methyltransferase Associated |
| WNT | Wingless |
| WRN | Werner syndrome protein |
| WTAP | Wilms Tumor 1-Associated Protein |
| WWOX | WW Domain Containing Oxidoreductase |
| Xi | Inactive X chromosome |
| XIC | X Chromosome Inactivation |
| Xic | X-Inactivation Center |
| XRCC1 | X-Ray Repair Cross-Complementing 1 |
| YTH | YT521-B Homology |
| YTHDF2 | YTH N6-Methyladenosine RNA binding protein 2 |
| YY1 | Yin Yang 1 |
| ZC3H3 | Zinc finger CCCH domain-containing protein 13 |
References
- Wang, J.; Zhang, X.; Chen, W.; Hu, X.; Li, J.; Liu, C. Regulatory Roles of Long Noncoding RNAs Implicated in Cancer Hallmarks. Int. J. Cancer 2020, 146, 906–916. [Google Scholar] [CrossRef] [PubMed]
- Stresemann, C.; Brueckner, B.; Musch, T.; Stopper, H.; Lyko, F. Functional Diversity of DNA Methyltransferase Inhibitors in Human Cancer Cell Lines. Cancer Res. 2006, 66, 2794–2800. [Google Scholar] [CrossRef]
- Ruddon, R.W. Cancer Biology; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- G C O Global Cancer Observatory. Available online: https://gco.iarc.fr/ (accessed on 16 September 2021).
- Hanahan, D.; Weinberg, R.A. Biological Hallmarks of Cancer. In Holland-Frei Cancer Medicine; Bast, R.C., Croce, C.M., Hait, W., Hong, W.K., Kufe, D.W., Pollock, R.E., Weichselbaum, R.R., Holland, J.F., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 9–16. [Google Scholar]
- Lutz, W.K. Endogenous Genotoxic Agents and Processes as a Basis of Spontaneous Carcinogenesis. Mutat. Res. Genet. Toxicol. 1990, 238, 287–295. [Google Scholar] [CrossRef]
- Bouyahya, A.; El Menyiy, N.; Oumeslakht, L.; El Allam, A.; Balahbib, A.; Rauf, A.; Muhammad, N.; Kuznetsova, E.; Derkho, M.; Thiruvengadam, M.; et al. Preclinical and Clinical Antioxidant Effects of Natural Compounds against Oxidative Stress-Induced Epigenetic Instability in Tumor Cells. Antioxidants 2021, 10, 1553. [Google Scholar] [CrossRef] [PubMed]
- Srikrishna, G.; Freeze, H.H. Endogenous Damage-Associated Molecular Pattern Molecules at the Crossroads of Inflammation and Cancer. Neoplasia 2009, 11, 615–628. [Google Scholar] [CrossRef]
- Smetana, K.; Lacina, L.; Szabo, P.; Dvořánková, B.; Brož, P.; Šedo, A. Ageing as an Important Risk Factor for Cancer. Anticancer Res. 2016, 36, 5009–5017. [Google Scholar] [CrossRef]
- Jena, N.R. DNA Damage by Reactive Species: Mechanisms, Mutation and Repair. J. Biosci. 2012, 37, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Ayesha, N.; Aboulaghras, S.; Jahangeer, M.; Riasat, A.; Ramzan, R.; Fatima, R.; Akram, M.; Balahbib, A.; Bouyahya, A.; Sepiashvili, E.; et al. Physiopathology and Effectiveness of Therapeutic Vaccines against Human Papillomavirus. Environ. Sci. Pollut. Res. 2021, 28, 47752–47772. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in Cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Knudson, A.G. Two Genetic Hits (More or Less) to Cancer. Nat. Rev. Cancer 2001, 1, 157–162. [Google Scholar] [CrossRef]
- Deininger, P. Genetic Instability in Cancer: Caretaker and Gatekeeper Genes. Ochsner J. 1999, 1, 206–209. [Google Scholar]
- Pitot, H.C. The Molecular Biology of Carcinogenesis. Cancer 1993, 72, 962–970. [Google Scholar] [CrossRef]
- Miranda Furtado, C.L.; Dos Santos Luciano, M.C.; Silva Santos, R.D.; Furtado, G.P.; Moraes, M.O.; Pessoa, C. Epidrugs: Targeting Epigenetic Marks in Cancer Treatment. Epigenetics 2019, 14, 1164–1176. [Google Scholar] [CrossRef]
- Zhao, X.; Zheng, F.; Li, Y.; Hao, J.; Tang, Z.; Tian, C.; Yang, Q.; Zhu, T.; Diao, C.; Zhang, C. BPTF Promotes Hepatocellular Carcinoma Growth by Modulating HTERT Signaling and Cancer Stem Cell Traits. Redox Biol. 2019, 20, 427–441. [Google Scholar] [CrossRef]
- Li, E.; Zhang, Y. DNA Methylation in Mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.B.; Lagas, J.S.; Broestl, L.; Sponagel, J.; Rockwell, N.; Rhee, G.; Rosen, S.F.; Chen, S.; Klein, R.S.; Imoukhuede, P. Sex Differences in Cancer Mechanisms. Biol. Sex Differ. 2020, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Dunford, A.; Weinstock, D.M.; Savova, V.; Schumacher, S.E.; Cleary, J.P.; Yoda, A.; Sullivan, T.J.; Hess, J.M.; Gimelbrant, A.A.; Beroukhim, R. Tumor-Suppressor Genes That Escape from X-Inactivation Contribute to Cancer Sex Bias. Nat. Genet. 2017, 49, 10–16. [Google Scholar] [CrossRef]
- Panning, B. X-Chromosome Inactivation: The Molecular Basis of Silencing. J. Biol. 2008, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, E.; Kirby, J.E.; Brown, D.E.; Mercier, F.E.; Sadreyev, R.I.; Scadden, D.T.; Lee, J.T. Xist RNA Is a Potent Suppressor of Hematologic Cancer in Mice. Cell 2013, 152, 727–742. [Google Scholar] [CrossRef]
- Marjanovic, N.D.; Weinberg, R.A.; Chaffer, C.L. Cell Plasticity and Heterogeneity in Cancer. Clin. Chem. 2013, 59, 168–179. [Google Scholar] [CrossRef]
- Nagao, M.; Sugimura, T. Carcinogenic Factors in Food with Relevance to Colon Cancer Development. Mutat. Res. Mol. Mech. Mutagen. 1993, 290, 43–51. [Google Scholar] [CrossRef]
- Nasir, A.; Bullo, M.M.H.; Ahmed, Z.; Imtiaz, A.; Yaqoob, E.; Jadoon, M.; Ahmed, H.; Afreen, A.; Yaqoob, S. Nutrigenomics: Epigenetics and Cancer Prevention: A Comprehensive Review. Crit. Rev. Food Sci. Nutr. 2020, 60, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.T.; Hord, N.G. The Role of Dietary Factors in Cancer Prevention: Beyond Fruits and Vegetables. Nutr. Clin. Pract. 2005, 20, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Marmorstein, R.; Zhou, M.-M. Writers and Readers of Histone Acetylation: Structure, Mechanism, and Inhibition. Cold Spring Harb. Perspect. Biol. 2014, 6, a018762. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.D. DNA Methylation and Human Disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Reyngold, M.; Chan, T.A. DNA methylation. In Molecular Oncology: Causes of Cancer and Targets for Treatment; Cambridge University Press: Cambridge, UK, 2018. [Google Scholar]
- Laget, S.; Defossez, P.-A. Le Double Jeu de l’épigénétique-Cible et Acteur Du Cancer. Médecine/Sciences 2008, 24, 725–730. [Google Scholar]
- Robertson, K.D.; Uzvolgyi, E.; Liang, G.; Talmadge, C.; Sumegi, J.; Gonzales, F.A.; Jones, P.A. The Human DNA Methyltransferases (DNMTs) 1, 3a and 3b: Coordinate MRNA Expression in Normal Tissues and Overexpression in Tumors. Nucl. Acids Res. 1999, 27, 2291–2298. [Google Scholar] [CrossRef]
- Bird, A. DNA Methylation Patterns and Epigenetic Memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Nan, X.; Ng, H.-H.; Johnson, C.A.; Laherty, C.D.; Turner, B.M.; Eisenman, R.N.; Bird, A. Transcriptional Repression by the Methyl-CpG-Binding Protein MeCP2 Involves a Histone Deacetylase Complex. Nature 1998, 393, 386–389. [Google Scholar] [CrossRef]
- Paulsen, M.; Ferguson-Smith, A.C. DNA Methylation in Genomic Imprinting, Development, and Disease. J. Pathol. 2001, 195, 97–110. [Google Scholar] [CrossRef]
- Klose, R.J.; Bird, A.P. Genomic DNA Methylation: The Mark and Its Mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Deltour, S.; Chopin, V.; Leprince, D. Modifications Épigénétiques et Cancer. Médecine/Sciences 2005, 21, 405–411. [Google Scholar] [CrossRef]
- Chen, R.Z.; Pettersson, U.; Beard, C.; Jackson-Grusby, L.; Jaenisch, R. DNA Hypomethylation Leads to Elevated Mutation Rates. Nature 1998, 395, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Gopisetty, G.; Ramachandran, K.; Singal, R. DNA Methylation and Apoptosis. Mol. Immunol. 2006, 43, 1729–1740. [Google Scholar] [CrossRef]
- Greger, V.; Passarge, E.; Höpping, W.; Messmer, E.; Horsthemke, B. Epigenetic Changes May Contribute to the Formation and Spontaneous Regression of Retinoblastoma. Hum. Genet. 1989, 83, 155–158. [Google Scholar] [CrossRef] [PubMed]
- Pasculli, B.; Barbano, R.; Parrella, P. Epigenetics of Breast Cancer: Biology and Clinical Implication in the Era of Precision Medicine. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2018; Volume 51, pp. 22–35. [Google Scholar]
- Zhao, S.G.; Chen, W.S.; Li, H.; Foye, A.; Zhang, M.; Sjöström, M.; Aggarwal, R.; Playdle, D.; Liao, A.; Alumkal, J.J. The DNA Methylation Landscape of Advanced Prostate Cancer. Nat. Genet. 2020, 52, 778–789. [Google Scholar] [CrossRef]
- Arantes, L.; De Carvalho, A.C.; Melendez, M.E.; Carvalho, A.L.; Goloni-Bertollo, E.M. Methylation as a Biomarker for Head and Neck Cancer. Oral Oncol. 2014, 50, 587–592. [Google Scholar] [CrossRef]
- Wong, K.K. DNMT1: A Key Drug Target in Triple-Negative Breast Cancer. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 72, pp. 198–213. [Google Scholar]
- Zhou, Z.; Lei, Y.; Wang, C. Analysis of Aberrant Methylation in DNA Repair Genes during Malignant Transformation of Human Bronchial Epithelial Cells Induced by Cadmium. Toxicol. Sci. 2012, 125, 412–417. [Google Scholar] [CrossRef]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting Epigenetic Regulators for Cancer Therapy: Mechanisms and Advances in Clinical Trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.; Huang, Y.; Jankowska, A.M.; Pape, U.J.; Tahiliani, M.; Bandukwala, H.S.; An, J.; Lamperti, E.D.; Koh, K.P.; Ganetzky, R. Impaired Hydroxylation of 5-Methylcytosine in Myeloid Cancers with Mutant TET2. Nature 2010, 468, 839–843. [Google Scholar] [CrossRef]
- Yamazaki, J.; Taby, R.; Vasanthakumar, A.; Macrae, T.; Ostler, K.R.; Shen, L.; Kantarjian, H.M.; Estecio, M.R.; Jelinek, J.; Godley, L.A. Effects of TET2 Mutations on DNA Methylation in Chronic Myelomonocytic Leukemia. Epigenetics 2012, 7, 201–207. [Google Scholar] [CrossRef]
- Lin, J.; Yao, D.; Qian, J.; Chen, Q.; Qian, W.; Li, Y.; Yang, J.; Wang, C.; Chai, H.; Qian, Z. IDH1 and IDH2 Mutation Analysis in Chinese Patients with Acute Myeloid Leukemia and Myelodysplastic Syndrome. Ann. Hematol. 2012, 91, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Kosmider, O.; Gelsi-Boyer, V.; Slama, L.; Dreyfus, F.; Beyne-Rauzy, O.; Quesnel, B.; Hunault-Berger, M.; Slama, B.; Vey, N.; Lacombe, C. Mutations of IDH1 and IDH2 Genes in Early and Accelerated Phases of Myelodysplastic Syndromes and MDS/Myeloproliferative Neoplasms. Leukemia 2010, 24, 1094–1096. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wahab, O.; Figueroa, M.E. Interpreting New Molecular Genetics in Myelodysplastic Syndromes. Hematol. Am. Soc. Hematol. Educ. 2012, 2012, 56–64. [Google Scholar] [CrossRef]
- Weissmann, S.; Alpermann, T.; Grossmann, V.; Kowarsch, A.; Nadarajah, N.; Eder, C.; Dicker, F.; Fasan, A.; Haferlach, C.; Haferlach, T. Landscape of TET2 Mutations in Acute Myeloid Leukemia. Leukemia 2012, 26, 934–942. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA Hypomethylation in Cancer Cells. Epigenomics 2009, 1, 239–259. [Google Scholar] [CrossRef]
- Zhang, W.; Klinkebiel, D.; Barger, C.J.; Pandey, S.; Guda, C.; Miller, A.; Akers, S.N.; Odunsi, K.; Karpf, A.R. Global DNA Hypomethylation in Epithelial Ovarian Cancer: Passive Demethylation and Association with Genomic Instability. Cancers 2020, 12, 764. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Song, R.; Gong, Y.; Zhao, H. Global DNA Hypomethylation in Leukocytes Associated with Glioma Risk. Oncotarget 2017, 8, 63223. [Google Scholar] [CrossRef][Green Version]
- De Cubas, A.A.; Dunker, W.; Zaninovich, A.; Hongo, R.A.; Bhatia, A.; Panda, A.; Beckermann, K.E.; Bhanot, G.; Ganesan, S.; Karijolich, J. DNA Hypomethylation Promotes Transposable Element Expression and Activation of Immune Signaling in Renal Cell Cancer. JCI Insight 2020, 5, 11. [Google Scholar] [CrossRef]
- Cho, M.; Uemura, H.; Kim, S.C.; Kawada, Y.; Yoshida, K.; Hirao, Y.; Konishi, N.; Saga, S.; Yoshikawa, K. Hypomethylation of the MN/CA9 Promoter and Upregulated MN/CA9 Expression in Human Renal Cell Carcinoma. Br. J. Cancer 2001, 85, 563–567. [Google Scholar] [CrossRef]
- Tang, J.; Pan, R.; Xu, L.; Ma, Q.; Ying, X.; Zhao, J.; Zhao, H.; Miao, L.; Xu, Y.; Duan, S. IL10 Hypomethylation Is Associated with the Risk of Gastric Cancer. Oncol. Lett. 2021, 21, 241. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetic Lesions Causing Genetic Lesions in Human Cancer: Promoter Hypermethylation of DNA Repair Genes. Eur. J. Cancer 2000, 36, 2294–2300. [Google Scholar] [CrossRef]
- Csepany, T.; Lin, A.; Baldick, C.J., Jr.; Beemon, K. Sequence Specificity of MRNA N6-Adenosine Methyltransferase. J. Biol. Chem. 1990, 265, 20117–20122. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of MRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C. Structural Basis of N 6-Adenosine Methylation by the METTL3–METTL14 Complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S. Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Zhong, L.; Liao, D.; Zhang, M.; Zeng, C.; Li, X.; Zhang, R.; Ma, H.; Kang, T. YTHDF2 Suppresses Cell Proliferation and Growth via Destabilizing the EGFR MRNA in Hepatocellular Carcinoma. Cancer Lett. 2019, 442, 252–261. [Google Scholar] [CrossRef]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The Role of Hypoxia in Cancer Progression, Angiogenesis, Metastasis, and Resistance to Therapy. Hypoxia 2015, 3, 83. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, H.; Liu, J.; Zhao, Z.; Wang, J.; Lu, Z.; Hu, B.; Zhou, J.; Zhao, Z.; Feng, M. YTHDF2 Reduction Fuels Inflammation and Vascular Abnormalization in Hepatocellular Carcinoma. Mol. Cancer 2019, 18, 163. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Hergeth, S.P.; Schneider, R. The H1 Linker Histones: Multifunctional Proteins beyond the Nucleosomal Core Particle. EMBO Rep. 2015, 16, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Bowman, G.D.; Poirier, M.G. Post-Translational Modifications of Histones That Influence Nucleosome Dynamics. Chem. Rev. 2015, 115, 2274–2295. [Google Scholar] [CrossRef]
- Luan, Y.; Ngo, L.; Han, Z.; Wang, X.; Qu, M.; Zheng, Y.G. Histone acetyltransferases: Enzymes, assays, and inhibitors. In Epigenetic Technological Applications; Elsevier: Amsterdam, The Netherlands, 2015; pp. 291–317. [Google Scholar]
- Castelli, G.; Pelosi, E.; Testa, U. Targeting Histone Methyltransferase and Demethylase in Acute Myeloid Leukemia Therapy. OncoTargets Ther. 2018, 11, 131. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Yoshida, M. Cold Spring Harbor Perspect. Biology 2014, 6, a018713. [Google Scholar]
- Glozak, M.A.; Sengupta, N.; Zhang, X.; Seto, E. Acetylation and Deacetylation of Non-Histone Proteins. Gene 2005, 363, 15–23. [Google Scholar] [CrossRef]
- Figueroa, M.E.; Chen, S.-C.; Andersson, A.K.; Phillips, L.A.; Li, Y.; Sotzen, J.; Kundu, M.; Downing, J.R.; Melnick, A.; Mullighan, C.G. Integrated Genetic and Epigenetic Analysis of Childhood Acute Lymphoblastic Leukemia. J. Clin. Invest. 2013, 123, 3099–3111. [Google Scholar] [CrossRef] [PubMed]
- Halkidou, K.; Gaughan, L.; Cook, S.; Leung, H.Y.; Neal, D.E.; Robson, C.N. Upregulation and Nuclear Recruitment of HDAC1 in Hormone Refractory Prostate Cancer. Prostate 2004, 59, 177–189. [Google Scholar] [CrossRef]
- Yu, H.; Li, S. Role of LINC00152 in Non-Small Cell Lung Cancer. J. Zhejiang Univ. Sci. B 2020, 21, 179–191. [Google Scholar] [CrossRef]
- Jung, K.H.; Noh, J.H.; Kim, J.K.; Eun, J.W.; Bae, H.J.; Xie, H.J.; Chang, Y.G.; Kim, M.G.; Park, H.; Lee, J.Y. HDAC2 Overexpression Confers Oncogenic Potential to Human Lung Cancer Cells by Deregulating Expression of Apoptosis and Cell Cycle Proteins. J. Cell. Biochem. 2012, 113, 2167–2177. [Google Scholar] [CrossRef]
- Zhao, H.; Yu, Z.; Zhao, L.; He, M.; Ren, J.; Wu, H.; Chen, Q.; Yao, W.; Wei, M. HDAC2 Overexpression Is a Poor Prognostic Factor of Breast Cancer Patients with Increased Multidrug Resistance-Associated Protein Expression Who Received Anthracyclines Therapy. Jpn. J. Clin. Oncol. 2016, 46, 893–902. [Google Scholar] [CrossRef]
- Shan, W.; Jiang, Y.; Yu, H.; Huang, Q.; Liu, L.; Guo, X.; Li, L.; Mi, Q.; Zhang, K.; Yang, Z. HDAC2 Overexpression Correlates with Aggressive Clinicopathological Features and DNA-Damage Response Pathway of Breast Cancer. Am. J. Cancer Res. 2017, 7, 1213. [Google Scholar] [PubMed]
- Song, Y.; Manson, J.E.; Buring, J.E.; Sesso, H.D.; Liu, S. Associations of Dietary Flavonoids with Risk of Type 2 Diabetes, and Markers of Insulin Resistance and Systemic Inflammation in Women: A Prospective Study and Cross-Sectional Analysis. J. Am. Coll. Nutr. 2005, 24, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, E.; Licht, J.D. Deregulation of H3K27 Methylation in Cancer. Nat. Genet. 2010, 42, 100–101. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, M.; Fukuda, A.; Kawai, M.; Araki, O.; Seno, H. The Role of the SWI/SNF Chromatin Remodeling Complex in Pancreatic Ductal Adenocarcinoma. Cancer Sci. 2021, 112, 490. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The Biology of Chromatin Remodeling Complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Mayes, K.; Qiu, Z.; Alhazmi, A.; Landry, J.W. ATP-Dependent Chromatin Remodeling Complexes as Novel Targets for Cancer Therapy. Adv. Cancer Res. 2014, 121, 183–233. [Google Scholar]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of Action and Regulation of ATP-Dependent Chromatin-Remodelling Complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Dykhuizen, E.C.; Hargreaves, D.C.; Miller, E.L.; Cui, K.; Korshunov, A.; Kool, M.; Pfister, S.; Cho, Y.-J.; Zhao, K.; Crabtree, G.R. BAF Complexes Facilitate Decatenation of DNA by Topoisomerase IIα. Nature 2013, 497, 624–627. [Google Scholar] [CrossRef]
- Erdel, F.; Rippe, K. Chromatin Remodelling in Mammalian Cells by ISWI-Type Complexes–Where, When and Why? FEBS J. 2011, 278, 3608–3618. [Google Scholar] [CrossRef]
- Dai, M.; Lu, J.-J.; Guo, W.; Yu, W.; Wang, Q.; Tang, R.; Tang, Z.; Xiao, Y.; Li, Z.; Sun, W. BPTF Promotes Tumor Growth and Predicts Poor Prognosis in Lung Adenocarcinomas. Oncotarget 2015, 6, 33878. [Google Scholar] [CrossRef]
- Dai, M.; Hu, S.; Liu, C.-F.; Jiang, L.; Yu, W.; Li, Z.-L.; Guo, W.; Tang, R.; Dong, C.-Y.; Wu, T.-H. BPTF Cooperates with P50 NF-ΚB to Promote COX-2 Expression and Tumor Cell Growth in Lung Cancer. Am. J. Transl. Res. 2019, 11, 7398. [Google Scholar]
- Mayes, K.; Elsayed, Z.; Alhazmi, A.; Waters, M.; Alkhatib, S.G.; Roberts, M.; Song, C.; Peterson, K.; Chan, V.; Ailaney, N. BPTF Inhibits NK Cell Activity and the Abundance of Natural Cytotoxicity Receptor Co-Ligands. Oncotarget 2017, 8, 64344. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.Y.; Wade, P.A. Cancer Biology and NuRD: A Multifaceted Chromatin Remodelling Complex. Nat. Rev. Cancer 2011, 11, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Günther, K.; Rust, M.; Leers, J.; Boettger, T.; Scharfe, M.; Jarek, M.; Bartkuhn, M.; Renkawitz, R. Differential Roles for MBD2 and MBD3 at Methylated CpG Islands, Active Promoters and Binding to Exon Sequences. Nucl. Acids Res. 2013, 41, 3010–3021. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.N.; Gaskill, K.; Calloway, C.; Ding, W.-Q. The Role of Metastasis-Associated Protein 1 (MTA1) in Breast Cancer Exosome-Mediated Intercellular Communication. FASEB J. 2017, 31, 178.6. [Google Scholar]
- Pakala, S.B.; Rayala, S.K.; Wang, R.-A.; Ohshiro, K.; Mudvari, P.; Reddy, S.D.N.; Zheng, Y.; Pires, R.; Casimiro, S.; Pillai, M.R. MTA1 Promotes STAT3 Transcription and Pulmonary Metastasis in Breast Cancer. Cancer Res. 2013, 73, 3761–3770. [Google Scholar] [CrossRef] [PubMed]
- Salot, S.; Gude, R. MTA1-Mediated Transcriptional Repression of SMAD7 in Breast Cancer Cell Lines. Eur. J. Cancer 2013, 49, 492–499. [Google Scholar] [CrossRef]
- Morrison, A.J.; Shen, X. Chromatin Remodelling beyond Transcription: The INO80 and SWR1 Complexes. Nat. Rev. Mol. Cell Biol. 2009, 10, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Conaway, R.C.; Conaway, J.W. The INO80 Chromatin Remodeling Complex in Transcription, Replication and Repair. Trends Biochem. Sci. 2009, 34, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Runge, J.S.; Raab, J.R.; Magnuson, T. Identification of Two Distinct Classes of the Human INO80 Complex Genome-Wide. G3 Genes Genomes Genet. 2018, 8, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, B.; Wang, L.; Li, P.; Bennett, B.D.; Snyder, R.; Garantziotis, S.; Fargo, D.C.; Cox, A.D.; Chen, L. INO80 Is Required for Oncogenic Transcription and Tumor Growth in Non-Small Cell Lung Cancer. Oncogene 2017, 36, 1430–1439. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, L.; Zhang, S.; Bennett, B.D.; He, F.; Zhang, Y.; Xiong, C.; Han, L.; Diao, L.; Li, P. INO80 Governs Superenhancer-Mediated Oncogenic Transcription and Tumor Growth in Melanoma. Genes Dev. 2016, 30, 1440–1453. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Liu, J.; Chen, A.; Lyu, J.; Ai, G.; Zeng, Q.; Sun, Y.; Chen, C.; Wang, J.; Qiu, J. Ino80 Promotes Cervical Cancer Tumorigenesis by Activating Nanog Expression. Oncotarget 2016, 7, 72250. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Liu, F.; Gao, J.; Lu, H.; Cai, J.; Zhao, X.; Sun, Y. Multilevel Regulation of RUVBL2 Expression Predicts Poor Prognosis in Hepatocellular Carcinoma. Cancer Cell Int. 2019, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bilgiç, F.; Gerçeker, E.; Boyacıoğlu, S.Ö.; Kasap, E.; Demirci, U.; Yıldırım, H.; Baykan, A.R.; Yüceyar, H. Potential Role of Chromatin Remodeling Factor Genes in Atrophic Gastritis/Gastric Cancer Risk. Turk. J. Gastroenterol. 2018, 29, 427. [Google Scholar] [CrossRef] [PubMed]
- Comfort, N. Genetics: We Are the 98%. Nature 2015, 520, 615–616. [Google Scholar] [CrossRef]
- Esteller, M. Non-Coding RNAs in Human Disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Piletič, K.; Kunej, T. MicroRNA Epigenetic Signatures in Human Disease. Arch. Toxicol. 2016, 90, 2405–2419. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M. Human MicroRNA Genes Are Frequently Located at Fragile Sites and Genomic Regions Involved in Cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Place, R.F.; Li, L.-C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 Induces Expression of Genes with Complementary Promoter Sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef]
- Humphries, B.; Wang, Z.; Yang, C. MicroRNA Regulation of Epigenetic Modifiers in Breast Cancer. Cancers 2019, 11, 897. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C. MicroRNA-29 Family Reverts Aberrant Methylation in Lung Cancer by Targeting DNA Methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Henson, R.; Wehbe–Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Frankel, L.B.; Christoffersen, N.R.; Jacobsen, A.; Lindow, M.; Krogh, A.; Lund, A.H. Programmed Cell Death 4 (PDCD4) Is an Important Functional Target of the MicroRNA MiR-21 in Breast Cancer Cells. J. Biol. Chem. 2008, 283, 1026–1033. [Google Scholar] [CrossRef]
- Khan, A.Q.; Ahmed, E.I.; Elareer, N.R.; Junejo, K.; Steinhoff, M.; Uddin, S. Role of MiRNA-Regulated Cancer Stem Cells in the Pathogenesis of Human Malignancies. Cells 2019, 8, 840. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long Non-Coding RNAs: Mechanism of Action and Functional Utility. Non-Coding RNA Res. 2016, 1, 43–50. [Google Scholar] [CrossRef]
- Romero-Barrios, N.; Legascue, M.F.; Benhamed, M.; Ariel, F.; Crespi, M. Splicing Regulation by Long Noncoding RNAs. Nucl. Acids Res. 2018, 46, 2169–2184. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.C.; Tucker, T.; Thorogood, N.P.; Brown, C.J. Mechanisms of X-Chromosome Inactivation. Front Biosci. 2006, 11, 852–866. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Arai, S.; Song, X.; Reichart, D.; Du, K.; Pascual, G.; Tempst, P.; Rosenfeld, M.G.; Glass, C.K.; Kurokawa, R. Induced NcRNAs Allosterically Modify RNA-Binding Proteins in Cis to Inhibit Transcription. Nature 2008, 454, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Yoon, J.-H.; You, B.-H.; Park, C.H.; Kim, Y.J.; Nam, J.-W.; Lee, S.K. The Long Noncoding RNA LUCAT1 Promotes Tumorigenesis by Controlling Ubiquitination and Stability of DNA Methyltransferase 1 in Esophageal Squamous Cell Carcinoma. Cancer Lett. 2018, 417, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Tano, K.; Mizuno, R.; Okada, T.; Rakwal, R.; Shibato, J.; Masuo, Y.; Ijiri, K.; Akimitsu, N. MALAT-1 Enhances Cell Motility of Lung Adenocarcinoma Cells by Influencing the Expression of Motility-Related Genes. FEBS Lett. 2010, 584, 4575–4580. [Google Scholar] [CrossRef]
- Wang, K.; Zhao, Y.; Wang, Y.-M. LncRNA MALAT1 Promotes Survival of Epithelial Ovarian Cancer Cells by Downregulating MiR-145-5p. Cancer Manag. Res. 2020, 12, 11359. [Google Scholar] [CrossRef]
- Pei, C.; Gong, X.; Zhang, Y. LncRNA MALAT-1 Promotes Growth and Metastasis of Epithelial Ovarian Cancer via Sponging Microrna-22. Am. J. Transl. Res. 2020, 12, 6977. [Google Scholar] [PubMed]
- Sun, Z.; Wang, H.; Wang, J.; Zhou, L.; Yang, P. Chemical Composition and Anti-Inflammatory, Cytotoxic and Antioxidant Activities of Essential Oil from Leaves of Mentha Piperita Grown in China. PLoS ONE 2014, 9, e114767. [Google Scholar] [CrossRef]
- Han, Y.; Liu, Y.; Gui, Y.; Cai, Z. Long Intergenic Non-Coding RNA TUG1 Is Overexpressed in Urothelial Carcinoma of the Bladder. J. Surg. Oncol. 2013, 107, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, J.; Qiu, M.; Xu, L.; Li, M.; Jiang, F.; Yin, R.; Xu, L. Upregulation of the Long Noncoding RNA TUG1 Promotes Proliferation and Migration of Esophageal Squamous Cell Carcinoma. Tumor Biol. 2015, 36, 1643–1651. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Lu, Y.-L.; Yang, Y.; Hu, L.-B.; Bai, Y.; Li, R.-Q.; Zhang, G.-Y.; Li, J.; Bi, C.-W.; Yang, L.-B. Overexpression of LncRNA ANRIL Promoted the Proliferation and Migration of Prostate Cancer Cells via Regulating Let-7a/TGF-Β1/Smad Signaling Pathway. Cancer Biomark. 2018, 21, 613–620. [Google Scholar] [CrossRef]
- Chen, C.; Wei, M.; Wang, C.; Sun, D.; Liu, P.; Zhong, X.; Yu, W. Long Noncoding RNA KCNQ1OT1 Promotes Colorectal Carcinogenesis by Enhancing Aerobic Glycolysis via Hexokinase-2. Aging 2020, 12, 11685. [Google Scholar] [CrossRef]
- Li, H.; Huang, H.; Li, S.; Mei, H.; Cao, T.; Lu, Q. Long Non-Coding RNA ADAMTS9-AS2 Inhibits Liver Cancer Cell Proliferation, Migration and Invasion. Exp. Ther. Med. 2021, 21, 559. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhi, X.; Wang, L.; Wang, W.; Li, Z.; Tang, J.; Wang, J.; Zhang, Q.; Xu, Z. Linc00152 Promotes Proliferation in Gastric Cancer through the EGFR-Dependent Pathway. J. Exp. Clin. Cancer Res. 2015, 34, 135. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zeng, J.; Liu, H.; Wang, T.; Yu, Z.; Chen, J. Role of HDAC1 in the Progression of Gastric Cancer and the Correlation with LncRNAs. Oncol. Lett. 2019, 17, 3296–3304. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Nie, X.; Zhang, W.; Gou, R.; Hu, Y.; Qi, Y.; Li, X.; Liu, Q.; Liu, J.; Lin, B. Identification of Long Noncoding RNA RP11-89K21. 1 and RP11-357H14. 17 as Prognostic Signature of Endometrial Carcinoma via Integrated Bioinformatics Analysis. Cancer Cell Int. 2020, 20, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Mo, Y.; Xue, L.; Shao, N.; Cao, J. Long Non-Coding RNA SUMO1P3 Promotes Tumour Progression by Regulating Cell Proliferation and Invasion in Glioma. Exp. Ther. Med. 2021, 21, 491. [Google Scholar] [CrossRef]
- Belwal, T.; Singh, G.; Jeandet, P.; Pandey, A.; Giri, L.; Ramola, S.; Bhatt, I.D.; Venskutonis, P.R.; Georgiev, M.I.; Clément, C. Anthocyanins, Multi-Functional Natural Products of Industrial Relevance: Recent Biotechnological Advances. Biotechnol. Adv. 2020, 43, 107600. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cao, X.; Xiong, N.; Wang, H.; Huang, J.; Sun, S.; Wang, T. Morin Exerts Neuroprotective Actions in Parkinson Disease Models in vitro and in vivo. Acta Pharmacol. Sin. 2010, 31, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Saclier, M.; Bonfanti, C.; Antonini, S.; Angelini, G.; Mura, G.; Zanaglio, F.; Taglietti, V.; Romanello, V.; Sandri, M.; Tonelli, C.; et al. Nutritional Intervention with Cyanidin Hinders the Progression of Muscular Dystrophy. Cell Death Dis. 2020, 11, 127. [Google Scholar] [CrossRef] [PubMed]
- Dormán, G.; Flachner, B.; Hajdú, I.; András, C.D. Chapter 21—Target Identification and Polypharmacology of Nutraceuticals. In Nutraceuticals; Gupta, R.C., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 263–286. ISBN 978-0-12-802147-7. [Google Scholar]
- Paluszczak, J.; Krajka-Kuźniak, V.; Baer-Dubowska, W. The Effect of Dietary Polyphenols on the Epigenetic Regulation of Gene Expression in MCF7 Breast Cancer Cells. Toxicol. Lett. 2010, 192, 119–125. [Google Scholar] [CrossRef]
- Turturică, M.; Stănciuc, N.; Bahrim, G.; Râpeanu, G. Effect of Thermal Treatment on Phenolic Compounds from Plum (Prunus Domestica) Extracts–A Kinetic Study. J. Food Eng. 2016, 171, 200–207. [Google Scholar] [CrossRef]
- Karthi, N.; Karthiga, A.; Kalaiyarasu, T.; Stalin, A.; Manju, V.; Singh, S.K.; Cyril, R.; Lee, S.-M. Exploration of Cell Cycle Regulation and Modulation of the DNA Methylation Mechanism of Pelargonidin: Insights from the Molecular Modeling Approach. Comput. Biol. Chem. 2017, 70, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, W.; Wang, C.; Wu, R.; Yin, R.; Kuo, H.-C.; Wang, L.; Kong, A.-N. Pelargonidin Reduces the TPA Induced Transformation of Mouse Epidermal Cells—Potential Involvement of Nrf2 Promoter Demethylation. Chem. Biol. Interact. 2019, 309, 108701. [Google Scholar] [CrossRef] [PubMed]
- Bonesi, M.; Loizzo, M.R.; Menichini, F.; Tundis, R. Chapter 23—Flavonoids in Treating Psoriasis. In Immunity and Inflammation in Health and Disease; Chatterjee, S., Jungraithmayr, W., Bagchi, D., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 281–294. ISBN 978-0-12-805417-8. [Google Scholar]
- Hafeez, B.B.; Siddiqui, I.A.; Asim, M.; Malik, A.; Afaq, F.; Adhami, V.M.; Saleem, M.; Din, M.; Mukhtar, H. A Dietary Anthocyanidin Delphinidin Induces Apoptosis of Human Prostate Cancer PC3 Cells in vitro and in vivo: Involvement of Nuclear Factor-ΚB Signaling. Cancer Res. 2008, 68, 8564–8572. [Google Scholar] [CrossRef]
- Kuo, H.-C.D.; Wu, R.; Li, S.; Yang, A.Y.; Kong, A.-N. Anthocyanin Delphinidin Prevents Neoplastic Transformation of Mouse Skin JB6 P+ Cells: Epigenetic Re-Activation of Nrf2-ARE Pathway. AAPS J. 2019, 21, 83. [Google Scholar] [CrossRef] [PubMed]
- Zhaohui, W.; Yingli, N.; Hongli, L.; Haijing, W.; Xiaohua, Z.; Chao, F.; Liugeng, W.; Hui, Z.; Feng, T.; Linfeng, Y.; et al. Amentoflavone Induces Apoptosis and Suppresses Glycolysis in Glioma Cells by Targeting MiR-124-3p. Neurosci. Lett. 2018, 686, 1–9. [Google Scholar] [CrossRef]
- Weng, J.-R.; Lai, I.-L.; Yang, H.-C.; Lin, C.-N.; Bai, L.-Y. Identification of Kazinol Q, a Natural Product from Formosan Plants, as an Inhibitor of DNA Methyltransferase: Identification of Kazinol as a Novel DNA Methyltransferase Inhibitor. Phytother. Res. 2014, 28, 49–54. [Google Scholar] [CrossRef]
- Lee, W.J.; Shim, J.-Y.; Zhu, B.T. Mechanisms for the Inhibition of DNA Methyltransferases by Tea Catechins and Bioflavonoids. Mol. Pharmacol. 2005, 68, 1018–1030. [Google Scholar] [CrossRef]
- Mittal, A.; Piyathilake, C.; Hara, Y.; Katiyar, S.K. Exceptionally High Protection of Photocarcinogenesis by Topical Application of (-)-Epi Gal Locatechin-3-Gal Late in Hydrophilic Cream in SKH-1 Hairless Mouse Model: Relationship to Inhibition of UVB-Induced Global DNA Hypomethylation. Neoplasia 2003, 5, 555–565. [Google Scholar] [CrossRef]
- Chuang, J.C.; Yoo, C.B.; Kwan, J.M.; Li, T.W.H.; Liang, G.; Yang, A.S.; Jones, P.A. Comparison of Biological Effects of Non-Nucleoside DNA Methylation Inhibitors versus 5-Aza-2′-Deoxycytidine. Mol. Cancer Ther. 2005, 4, 1515–1520. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.Z.; Jin, Z.; Wang, Y.; Liao, J.; Yang, G.-Y.; Wang, L.-D.; Yang, C.S. Promoter Hypermethylation and Inactivation of O6-Methylguanine-DNA Methyltransferase in Esophageal Squamous Cell Carcinomas and Its Reactivation in Cell Lines. Int. J. Oncol. 2005, 26, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Yamada, H.; Sugimura, H.; Tsuneyoshi, T. Suppressive Effect of Epigallocatechin Gallate (EGCg) on DNA Methylation in Mice: Detection by Methylation Sensitive Restriction Endonuclease Digestion and PCR. J. Food Agric. Environ. 2005, 3, 73–76. [Google Scholar]
- Berletch, J.B.; Liu, C.; Love, W.K.; Andrews, L.G.; Katiyar, S.K.; Tollefsbol, T.O. Epigenetic and Genetic Mechanisms Contribute to Telomerase Inhibition by EGCG. J. Cell. Biochem. 2008, 103, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Long, N.K.; Makita, H.; Toida, M.; Yamashita, T.; Hatakeyama, D.; Hara, A.; Mori, H.; Shibata, T. Effects of Green Tea Polyphenol on Methylation Status of RECK Gene and Cancer Cell Invasion in Oral Squamous Cell Carcinoma Cells. Br. J. Cancer 2008, 99, 647–654. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Adhikary, G.; Eckert, R.L. The Bmi-1 Polycomb Protein Antagonizes the (−)-Epigallocatechin-3-Gallate-Dependent Suppression of Skin Cancer Cell Survival. Carcinogenesis 2010, 31, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.H.I.; Xu, Z.; Hung, M.-S.; Lin, Y.-C.; Wang, T.; Gong, M.I.N.; Zhi, X.; Jablon, D.M.; You, L. Promoter Demethylation of WIF-1 by Epigallocatechin-3-Gallate in Lung Cancer Cells. Anticancer Res. 2009, 29, 2025–2030. [Google Scholar]
- Medina-Franco, J.L.; López-Vallejo, F.; Kuck, D.; Lyko, F. Natural Products as DNA Methyltransferase Inhibitors: A Computer-Aided Discovery Approach. Mol. Divers. 2011, 15, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Meeran, S.M.; Patel, S.N.; Chan, T.-H.; Tollefsbol, T.O. A Novel Prodrug of Epigallocatechin-3-Gallate: Differential Epigenetic HTERT Repression in Human Breast Cancer Cells. Cancer Prev. Res. 2011, 4, 1243–1254. [Google Scholar] [CrossRef]
- Nandakumar, V.; Vaid, M.; Katiyar, S.K. (-)-Epigallocatechin-3-Gallate Reactivates Silenced Tumor Suppressor Genes, Cip1/P21 and P16INK4a, by Reducing DNA Methylation and Increasing Histones Acetylation in Human Skin Cancer Cells. Carcinogenesis 2011, 32, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Min, N.Y.; Kim, J.-H.; Choi, J.-H.; Liang, W.; Ko, Y.J.; Rhee, S.; Bang, H.; Ham, S.W.; Park, A.J.; Lee, K.-H. Selective Death of Cancer Cells by Preferential Induction of Reactive Oxygen Species in Response to (−)-Epigallocatechin-3-Gallate. Biochem. Biophys. Res. Commun. 2012, 421, 91–97. [Google Scholar] [CrossRef]
- Achour, M.; Mousli, M.; Alhosin, M.; Ibrahim, A.; Peluso, J.; Muller, C.D.; Schini-Kerth, V.B.; Hamiche, A.; Dhe-Paganon, S.; Bronner, C. Epigallocatechin-3-Gallate up-Regulates Tumor Suppressor Gene Expression via a Reactive Oxygen Species-Dependent down-Regulation of UHRF1. Biochem. Biophys. Res. Commun. 2013, 430, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Landen, C.N.; Li, Y.; Alvarez, R.D.; Tollefsbol, T.O. Epigallocatechin Gallate and Sulforaphane Combination Treatment Induce Apoptosis in Paclitaxel-Resistant Ovarian Cancer Cells through HTERT and Bcl-2 down-Regulation. Exp. Cell Res. 2013, 319, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Moseley, V.R.; Morris, J.; Knackstedt, R.W.; Wargovich, M.J. Green Tea Polyphenol Epigallocatechin 3-Gallate, Contributes to the Degradation of DNMT3A and HDAC3 in HCT 116 Human Colon Cancer Cells. Anticancer Res. 2013, 33, 5325–5333. [Google Scholar] [PubMed]
- Mirza, S.; Sharma, G.; Parshad, R.; Gupta, S.D.; Pandya, P.; Ralhan, R. Expression of DNA Methyltransferases in Breast Cancer Patients and to Analyze the Effect of Natural Compounds on DNA Methyltransferases and Associated Proteins. J. Breast Cancer 2013, 16, 23. [Google Scholar] [CrossRef] [PubMed]
- Groh, I.A.M.; Chen, C.; Lüske, C.; Cartus, A.T.; Esselen, M. Plant Polyphenols and Oxidative Metabolites of the Herbal Alkenylbenzene Methyleugenol Suppress Histone Deacetylase Activity in Human Colon Carcinoma Cells. J. Nutr. Metab. 2013, 2013, 821082. [Google Scholar] [CrossRef]
- Wu, D.-S.; Shen, J.-Z.; Yu, A.-F.; Fu, H.-Y.; Zhou, H.-R.; Shen, S.-F. Epigallocatechin-3-Gallate and Trichostatin A Synergistically Inhibit Human Lymphoma Cell Proliferation through Epigenetic Modification of P16INK4a. Oncol. Rep. 2013, 30, 2969–2975. [Google Scholar] [CrossRef][Green Version]
- Appari, M.; Babu, K.R.; Kaczorowski, A.; Gros, W.; Her, I. Sulforaphane, Quercetin and Catechins Complement Each Other in Elimination of Advanced Pancreatic Cancer by MiR-Let-7 Induction and K-Ras Inhibition. Int. J. Oncol. 2014, 45, 1391–1400. [Google Scholar] [CrossRef]
- Deb, G.; Thakur, V.S.; Limaye, A.M.; Gupta, S. Epigenetic Induction of Tissue Inhibitor of Matrix Metalloproteinase-3 by Green Tea Polyphenols in Breast Cancer Cells. Mol. Carcinog. 2015, 54, 485–499. [Google Scholar] [CrossRef]
- Saldanha, S.N.; Kala, R.; Tollefsbol, T.O. Molecular Mechanisms for Inhibition of Colon Cancer Cells by Combined Epigenetic-Modulating Epigallocatechin Gallate and Sodium Butyrate. Exp. Cell Res. 2014, 324, 40–53. [Google Scholar] [CrossRef]
- Chen, L.-L.; Han, W.-F.; Geng, Y.; Su, J.-S. A Genome-Wide Study of DNA Methylation Modified by Epigallocatechin-3-Gallate in the CAL-27 Cell Line. Mol. Med. Rep. 2015, 12, 5886–5890. [Google Scholar] [CrossRef]
- Khan, M.A.; Hussain, A.; Sundaram, M.K.; Alalami, U.; Gunasekera, D.; Ramesh, L.; Hamza, A.; Quraishi, U. (−)-Epigallocatechin-3-Gallate Reverses the Expression of Various Tumor-Suppressor Genes by Inhibiting DNA Methyltransferases and Histone Deacetylases in Human Cervical Cancer Cells. Oncol. Rep. 2015, 33, 1976–1984. [Google Scholar] [CrossRef]
- Tyagi, T.; Treas, J.N.; Mahalingaiah, P.K.S.; Singh, K.P. Potentiation of Growth Inhibition and Epigenetic Modulation by Combination of Green Tea Polyphenol and 5-Aza-2′-Deoxycytidine in Human Breast Cancer Cells. Breast Cancer Res. Treat. 2015, 149, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Han, L.; Zhou, Y.; Sun, S. Green Tea Polyphenol EGCG Reverse Cisplatin Resistance of A549/DDP Cell Line through Candidate Genes Demethylation. Biomed. Pharmacother. 2015, 69, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Borutinskaitė, V.; Virkšaitė, A.; Gudelytė, G.; Navakauskienė, R. Green Tea Polyphenol EGCG Causes Anti-Cancerous Epigenetic Modulations in Acute Promyelocytic Leukemia Cells. Leuk. Lymphoma 2018, 59, 469–478. [Google Scholar] [CrossRef]
- Lewis, K.; Jordan, H.; Tollefsbol, T. Effects of SAHA and EGCG on Growth Potentiation of Triple-Negative Breast Cancer Cells. Cancers 2018, 11, 23. [Google Scholar] [CrossRef]
- Lubecka, K.; Kaufman-Szymczyk, A.; Cebula-Obrzut, B.; Smolewski, P.; Szemraj, J.; Fabianowska-Majewska, K. Novel Clofarabine-Based Combinations with Polyphenols Epigenetically Reactivate Retinoic Acid Receptor Beta, Inhibit Cell Growth, and Induce Apoptosis of Breast Cancer Cells. Int. J. Mol. Sci. 2018, 19, 3970. [Google Scholar] [CrossRef]
- Pal, D.; Sur, S.; Roy, R.; Mandal, S.; Kumar Panda, C. Epigallocatechin Gallate in Combination with Eugenol or Amarogentin Shows Synergistic Chemotherapeutic Potential in Cervical Cancer Cell Line: PAL. J. Cell. Physiol. 2019, 234, 825–836. [Google Scholar] [CrossRef]
- Vitkeviciene, A.; Baksiene, S.; Borutinskaite, V.; Navakauskiene, R. Epigallocatechin-3-Gallate and BIX-01294 Have Different Impact on Epigenetics and Senescence Modulation in Acute and Chronic Myeloid Leukemia Cells. Eur. J. Pharmacol. 2018, 838, 32–40. [Google Scholar] [CrossRef]
- Deb, G.; Shankar, E.; Thakur, V.S.; Ponsky, L.E.; Bodner, D.R.; Fu, P.; Gupta, S. Green Tea-Induced Epigenetic Reactivation of Tissue Inhibitor of Matrix Metalloproteinase-3 Suppresses Prostate Cancer Progression through Histone-Modifying Enzymes: DEB. Mol. Carcinog. 2019, 58, 1194–1207. [Google Scholar] [CrossRef]
- Sheng, J.; Shi, W.; Guo, H.; Long, W.; Wang, Y.; Qi, J.; Liu, J.; Xu, Y. The Inhibitory Effect of (−)-Epigallocatechin-3-Gallate on Breast Cancer Progression via Reducing SCUBE2 Methylation and DNMT Activity. Molecules 2019, 24, 2899. [Google Scholar] [CrossRef]
- Ciesielski, O.; Biesiekierska, M.; Balcerczyk, A. Epigallocatechin-3-Gallate (EGCG) Alters Histone Acetylation and Methylation and Impacts Chromatin Architecture Profile in Human Endothelial Cells. Molecules 2020, 25, 2326. [Google Scholar] [CrossRef]
- Steed, K.L.; Jordan, H.R.; Tollefsbol, T.O. SAHA and EGCG Promote Apoptosis in Triple-Negative Breast Cancer Cells, Possibly Through the Modulation of CIAP2. Anticancer Res. 2020, 40, 9–26. [Google Scholar] [CrossRef]
- Hermawan, A.; Putri, H.; Utomo, R.Y. Comprehensive Bioinformatics Study Reveals Targets and Molecular Mechanism of Hesperetin in Overcoming Breast Cancer Chemoresistance. Mol. Divers. 2020, 24, 933–947. [Google Scholar] [CrossRef]
- Lewinska, A.; Siwak, J.; Rzeszutek, I.; Wnuk, M. Diosmin Induces Genotoxicity and Apoptosis in DU145 Prostate Cancer Cell Line. Toxicol. In Vitro 2015, 29, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Bedmar, Z.; Anter, J.; Alonso-Moraga, A.; Martín de las Mulas, J.; Millán-Ruiz, Y.; Guil-Luna, S. Demethylating and Anti-Hepatocarcinogenic Potential of Hesperidin, a Natural Polyphenol of Citrus Juices. Mol. Carcinog. 2017, 56, 1653–1662. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Wen, L.; Peng, R.; Li, H.; Liu, H.; Peng, H.; Sun, Y.; Wu, T.; Chen, L.; Duan, Q.; et al. Naringenin Ameliorated Kidney Injury through Let-7a/TGFBR1 Signaling in Diabetic Nephropathy. J. Diabetes Res. 2016, 2016, 8738760. [Google Scholar] [CrossRef] [PubMed]
- Curti, V.; Di Lorenzo, A.; Rossi, D.; Martino, E.; Capelli, E.; Collina, S.; Daglia, M. Enantioselective Modulatory Effects of Naringenin Enantiomers on the Expression Levels of MiR-17-3p Involved in Endogenous Antioxidant Defenses. Nutrients 2017, 9, 215. [Google Scholar] [CrossRef] [PubMed]
- Kuang, H.; Tang, Z.; Zhang, C.; Wang, Z.; Li, W.; Yang, C.; Wang, Q.; Yang, B.; Kong, A.-N. Taxifolin Activates the Nrf2 Anti-Oxidative Stress Pathway in Mouse Skin Epidermal JB6 P+ Cells through Epigenetic Modifications. Int. J. Mol. Sci. 2017, 18, 1546. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Chang, H.; Gu, Y.; Chen, J.; Yi, L.; Xie, Q.; Zhu, J.; Zhang, Q.; Mi, M. 3, 6-Dihydroxyflavone Suppresses Breast Carcinogenesis by Epigenetically Regulating MiR-34a and MiR-21. Cancer Prev. Res. 2015, 8, 509–517. [Google Scholar] [CrossRef]
- Peng, X.; Chang, H.; Chen, J.; Zhang, Q.; Yu, X.; Mi, M. 3,6-Dihydroxyflavone Regulates MicroRNA-34a through DNA Methylation. BMC Cancer 2017, 17, 619. [Google Scholar] [CrossRef]
- Kanwal, R.; Datt, M.; Liu, X.; Gupta, S. Dietary Flavones as Dual Inhibitors of DNA Methyltransferases and Histone Methyltransferases. PLoS ONE 2016, 11, e0162956. [Google Scholar] [CrossRef]
- Pandey, M.; Kaur, P.; Shukla, S.; Abbas, A.; Fu, P.; Gupta, S. Plant Flavone Apigenin Inhibits HDAC and Remodels Chromatin to Induce Growth Arrest and Apoptosis in Human Prostate Cancer Cells: In Vitro and In Vivo Study. Mol. Carcinog. 2012, 51, 952–962. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Gonzalez, X.; Fuentes, F.; Su, Z.-Y.; Kong, A.-N.T. Apigenin Reactivates Nrf2 Anti-Oxidative Stress Signaling in Mouse Skin Epidermal JB6 P + Cells Through Epigenetics Modifications. AAPS J. 2014, 16, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.-H.; Chien, M.-H.; Lin, W.-L.; Wen, Y.-C.; Chow, J.-M.; Chen, C.-K.; Kuo, T.-C.; Lee, W.-J. Inhibition of MDA-MB-231 Breast Cancer Cell Proliferation and Tumor Growth by Apigenin through Induction of G2/M Arrest and Histone H3 Acetylation-Mediated P21 WAF1/CIP1 Expression: Apigenin Inhibits MDA-MB-231 Breast Cancer Cell. Environ. Toxicol. 2017, 32, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; He, K.; Huang, L.; Zhang, L.; Liu, A.; Zhang, J. Casticin Inhibits the Activity of Transcription Factor Sp1 and the Methylation of RECK in MGC803 Gastric Cancer Cells. Exp. Ther. Med. 2017, 13, 745–750. [Google Scholar] [CrossRef]
- Li, X.; Wang, L.; Cao, X.; Zhou, L.; Xu, C.; Cui, Y.; Qiu, Y.; Cao, J. Casticin Inhibits Stemness of Hepatocellular Carcinoma Cells via Disrupting the Reciprocal Negative Regulation between DNMT1 and MiR-148a-3p. Toxicol. Appl. Pharmacol. 2020, 396, 114998. [Google Scholar] [CrossRef]
- Lewinska, A.; Adamczyk-Grochala, J.; Kwasniewicz, E.; Deregowska, A.; Wnuk, M. Diosmin-Induced Senescence, Apoptosis and Autophagy in Breast Cancer Cells of Different P53 Status and ERK Activity. Toxicol. Lett. 2017, 265, 117–130. [Google Scholar] [CrossRef]
- Liang, X.; Xu, C.; Cao, X.; Wang, W. Isovitexin Suppresses Cancer Stemness Property And Induces Apoptosis Of Osteosarcoma Cells By Disruption Of The DNMT1/MiR-34a/Bcl-2 Axis. Cancer Manag. Res. 2019, 11, 8923–8936. [Google Scholar] [CrossRef]
- Attoub, S.; Hassan, A.H.; Vanhoecke, B.; Iratni, R.; Takahashi, T.; Gaben, A.-M.; Bracke, M.; Awad, S.; John, A.; Kamalboor, H.A.; et al. Inhibition of Cell Survival, Invasion, Tumor Growth and Histone Deacetylase Activity by the Dietary Flavonoid Luteolin in Human Epithelioid Cancer Cells. Eur. J. Pharmacol. 2011, 651, 18–25. [Google Scholar] [CrossRef]
- Dong, X.; Zhang, J.; Yang, F.; Wu, J.; Cai, R.; Wang, T.; Zhang, J. Effect of Luteolin on the Methylation Status of the OPCML Gene and Cell Growth in Breast Cancer Cells. Exp. Ther. Med. 2018, 16, 3186–3194. [Google Scholar] [CrossRef]
- Kang, K.A.; Piao, M.J.; Hyun, Y.J.; Zhen, A.X.; Cho, S.J.; Ahn, M.J.; Yi, J.M.; Hyun, J.W. Luteolin Promotes Apoptotic Cell Death via Upregulation of Nrf2 Expression by DNA Demethylase and the Interaction of Nrf2 with P53 in Human Colon Cancer Cells. Exp. Mol. Med. 2019, 51, 1–14. [Google Scholar] [CrossRef]
- Krifa, M.; Leloup, L.; Ghedira, K.; Mousli, M.; Chekir-Ghedira, L. Luteolin Induces Apoptosis in BE Colorectal Cancer Cells by Downregulating Calpain, UHRF1, and DNMT1 Expressions. Nutr. Cancer 2014, 66, 1220–1227. [Google Scholar] [CrossRef]
- Selvi, R.B.; Swaminathan, A.; Chatterjee, S.; Shanmugam, M.K.; Li, F.; Ramakrishnan, G.B.; Siveen, K.S.; Chinnathambi, A.; Zayed, M.E.; Alharbi, S.A. Inhibition of P300 Lysine Acetyltransferase Activity by Luteolin Reduces Tumor Growth in Head and Neck Squamous Cell Carcinoma (HNSCC) Xenograft Mouse Model. Oncotarget 2015, 6, 43806. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Wu, R.; Xiao, X.; Yang, C.; Yang, Y.; Wang, C.; Lin, L.; Kong, A. The Dietary Flavone Luteolin Epigenetically Activates the Nrf2 Pathway and Blocks Cell Transformation in Human Colorectal Cancer HCT116 Cells. J. Cell. Biochem. 2018, 119, 9573–9582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, S.; Li, J.; Yin, F.; Hua, Y.; Wang, Z.; Lin, B.; Wang, H.; Zou, D.; Zhou, Z.; et al. Natural Product Pectolinarigenin Inhibits Osteosarcoma Growth and Metastasis via SHP-1-Mediated STAT3 Signaling Inhibition. Cell Death Dis. 2016, 7, e2421. [Google Scholar] [CrossRef]
- Anestopoulos, I.; Sfakianos, A.; Franco, R.; Chlichlia, K.; Panayiotidis, M.; Kroll, D.; Pappa, A. A Novel Role of Silibinin as a Putative Epigenetic Modulator in Human Prostate Carcinoma. Molecules 2016, 22, 62. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.T.; Sávio, A.L.V.; de Castro Marcondes, J.P.; Barros, T.M.; Barbosa, L.C.; Salvadori, D.M.F.; da Silva, G.N. Cytotoxic and Toxicogenomic Effects of Silibinin in Bladder Cancer Cells with Different TP53 Status. J. Biosci. 2017, 42, 91–101. [Google Scholar] [CrossRef]
- Kauntz, H.; Bousserouel, S.; Gossé, F.; Raul, F. Epigenetic Effects of the Natural Flavonolignan Silibinin on Colon Adenocarcinoma Cells and Their Derived Metastatic Cells. Oncol. Lett. 2013, 5, 1273–1277. [Google Scholar] [CrossRef] [PubMed]
- Mateen, S.; Raina, K.; Agarwal, C.; Agarwal, R. Inhibition of Epigenetic Chromatin-Modification Enzymes: Histone Deacetylases and DNA Methyltransferases by Silibinin in Human NSCLC H1299 Cells; AACR: Philadelphia, PA, USA, 2010; ISBN 0008-5472. [Google Scholar]
- Zeng, H.; Huang, P.; Wang, X.; Wu, J.; Wu, M.; Huang, J. Galangin-Induced down-Regulation of BACE1 by Epigenetic Mechanisms in SH-SY5Y Cells. Neuroscience 2015, 294, 172–181. [Google Scholar] [CrossRef]
- Berger, A.; Venturelli, S.; Kallnischkies, M.; Böcker, A.; Busch, C.; Weiland, T.; Noor, S.; Leischner, C.; Weiss, T.S.; Lauer, U.M.; et al. Kaempferol, a New Nutrition-Derived Pan-Inhibitor of Human Histone Deacetylases. J. Nutr. Biochem. 2013, 24, 977–985. [Google Scholar] [CrossRef]
- Kim, T.W.; Lee, S.Y.; Kim, M.; Cheon, C.; Ko, S.-G. Kaempferol Induces Autophagic Cell Death via IRE1-JNK-CHOP Pathway and Inhibition of G9a in Gastric Cancer Cells. Cell Death Dis. 2018, 9, 875. [Google Scholar] [CrossRef]
- Qiu, W.; Lin, J.; Zhu, Y.; Zhang, J.; Zeng, L.; Su, M.; Tian, Y. Kaempferol Modulates DNA Methylation and Downregulates DNMT3B in Bladder Cancer. Cell. Physiol. Biochem. 2017, 41, 1325–1335. [Google Scholar] [CrossRef] [PubMed]
- Nowak, E.; Sypniewski, D.; Bednarek, I. Morin Exerts Anti-Metastatic, Anti-Proliferative and Anti-Adhesive Effect in Ovarian Cancer Cells: An in Vitro Studies. Mol. Biol. Rep. 2020, 47, 1965–1978. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Wang, C.; Lu, C.; Zhao, B.; Cui, Y.; Shi, X.; Ma, X. Quercetin Is Able to Demethylate the P16INK4a Gene Promoter. Chemotherapy 2009, 55, 6–10. [Google Scholar] [CrossRef]
- Tao, S.; He, H.; Chen, Q. Quercetin Inhibits Proliferation and Invasion Acts by Up-Regulating MiR-146a in Human Breast Cancer Cells. Mol. Cell. Biochem. 2015, 402, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guo, Q.; Chen, J.; Chen, Z. Quercetin Enhances Cisplatin Sensitivity of Human Osteosarcoma Cells by Modulating MicroRNA-217-KRAS Axis. Mol. Cells 2015, 38, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Kedhari Sundaram, M.; Hussain, A.; Haque, S.; Raina, R.; Afroze, N. Quercetin Modifies 5′CpG Promoter Methylation and Reactivates Various Tumor Suppressor Genes by Modulating Epigenetic Marks in Human Cervical Cancer Cells. J. Cell. Biochem. 2019, 120, 18357–18369. [Google Scholar] [CrossRef]
- Kundur, S.; Prayag, A.; Selvakumar, P.; Nguyen, H.; McKee, L.; Cruz, C.; Srinivasan, A.; Shoyele, S.; Lakshmikuttyamma, A. Synergistic Anticancer Action of Quercetin and Curcumin against Triple-negative Breast Cancer Cell Lines. J. Cell. Physiol. 2019, 234, 11103–11118. [Google Scholar] [CrossRef]
- Tseng Quercetin Induces FasL-Related Apoptosis, in Part, through Promotion of Histone H3 Acetylation in Human Leukemia HL-60 Cells. Oncol. Rep. 2011, 25, 583–591. [CrossRef]
- Lee, M.; Son, M.; Ryu, E.; Shin, Y.S.; Kim, J.G.; Kang, B.W.; Sung, G.-H.; Cho, H.; Kang, H. Quercetin-Induced Apoptosis Prevents EBV Infection. Oncotarget 2015, 6, 12603. [Google Scholar] [CrossRef]
- Liu, C.-M.; Ma, J.-Q.; Xie, W.-R.; Liu, S.-S.; Feng, Z.-J.; Zheng, G.-H.; Wang, A.-M. Quercetin Protects Mouse Liver against Nickel-Induced DNA Methylation and Inflammation Associated with the Nrf2/HO-1 and P38/STAT1/NF-ΚB Pathway. Food Chem. Toxicol. 2015, 82, 19–26. [Google Scholar] [CrossRef]
- Priyadarsini, R.V.; Vinothini, G.; Murugan, R.S.; Manikandan, P.; Nagini, S. The Flavonoid Quercetin Modulates the Hallmark Capabilities of Hamster Buccal Pouch Tumors. Nutr. Cancer 2011, 63, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, P.A.; Braune, A.; Hölzlwimmer, G.; Quintanilla-Fend, L.; Haller, D. Quercetin Inhibits TNF-Induced NF-κ B Transcription Factor Recruitment to Proinflammatory Gene Promoters in Murine Intestinal Epithelial Cells. J. Nutr. 2007, 137, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Kumar, L.; Mohanty, S.K.; Maikhuri, J.P.; Rajender, S.; Gupta, G. Sensitization of Androgen Refractory Prostate Cancer Cells to Anti-Androgens through Re-Expression of Epigenetically Repressed Androgen Receptor—Synergistic Action of Quercetin and Curcumin. Mol. Cell. Endocrinol. 2016, 431, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.-G.; Wang, J.-L.; Yang, S.-L.; Wu, J.-L. Aberrant Epigenetic Alteration in Eca9706 Cells Modulated by Nanoliposomal Quercetin Combined with Butyrate Mediated via Epigenetic-NF-ΚB Signaling. Asian Pac. J. Cancer Prev. 2014, 15, 4539–4543. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fang, M.Z.; Chen, D.; Sun, Y.; Jin, Z.; Christman, J.K.; Yang, C.S. Reversal of Hypermethylation and Reactivation of P16INK4a, RARβ, and MGMT Genes by Genistein and Other Isoflavones from Soy. Clin. Cancer Res. 2005, 11, 7033–7041. [Google Scholar] [CrossRef]
- Vandegehuchte, M.B.; Lemière, F.; Vanhaecke, L.; Vanden Berghe, W.; Janssen, C.R. Direct and Transgenerational Impact on Daphnia Magna of Chemicals with a Known Effect on DNA Methylation. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2010, 151, 278–285. [Google Scholar] [CrossRef]
- Dagdemir, A.; Durif, J.; Ngollo, M.; Bignon, Y.-J.; Bernard-Gallon, D. Histone Lysine Trimethylation or Acetylation Can Be Modulated by Phytoestrogen, Estrogen or Anti-HDAC in Breast Cancer Cell Lines. Epigenomics 2013, 5, 51–63. [Google Scholar] [CrossRef]
- Vardi, A.; Bosviel, R.; Rabiau, N.; Adjakly, M.; Satih, S.; Dechelotte, P.; Boiteux, J.-P.; Fontana, L.; Bignon, Y.-J.; Guy, L. Soy Phytoestrogens Modify DNA Methylation of GSTP1, RASSF1A, EPH2 and BRCA1 Promoter in Prostate Cancer Cells. In Vivo 2010, 24, 393–400. [Google Scholar]
- Tabares-Guevara, J.H.; Lara-Guzmán, O.J.; Londoño-Londoño, J.A.; Sierra, J.A.; León-Varela, Y.M.; Álvarez-Quintero, R.M.; Osorio, E.J.; Ramirez-Pineda, J.R. Natural Biflavonoids Modulate Macrophage–Oxidized LDL Interaction in vitro and Promote Atheroprotection in vivo. Front. Immunol. 2017, 8, 923. [Google Scholar] [CrossRef]
- Kim, H.K.; Son, K.H.; Chang, H.W.; Kang, S.S.; Kim, H.P. Amentoflavone, a Plant Biflavone: A New Potential Anti-Inflammatory Agent. Arch. Pharm. Res. 1998, 21, 406–410. [Google Scholar] [CrossRef]
- Mazimba, O.; Keroletswe, N. Flavans: Synthetic Strategies: A Review. Int. Res. J. Pure Appl. Chem. 2015, 8, 112–146. [Google Scholar] [CrossRef]
- Wei, B.-L.; Chen, Y.-C.; Hsu, H.-Y. Kazinol Q from Broussonetia Kazinoki Enhances Cell Death Induced by Cu (Ll) through Increased Reactive Oxygen Species. Molecules 2011, 16, 3212–3221. [Google Scholar] [CrossRef]
- Bonetti, F.; Brombo, G.; Zuliani, G. Chapter 19—Nootropics, Functional Foods, and Dietary Patterns for Prevention of Cognitive Decline. In Nutrition and Functional Foods for Healthy Aging; Watson, R.R., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 211–232. ISBN 978-0-12-805376-8. [Google Scholar]
- Lan, X.; Han, X.; Li, Q.; Wang, J. (−)-Epicatechin, a Natural Flavonoid Compound, Protects Astrocytes Against Hemoglobin Toxicity via Nrf2 and AP-1 Signaling Pathways. Mol. Neurobiol. 2017, 54, 7898–7907. [Google Scholar] [CrossRef]
- Khan, M.K.; Dangles, O. A Comprehensive Review on Flavanones, the Major Citrus Polyphenols. J. Food Compos. Anal. 2014, 33, 85–104. [Google Scholar] [CrossRef]
- Muhammad, T.; Ikram, M.; Ullah, R.; Rehman, S.U.; Kim, M.O. Hesperetin, a Citrus Flavonoid, Attenuates LPS-Induced Neuroinflammation, Apoptosis and Memory Impairments by Modulating TLR4/NF-ΚB Signaling. Nutrients 2019, 11, 648. [Google Scholar] [CrossRef]
- Devi, K.P.; Rajavel, T.; Nabavi, S.F.; Setzer, W.N.; Ahmadi, A.; Mansouri, K.; Nabavi, S.M. Hesperidin: A Promising Anticancer Agent from Nature. Ind. Crops Prod. 2015, 76, 582–589. [Google Scholar] [CrossRef]
- Choi, J.; Lee, D.-H.; Jang, H.; Park, S.-Y.; Seol, J.-W. Naringenin Exerts Anticancer Effects by Inducing Tumor Cell Death and Inhibiting Angiogenesis in Malignant Melanoma. Int. J. Med. Sci. 2020, 17, 3049–3057. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, Y.; Zhang, L.; Chen, L.; Du, Y.; Ye, T.; Shi, X. Naringenin Exerts Anti-Angiogenic Effects in Human Endothelial Cells: Involvement of ERRα/VEGF/KDR Signaling Pathway. Fitoterapia 2016, 111, 78–86. [Google Scholar] [CrossRef]
- Weidmann, A.E. Dihydroquercetin: More than Just an Impurity? Eur. J. Pharmacol. 2012, 684, 19–26. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, X.; Cui, Y.; Zhou, H.; Xu, D.; Shan, T.; Zhang, F.; Guo, Y.; Chen, Y.; Wu, D. Taxifolin Protects against Cardiac Hypertrophy and Fibrosis during Biomechanical Stress of Pressure Overload. Toxicol. Appl. Pharmacol. 2015, 287, 168–177. [Google Scholar] [CrossRef]
- Manigandan, K.; Manimaran, D.; Jayaraj, R.L.; Elangovan, N.; Dhivya, V.; Kaphle, A. Taxifolin Curbs NF-ΚB-Mediated Wnt/β-Catenin Signaling via up-Regulating Nrf2 Pathway in Experimental Colon Carcinogenesis. Biochimie 2015, 119, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.H.; Jiang, Y.-M.; Shi, J.; Tomas-Barberan, F.A.; Datta, N.; Singanusong, R.; Chen, S.S. Flavonoids in Food and Their Health Benefits. Plant Foods Hum. Nutr. 2004, 59, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Sergeev, I.N.; Li, S.; Colby, J.; Ho, C.-T.; Dushenkov, S. Polymethoxylated Flavones Induce Ca2+-Mediated Apoptosis in Breast Cancer Cells. Life Sci. 2006, 80, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, P.K.; Kim, J.; Son, K.; Durai, P.; Kim, Y. 3,6-Dihydroxyflavone: A Potent Inhibitor with Anti-Inflammatory Activity Targeting Toll-like Receptor 2: TLR2/1 Inhibitor, 3,6-Dihydroxyflavone. Bull. Korean Chem. Soc. 2019, 40, 51–55. [Google Scholar] [CrossRef]
- Ali, F.; Rahul; Naz, F.; Jyoti, S.; Siddique, Y.H. Health Functionality of Apigenin: A Review. Int. J. Food Prop. 2017, 20, 1197–1238. [Google Scholar] [CrossRef]
- Shukla, S.; Gupta, S. Apigenin: A Promising Molecule for Cancer Prevention. Pharm. Res. 2010, 27, 962–978. [Google Scholar] [CrossRef]
- Forzato, C.; Vida, V.; Berti, F. Biosensors and Sensing Systems for Rapid Analysis of Phenolic Compounds from Plants: A Comprehensive Review. Biosensors 2020, 10, 105. [Google Scholar] [CrossRef]
- Gao, Y.; Snyder, S.A.; Smith, J.N.; Chen, Y.C. Anticancer Properties of Baicalein: A Review. Med. Chem. Res. 2016, 25, 1515–1523. [Google Scholar] [CrossRef]
- Azizul, N.H.; Ahmad, W.; Rosli, N.L. The Coastal Medicinal Plant Vitex Rotundifolia: A Mini-Review on Its Bioactive Compounds and Pharmacological Activity. Tradit. Med. Res. 2021, 6, 11. [Google Scholar]
- Kasala, E.R.; Bodduluru, L.N.; Madana, R.M.; Gogoi, R.; Barua, C.C. Chemopreventive and Therapeutic Potential of Chrysin in Cancer: Mechanistic Perspectives. Toxicol. Lett. 2015, 233, 214–225. [Google Scholar] [CrossRef]
- Khoo, B.Y.; Chua, S.L.; Balaram, P. Apoptotic Effects of Chrysin in Human Cancer Cell Lines. Int. J. Mol. Sci. 2010, 11, 2188–2199. [Google Scholar] [CrossRef]
- Thanapongsathorn, W.; Vajrabukka, T. Clinical Trial of Oral Diosmin (Daflon®) in the Treatment of Hemorrhoids. Dis. Colon Rectum 1992, 35, 1085–1088. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, K.; Xu, B. Molecular Targets of Vitexin and Isovitexin in Cancer Therapy: A Critical Review. Ann. N. Y. Acad. Sci. 2017, 1401, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Tuorkey, M.J. Molecular Targets of Luteolin in Cancer. Eur. J. Cancer Prev. 2016, 25, 65. [Google Scholar] [CrossRef] [PubMed]
- Bharti, S.; Rani, N.; Krishnamurthy, B.; Arya, D.S. Preclinical Evidence for the Pharmacological Actions of Naringin: A Review. Planta Med. 2014, 80, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Qi, Q.-L.; Wang, M.-T.; Li, Q.-Y. Therapeutic Potential of Naringin: An Overview. Pharm. Biol. 2016, 54, 3203–3210. [Google Scholar] [CrossRef]
- Lee, H.J.; Venkatarame Gowda Saralamma, V.; Kim, S.M.; Ha, S.E.; Raha, S.; Lee, W.S.; Kim, E.H.; Lee, S.J.; Heo, J.D.; Kim, G.S. Pectolinarigenin Induced Cell Cycle Arrest, Autophagy, and Apoptosis in Gastric Cancer Cell via PI3K/AKT/MTOR Signaling Pathway. Nutrients 2018, 10, 1043. [Google Scholar] [CrossRef]
- Bijak, M. Flavonolignans—compounds not only for liver treatment. Pol. Merkur. Lek. Organ Pol. Tow. Lek. 2017, 42, 34–37. [Google Scholar]
- Gazak, R.; Walterova, D.; Kren, V. Silybin and Silymarin-New and Emerging Applications in Medicine. Curr. Med. Chem. 2007, 14, 315–338. [Google Scholar] [CrossRef]
- Garg, G.; Khandelwal, A.; Blagg, B.S.J. Chapter Three—Anticancer Inhibitors of Hsp90 Function: Beyond the Usual Suspects. In Advances in Cancer Research; Isaacs, J., Whitesell, L., Eds.; Hsp90 in Cancer: Beyond the Usual Suspects; Academic Press: Cambridge, MA, USA, 2016; Volume 129, pp. 51–88. [Google Scholar]
- Lesser, S.; Wolffram, S. Oral Bioavailability of the Flavonol Quercetin a Review. Curr. Top. Nutraceutical Res. 2006, 4, 239. [Google Scholar]
- Kopustinskiene, D.M.; Jakstas, V.; Savickas, A.; Bernatoniene, J. Flavonoids as Anticancer Agents. Nutrients 2020, 12, 457. [Google Scholar] [CrossRef]
- Shukla, R.; Pandey, V.; Vadnere, G.P.; Lodhi, S. Role of flavonoids in management of inflammatory disorders. In Bioactive Food as Dietary Interventions for Arthritis and Related Inflammatory Diseases; Elsevier: Amsterdam, The Netherlands, 2019; pp. 293–322. [Google Scholar]
- Das, G.; Patra, J.K.; Gonçalves, S.; Romano, A.; Gutiérrez-Grijalva, E.P.; Heredia, J.B.; Talukdar, A.D.; Shome, S.; Shin, H.-S. Galangal, the Multipotent Super Spices: A Comprehensive Review. Trends Food Sci. Technol. 2020, 101, 50–62. [Google Scholar] [CrossRef]
- Ren, J.; Lu, Y.; Qian, Y.; Chen, B.; Wu, T.; Ji, G. Recent Progress Regarding Kaempferol for the Treatment of Various Diseases. Exp. Ther. Med. 2019, 18, 2759–2776. [Google Scholar] [CrossRef]
- Hwang-Bo, H.; Lee, W.S.; Nagappan, A.; Kim, H.J.; Panchanathan, R.; Park, C.; Chang, S.; Kim, N.D.; Leem, S.; Chang, Y.; et al. Morin Enhances Auranofin Anticancer Activity by Up-regulation of DR4 and DR5 and Modulation of Bcl-2 through Reactive Oxygen Species Generation in Hep3B Human Hepatocellular Carcinoma Cells. Phytother. Res. 2019, 33, 1384–1393. [Google Scholar] [CrossRef]
- Brodowska, K.M. Natural Flavonoids: Classification, Potential Role, and Application of Flavonoid Analogues. Eur. J. Biol. Res. 2017, 7, 108–123. [Google Scholar]
- Nabavi, S.F.; Russo, G.L.; Daglia, M.; Nabavi, S.M. Role of Quercetin as an Alternative for Obesity Treatment: You Are What You Eat! Food Chem. 2015, 179, 305–310. [Google Scholar] [CrossRef]
- Tolun, A.; Altintas, Z. Medicinal properties and functional components of beverages. In Functional and Medicinal Beverages; Elsevier: Amsterdam, The Netherlands, 2019; pp. 235–284. [Google Scholar]
- Yu, C.; Zhang, P.; Lou, L.; Wang, Y. Perspectives Regarding the Role of Biochanin A in Humans. Front. Pharmacol. 2019, 10, 793. [Google Scholar] [CrossRef]
- Sun, M.-Y.; Ye, Y.; Xiao, L.; Rahman, K.; Xia, W.; Zhang, H. Daidzein: A Review of Pharmacological Effects. Afr. J. Tradit. Complement. Altern. Med. 2016, 13, 117–132. [Google Scholar] [CrossRef]
- Setchell, K.D.; Clerici, C. Equol: History, Chemistry, and Formation. J. Nutr. 2010, 140, 1355S–1362S. [Google Scholar] [CrossRef]
- Saha, S.; Sadhukhan, P.; C Sil, P. Genistein: A Phytoestrogen with Multifaceted Therapeutic Properties. Mini Rev. Med. Chem. 2014, 14, 920–940. [Google Scholar] [CrossRef]
- Omari, N.; Bakrim, S.; Bakha, M.; Lorenzo, J.M.; Rebezov, M.; Shariati, M.A.; Aboulaghras, S.; Balahbib, A.; Khayrullin, M.; Bouyahya, A. Natural Bioactive Compounds Targeting Epigenetic Pathways in Cancer: A Review on Alkaloids, Terpenoids, Quinones, and Isothiocyanates. Nutrients 2021, 13, 3714. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Howard, L.R.; VillalóN, B. Flavonoids and Antioxidant Activity of Fresh Pepper (Capsicum annuum) Cultivars. J. Food Sci. 1995, 60, 473–476. [Google Scholar] [CrossRef]
- Chu, C.; Du, Y.; Yu, X.; Shi, J.; Yuan, X.; Liu, X.; Liu, Y.; Zhang, H.; Zhang, Z.; Yan, N. Dynamics of Antioxidant Activities, Metabolites, Phenolic Acids, Flavonoids, and Phenolic Biosynthetic Genes in Germinating Chinese Wild Rice (Zizania latifolia). Food Chem. 2020, 318, 126483. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Bruins, M.E.; de Bruijn, W.J.; Vincken, J.-P. A Comparison of the Phenolic Composition of Old and Young Tea Leaves Reveals a Decrease in Flavanols and Phenolic Acids and an Increase in Flavonols upon Tea Leaf Maturation. J. Food Compos. Anal. 2020, 86, 103385. [Google Scholar] [CrossRef]
- Choi, S.K.; Pandiyan, K.; Eun, J.W.; Yang, X.; Hong, S.H.; Nam, S.W.; Jones, P.A.; Liang, G.; You, J.S. Epigenetic Landscape Change Analysis during Human EMT Sheds Light on a Key EMT Mediator TRIM29. Oncotarget 2017, 8, 98322. [Google Scholar] [CrossRef][Green Version]
- Eroglu, O.; Celik, E.G.; Kaya, H.; Celen, M.; Karabicici, M.; Karacoban, E. Investigation of Methylation Profiles of TP53, Caspase 9, Caspase 8, Caspase 3 Genes Treated with DNA Methyl Transferase Inhibitor (DNMTi) Zebularine (ZEB) and Caffeic Acid Phenethyl Ester (CAPE) on MCF-7 and MDA-MB-231 Breast Cancer Cell Lines. J. Cancer Ther. 2019, 10, 69–85. [Google Scholar] [CrossRef][Green Version]
- Lee, W.J.; Zhu, B.T. Inhibition of DNA Methylation by Caffeic Acid and Chlorogenic Acid, Two Common Catechol-Containing Coffee Polyphenols. Carcinogenesis 2006, 27, 269–277. [Google Scholar] [CrossRef]
- Bora-Tatar, G.; Dayangaç-Erden, D.; Demir, A.S.; Dalkara, S.; Yelekçi, K.; Erdem-Yurter, H. Molecular Modifications on Carboxylic Acid Derivatives as Potent Histone Deacetylase Inhibitors: Activity and Docking Studies. Bioorg. Med. Chem. 2009, 17, 5219–5228. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, F.; Chen, L.; Yang, Y.; Cao, S.; Ye, Y.; Wang, X.; Mu, J.; Li, Z.; Li, L. Blockage of TGFβ-SMAD2 by Demethylation-Activated MiR-148a Is Involved in Caffeic Acid-Induced Inhibition of Cancer Stem Cell-like Properties in vitro and in vivo. FEBS Open Biol. 2015, 5, 466–475. [Google Scholar] [CrossRef]
- Weng, Y.-P.; Hung, P.-F.; Ku, W.-Y.; Chang, C.-Y.; Wu, B.-H.; Wu, M.-H.; Yao, J.-Y.; Yang, J.-R.; Lee, C.-H. The Inhibitory Activity of Gallic Acid against DNA Methylation: Application of Gallic Acid on Epigenetic Therapy of Human Cancers. Oncotarget 2018, 9, 361–374. [Google Scholar] [CrossRef]
- Kam, A.; Li, K.M.; Razmovski-Naumovski, V.; Nammi, S.; Chan, K.; Li, G.Q. Gallic Acid Protects against Endothelial Injury by Restoring the Depletion of DNA Methyltransferase 1 and Inhibiting Proteasome Activities. Int. J. Cardiol. 2014, 171, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Tsuneoka, M. Gallic Acid Derivatives Propyl Gallate and Epigallocatechin Gallate Reduce RRNA Transcription via Induction of KDM2A Activation. Biomolecules 2022, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Semba, R.D.; Ferrucci, L.; Bartali, B.; Urpí-Sarda, M.; Zamora-Ros, R.; Sun, K.; Cherubini, A.; Bandinelli, S.; Andres-Lacueva, C. Resveratrol Levels and All-Cause Mortality in Older Community-Dwelling Adults. JAMA Intern. Med. 2014, 174, 1077. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.Y.; Hudson, T.S.; Wang, T.-C.; Remsberg, C.M.; Davies, N.M.; Takahashi, Y.; Kim, Y.S.; Seifried, H.; Vinyard, B.T.; Perkins, S.N.; et al. Differential Effects of Resveratrol on Androgen-Responsive LNCaP Human Prostate Cancer Cells in vitro and in vivo. Carcinogenesis 2008, 29, 2001–2010. [Google Scholar] [CrossRef]
- Papoutsis, A.J.; Lamore, S.D.; Wondrak, G.T.; Selmin, O.I.; Romagnolo, D.F. Resveratrol Prevents Epigenetic Silencing of BRCA-1 by the Aromatic Hydrocarbon Receptor in Human Breast Cancer Cells. J. Nutr. 2010, 140, 1607–1614. [Google Scholar] [CrossRef]
- Chatterjee, B.; Ghosh, K.; Kanade, S.R. Resveratrol Modulates Epigenetic Regulators of Promoter Histone Methylation and Acetylation That Restores BRCA1, P53, P21 CIP1 in Human Breast Cancer Cell Lines. BioFactors 2019, 45, 818–829. [Google Scholar] [CrossRef]
- Fu, X.; Li, M.; Tang, C.; Huang, Z.; Najafi, M. Targeting of Cancer Cell Death Mechanisms by Resveratrol: A Review. Apoptosis 2021, 26, 561–573. [Google Scholar] [CrossRef]
- Stefanska, B.; Salamé, P.; Bednarek, A.; Fabianowska-Majewska, K. Comparative Effects of Retinoic Acid, Vitamin D and Resveratrol Alone and in Combination with Adenosine Analogues on Methylation and Expression of Phosphatase and Tensin Homologue Tumour Suppressor Gene in Breast Cancer Cells. Br. J. Nutr. 2012, 107, 781–790. [Google Scholar] [CrossRef]
- Qin, W.; Zhang, K.; Clarke, K.; Weiland, T.; Sauter, E.R. Methylation and MiRNA Effects of Resveratrol on Mammary Tumors vs. Normal Tissue. Nutr. Cancer 2014, 66, 270–277. [Google Scholar] [CrossRef]
- Ahmadirad, H.; Hajizadeh, M.R.H.; Mahmoodi, M.; Mirzaee, M.; Shahrbabaki, F.M.; Soltaninejad, M. Effect of Resveratrol the Expression of Some Genes Involved Epigenetic in Breast Cancer Cell Lines (MCF-7, MDA-MB-453). SSU J. 2017, 25, 526–536. [Google Scholar]
- Liu, C.; Yen, H.; Tsao, C.-W.; Su, H.-Y.; Lin, Y.-W. Resveratrol Inhibits Ovarian Cancer Cell Growth through Epigenetic Regulation of Wnt Antogonist SFRP5. FASEB J. 2017, 31, 790.10. [Google Scholar] [CrossRef]
- Izquierdo-Torres, E.; Hernández-Oliveras, A.; Meneses-Morales, I.; Rodríguez, G.; Fuentes-García, G.; Zarain-Herzberg, Á. Resveratrol Up-Regulates ATP2A3 Gene Expression in Breast Cancer Cell Lines through Epigenetic Mechanisms. Int. J. Biochem. Cell Biol. 2019, 113, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, H.; Wu, M.-L.; Wu, J.; Sun, Y.; Zhang, K.-L.; Liu, J. Resveratrol Reverses Retinoic Acid Resistance of Anaplastic Thyroid Cancer Cells via Demethylating CRABP2 Gene. Front. Endocrinol. 2019, 10, 734. [Google Scholar] [CrossRef] [PubMed]
- Beetch, M.; Lubecka, K.; Shen, K.; Flower, K.; Harandi-Zadeh, S.; Suderman, M.; Flanagan, J.M.; Stefanska, B. Stilbenoid-Mediated Epigenetic Activation of Semaphorin 3A in Breast Cancer Cells Involves Changes in Dynamic Interactions of DNA with DNMT3A and NF1C Transcription Factor. Mol. Nutr. Food Res. 2019, 63, 1801386. [Google Scholar] [CrossRef]
- Almeida, T.C.; Guerra, C.C.C.; De Assis, B.L.G.; Oliveira Aguiar Soares, R.D.; Garcia, C.C.M.; Lima, A.A.; Silva, G.N. Antiproliferative and Toxicogenomic Effects of Resveratrol in Bladder Cancer Cells with Different TP53 Status. Environ. Mol. Mutagen. 2019, 60, 740–751. [Google Scholar] [CrossRef]
- Kaufman-Szymczyk, A.; Majda, K.; Szulawska-Mroczek, A.; Fabianowska-Majewska, K.; Lubecka, K. Clofarabine-Phytochemical Combination Exposures in CML Cells Inhibit DNA Methylation Machinery, Upregulate Tumor Suppressor Genes and Promote Caspase-Dependent Apoptosis. Mol. Med. Rep. 2019, 20, 3597–3608. [Google Scholar] [CrossRef]
- Tili, E.; Michaille, J.-J.; Alder, H.; Volinia, S.; Delmas, D.; Latruffe, N.; Croce, C.M. Resveratrol Modulates the Levels of MicroRNAs Targeting Genes Encoding Tumor-Suppressors and Effectors of TGFβ Signaling Pathway in SW480 Cells. Biochem. Pharmacol. 2010, 80, 2057–2065. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Hsiao, W.-C.; Wright, D.E.; Hsu, C.-L.; Lo, Y.-C.; Wang Hsu, G.-S.; Kao, C.-F. Resveratrol Activates the Histone H2B Ubiquitin Ligase, RNF20, in MDA-MB-231 Breast Cancer Cells. J. Funct. Foods 2013, 5, 790–800. [Google Scholar] [CrossRef]
- Stefanska, B.; Rudnicka, K.; Bednarek, A.; Fabianowska-Majewska, K. Hypomethylation and Induction of Retinoic Acid Receptor Beta 2 by Concurrent Action of Adenosine Analogues and Natural Compounds in Breast Cancer Cells. Eur. J. Pharmacol. 2010, 638, 47–53. [Google Scholar] [CrossRef]
- Ryu, J.; Yoon, N.A.; Seong, H.; Jeong, J.Y.; Kang, S.; Park, N.; Choi, J.; Lee, D.H.; Roh, G.S.; Kim, H.J.; et al. Resveratrol Induces Glioma Cell Apoptosis through Activation of Tristetraprolin. Mol. Cells 2015, 38, 991–997. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Lin, C.-W.; Yu, C.-C.; Wang, B.-Y.; Huang, Y.-H.; Hsieh, Y.-C.; Kuo, Y.-L.; Chang, W.-W. Resveratrol Suppresses Myofibroblast Activity of Human Buccal Mucosal Fibroblasts through the Epigenetic Inhibition of ZEB1 Expression. Oncotarget 2016, 7, 12137–12149. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ying, Y.; Zhu, H.; Zhu, T.; Qu, C.; Jiang, J.; Fang, B. Curcumin-induced Promoter Hypermethylation of the Mammalian Target of Rapamycin Gene in Multiple Myeloma Cells. Oncol. Lett. 2018, 17, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Parashar, G.; Capalash, N. Promoter Methylation-Independent Reactivation of PAX1 by Curcumin and Resveratrol Is Mediated by UHRF1. Clin. Exp. Med. 2016, 16, 471–478. [Google Scholar] [CrossRef]
- Medina-Aguilar, R.; Pérez-Plasencia, C.; Marchat, L.A.; Gariglio, P.; García Mena, J.; Rodríguez Cuevas, S.; Ruíz-García, E.; Astudillo-de la Vega, H.; Hernández Juárez, J.; Flores-Pérez, A.; et al. Methylation Landscape of Human Breast Cancer Cells in Response to Dietary Compound Resveratrol. PLoS ONE 2016, 11, e0157866. [Google Scholar] [CrossRef]
- Lubecka, K.; Kurzava, L.; Flower, K.; Buvala, H.; Zhang, H.; Teegarden, D.; Camarillo, I.; Suderman, M.; Kuang, S.; Andrisani, O.; et al. Stilbenoids Remodel the DNA Methylation Patterns in Breast Cancer Cells and Inhibit Oncogenic NOTCH Signaling through Epigenetic Regulation of MAML2 Transcriptional Activity. Carcinogenesis 2016, 37, 656–668. [Google Scholar] [CrossRef]
- Fudhaili, A.; Yoon, N.; Kang, S.; Ryu, J.; Jeong, J.; Lee, D.; Kang, S. Resveratrol Epigenetically Regulates the Expression of Zinc finger Protein 36 in Non-small Cell Lung Cancer Cell Lines. Oncol. Rep. 2018, 41, 1377–1386. [Google Scholar] [CrossRef]
- Ji, S.; Zheng, Z.; Liu, S.; Ren, G.; Gao, J.; Zhang, Y.; Li, G. Resveratrol Promotes Oxidative Stress to Drive DLC1 Mediated Cellular Senescence in Cancer Cells. Exp. Cell Res. 2018, 370, 292–302. [Google Scholar] [CrossRef]
- Lee, H.; Zhang, P.; Herrmann, A.; Yang, C.; Xin, H.; Wang, Z.; Hoon, D.S.B.; Forman, S.J.; Jove, R.; Riggs, A.D.; et al. Acetylated STAT3 Is Crucial for Methylation of Tumor-Suppressor Gene Promoters and Inhibition by Resveratrol Results in Demethylation. Proc. Natl. Acad. Sci. USA 2012, 109, 7765–7769. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Guo, J.; Fan, Y.; Ma, Z.; Lu, Z.; Zhang, C.; Zhao, H.; Bie, X. Module and Individual Domain Deletions of NRPS to Produce Plipastatin Derivatives in Bacillus Subtilis. Microb. Cell Factories 2018, 17, 1–13. [Google Scholar] [CrossRef]
- Kala, R.; Shah, H.N.; Martin, S.L.; Tollefsbol, T.O. Epigenetic-Based Combinatorial Resveratrol and Pterostilbene Alters DNA Damage Response by Affecting SIRT1 and DNMT Enzyme Expression, Including SIRT1-Dependent γ-H2AX and Telomerase Regulation in Triple-Negative Breast Cancer. BMC Cancer 2015, 15, 672. [Google Scholar] [CrossRef]
- Lee, M.-J.; Tsai, Y.-J.; Lin, M.-Y.; You, H.-L.; Kalyanam, N.; Ho, C.-T.; Pan, M.-H. Calebin-A Induced Death of Malignant Peripheral Nerve Sheath Tumor Cells by Activation of Histone Acetyltransferase. Phytomedicine 2019, 57, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, Y.; Cui, G.; Zhou, J. Curcumin, a Potent Anti-Tumor Reagent, Is a Novel Histone Deacetylase Inhibitor Regulating B-NHL Cell Line Raji Proliferation. Acta Pharmacol. Sin. 2005, 26, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shu, W.; Chen, W.; Wu, Q.; Liu, H.; Cui, G. Curcumin, Both Histone Deacetylase and P300/CBP-Specific Inhibitor, Represses the Activity of Nuclear Factor Kappa B and Notch 1 in Raji Cells. Basic Clin. Pharmacol. Toxicol. 2007, 101, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Krauthauser, C.; Maduskuie, V.; Fawcett, P.T.; Olson, J.M.; Rajasekaran, S.A. Curcumin-Induced HDAC Inhibition and Attenuation of Medulloblastoma Growth in vitro and in vivo. BMC Cancer 2011, 11, 144. [Google Scholar] [CrossRef] [PubMed]
- Shu, L.; Khor, T.O.; Lee, J.-H.; Boyanapalli, S.S.S.; Huang, Y.; Wu, T.-Y.; Saw, C.L.-L.; Cheung, K.-L.; Kong, A.-N.T. Epigenetic CpG Demethylation of the Promoter and Reactivation of the Expression of Neurog1 by Curcumin in Prostate LNCaP Cells. AAPS J. 2011, 13, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Chen, J.; Shi, Y.; Jia, J.; Zhang, Y. Curcumin-Induced Histone Hypoacetylation: The Role of Reactive Oxygen Species. Biochem. Pharmacol. 2005, 69, 1205–1213. [Google Scholar] [CrossRef]
- Balasubramanyam, K.; Varier, R.A.; Altaf, M.; Swaminathan, V.; Siddappa, N.B.; Ranga, U.; Kundu, T.K. Curcumin, a Novel P300/CREB-Binding Protein-Specific Inhibitor of Acetyltransferase, Represses the Acetylation of Histone/Nonhistone Proteins and Histone Acetyltransferase-Dependent Chromatin Transcription. J. Biol. Chem. 2004, 279, 51163–51171. [Google Scholar] [CrossRef]
- Sanaei, M.; Kavoosi, F.; Arabloo, M. Effect of Curcumin in Comparison with Trichostatin A on the Reactivation of Estrogen Receptor Alpha Gene Expression, Cell Growth Inhibition and Apoptosis Induction in Hepatocellular Carcinoma Hepa 1-6 Cell LLine. Asian Pac. J. Cancer Prev. 2020, 21, 1045–1050. [Google Scholar] [CrossRef]
- Tong, R.; Wu, X.; Liu, Y.; Liu, Y.; Zhou, J.; Jiang, X.; Zhang, L.; He, X.; Ma, L. Curcumin-Induced DNA Demethylation in Human Gastric Cancer Cells Is Mediated by the DNA-Damage Response Pathway. Oxid. Med. Cell. Longev. 2020, 2020, 1–13. [Google Scholar] [CrossRef]
- Sun, M.; Estrov, Z.; Ji, Y.; Coombes, K.R.; Harris, D.H.; Kurzrock, R. Curcumin (Diferuloylmethane) Alters the Expression Profiles of MicroRNAs in Human Pancreatic Cancer Cells. Mol. Cancer Ther. 2008, 7, 464–473. [Google Scholar] [CrossRef]
- Mudduluru, G.; George-William, J.N.; Muppala, S.; Asangani, I.A.; Kumarswamy, R.; Nelson, L.D.; Allgayer, H. Curcumin Regulates MiR-21 Expression and Inhibits Invasion and Metastasis in Colorectal Cancer. Biosci. Rep. 2011, 31, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.; Arora, S.; Majid, S.; Shahryari, V.; Chen, Y.; Deng, G.; Yamamura, S.; Ueno, K.; Dahiya, R. Curcumin Modulates MicroRNA-203–Mediated Regulation of the Src-Akt Axis in Bladder Cancer. Cancer Prev. Res. 2011, 4, 1698–1709. [Google Scholar] [CrossRef]
- Du, L.; Xie, Z.; Wu, L.; Chiu, M.; Lin, J.; Chan, K.K.; Liu, S.; Liu, Z. Reactivation of RASSF1A in Breast Cancer Cells by Curcumin. Nutr. Cancer 2012, 64, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xie, Z.; Jones, W.; Pavlovicz, R.E.; Liu, S.; Yu, J.; Li, P.; Lin, J.; Fuchs, J.R.; Marcucci, G.; et al. Curcumin Is a Potent DNA Hypomethylation Agent. Bioorg. Med. Chem. Lett. 2009, 19, 706–709. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Frank, D.; Patnana, P.K.; Schütte, J.; Al-Matary, Y.; Liu, L.; Wei, L.; Dugas, M.; Varghese, J.; Nimmagadda, S.C.; et al. Curcumin as an Epigenetic Therapeutic Agent in Myelodysplastic Syndromes (MDS). Int. J. Mol. Sci. 2022, 23, 411. [Google Scholar] [CrossRef] [PubMed]
- Link, A.; Balaguer, F.; Shen, Y.; Lozano, J.J.; Leung, H.-C.E.; Boland, C.R.; Goel, A. Curcumin Modulates DNA Methylation in Colorectal Cancer Cells. PLoS ONE 2013, 8, e57709. [Google Scholar] [CrossRef]
- Abusnina, A.; Keravis, T.; Yougbaré, I.; Bronner, C.; Lugnier, C. Anti-Proliferative Effect of Curcumin on Melanoma Cells Is Mediated by PDE1A Inhibition That Regulates the Epigenetic Integrator UHRF1. Mol. Nutr. Food Res. 2011, 55, 1677–1689. [Google Scholar] [CrossRef]
- Yu, J.; Peng, Y.; Wu, L.-C.; Xie, Z.; Deng, Y.; Hughes, T.; He, S.; Mo, X.; Chiu, M.; Wang, Q.-E.; et al. Curcumin Down-Regulates DNA Methyltransferase 1 and Plays an Anti-Leukemic Role in Acute Myeloid Leukemia. PLoS ONE 2013, 8, e55934. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, C.; Lin, Z.; Guo, Y.; Shi, L.; Dong, P.; Lu, Z.; Gao, S.; Liao, Y.; Chen, B.; et al. Curcumin Up-Regulates Phosphatase and Tensin Homologue Deleted on Chromosome 10 through MicroRNA-Mediated Control of DNA Methylation—A Novel Mechanism Suppressing Liver Fibrosis. FEBS J. 2014, 281, 88–103. [Google Scholar] [CrossRef]
- Jiang, A.; Wang, X.; Shan, X.; Li, Y.; Wang, P.; Jiang, P.; Feng, Q. Curcumin Reactivates Silenced Tumor Suppressor Gene RARβ by Reducing DNA Methylation: Curcumin Reactivates RARβ by Reducing DNA Methylation. Phytother. Res. 2015, 29, 1237–1245. [Google Scholar] [CrossRef]
- Zamani, M.; Sadeghizadeh, M.; Behmanesh, M.; Najafi, F. Dendrosomal Curcumin Increases Expression of the Long Non-Coding RNA Gene MEG3 via up-Regulation of Epi-MiRs in Hepatocellular Cancer. Phytomedicine 2015, 22, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.-H.; Xu, Z.-L.; Dong, D.; He, S.-A.; Yu, H. Protective Effect of Anthocyanins Extract from Blueberry on TNBS-Induced IBD Model of Mice. Evid. Based Complement. Alternat. Med. 2011, 2011, 525462. [Google Scholar] [CrossRef]
- Sanaei, M.; Kavoosi, F. Effect of Curcumin and Trichostatin A on the Expression of DNA Methyltransfrase 1 in Hepatocellular Carcinoma Cell Line Hepa. Iran. J. Ped. Hemat. Oncol. 2018, 8, 10. [Google Scholar]
- Hua, W.-F.; Fu, Y.-S.; Liao, Y.-J.; Xia, W.-J.; Chen, Y.-C.; Zeng, Y.-X.; Kung, H.-F.; Xie, D. Curcumin Induces Down-Regulation of EZH2 Expression through the MAPK Pathway in MDA-MB-435 Human Breast Cancer Cells. Eur. J. Pharmacol. 2010, 637, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Khor, T.O.; Huang, Y.; Wu, T.-Y.; Shu, L.; Lee, J.; Kong, A.-N.T. Pharmacodynamics of Curcumin as DNA Hypomethylation Agent in Restoring the Expression of Nrf2 via Promoter CpGs Demethylation. Biochem. Pharmacol. 2011, 82, 1073–1078. [Google Scholar] [CrossRef]
- Guo, Y.; Shu, L.; Zhang, C.; Su, Z.-Y.; Kong, A.-N.T. Curcumin Inhibits Anchorage-Independent Growth of HT29 Human Colon Cancer Cells by Targeting Epigenetic Restoration of the Tumor Suppressor Gene DLEC1. Biochem. Pharmacol. 2015, 94, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Kumar, U.; Sharma, U.; Rathi, G. Reversal of Hypermethylation and Reactivation of Glutathione S -Transferase Pi 1 Gene by Curcumin in Breast Cancer Cell Line. Tumor Biol. 2017, 39, 101042831769225. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Huang, E.; Barringer, S.A.; Yousef, A.E. Factors Affecting Alicyclobacillus Acidoterrestris Growth and Guaiacol Production and Controlling Apple Juice Spoilage by Lauric Arginate and ϵ-Polylysine. LWT 2020, 119, 108883. [Google Scholar] [CrossRef]
- Kaleem, M.; Perwaiz, M.; Nur, S.M.; Abdulrahman, A.O.; Ahmad, W.; Al-Abbasi, F.A.; Kumar, V.; Kamal, M.A.; Anwar, F. Epigenetics of Triple-Negative Breast Cancer via Natural Compounds. Curr. Med. Chem. 2021. [Google Scholar] [CrossRef]
- El Omari, N.; Bakha, M.; Imtara, H.; Guaouguaoua, F.-E.; Balahbib, A.; Zengin, G.; Bouyahya, A. Anticancer Mechanisms of Phytochemical Compounds: Focusing on Epigenetic Targets. Environ. Sci. Pollut. Res. 2021, 28, 47869–47903. [Google Scholar] [CrossRef]
- Liu, C.; Yen, H.; Tsao, C.; Kuo, C.; Lin, Y. The Regulation of Curcumin on Cacinogenesis and Modulation through Wnt/β-Catenin Signaling in Ovarian Cancer Cell Line. FASEB J. 2018, 32, 787–789. [Google Scholar] [CrossRef]
- Yen, H.-Y.; Tsao, C.-W.; Lin, Y.-W.; Kuo, C.-C.; Tsao, C.-H.; Liu, C.-Y. Regulation of Carcinogenesis and Modulation through Wnt/β-Catenin Signaling by Curcumin in an Ovarian Cancer Cell Line. Sci. Rep. 2019, 9, 17267. [Google Scholar] [CrossRef] [PubMed]
- Al-Yousef, N.; Shinwari, Z.; Al-Shahrani, B.; Al-Showimi, M.; Al-Moghrabi, N. Curcumin Induces Re-Expression of BRCA1 and Suppression of γ Synuclein by Modulating DNA Promoter Methylation in Breast Cancer Cell Lines. Oncol. Rep. 2020, 43, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Rajashekaraiah, R.; Kumar, P.R.; Prakash, N.; Rao, G.S.; Devi, V.R.; Metta, M.; Narayanaswamy, H.D.; Swamy, M.N.; Satyanarayan, K.; Rao, S.; et al. Anticancer Efficacy of 6-Thioguanine Loaded Chitosan Nanoparticles with or without Curcumin. Int. J. Biol. Macromol. 2020, 148, 704–714. [Google Scholar] [CrossRef]
- Wang, R.-H.; Zheng, Y.; Kim, H.-S.; Xu, X.; Cao, L.; Lahusen, T.; Lee, M.-H.; Xiao, C.; Vassilopoulos, A.; Chen, W.; et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-Associated Tumorigenesis. Mol. Cell 2008, 32, 11–20. [Google Scholar] [CrossRef]
- Chen, X.; Li, W.; Xu, C.; Wang, J.; Zhu, B.; Huang, Q.; Chen, D.; Sheng, J.; Zou, Y.; Lee, Y.M.; et al. Comparative Profiling of Analog Targets: A Case Study on Resveratrol for Mouse Melanoma Metastasis Suppression. Theranostics 2018, 8, 3504–3516. [Google Scholar] [CrossRef]
- Gao, Y.; Tollefsbol, T. Combinational Proanthocyanidins and Resveratrol Synergistically Inhibit Human Breast Cancer Cells and Impact Epigenetic–Mediating Machinery. Int. J. Mol. Sci. 2018, 19, 2204. [Google Scholar] [CrossRef]
- Liu, Z. Abstract 2014: Modulation of DNA Methyltransferases in Leukemia Cells by Curcumin and Its Associated Anti-Leukemia Activities |Cancer Research. Available online: https://cancerres.aacrjournals.org/content/71/8_Supplement/2014.short (accessed on 21 June 2021).
- Lewinska, A.; Adamczyk, J.; Pajak, J.; Stoklosa, S.; Kubis, B.; Pastuszek, P.; Slota, E.; Wnuk, M. Curcumin-Mediated Decrease in the Expression of Nucleolar Organizer Regions in Cervical Cancer (HeLa) Cells. Mutat. Res. Toxicol. Environ. Mutagen. 2014, 771, 43–52. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, J.; Hu, Y.; Wang, J.; Yuan, C. Curcumin Inhibits Growth of Human Breast Cancer Cells through Demethylation of DLC1 Promoter. Mol. Cell. Biochem. 2017, 425, 47–58. [Google Scholar] [CrossRef]
- Wu, H.; Zhou, J.; Zeng, C.; Wu, D.; Mu, Z.; Chen, B.; Xie, Y.; Ye, Y.; Liu, J. Curcumin Increases Exosomal TCF21 Thus Suppressing Exosome-Induced Lung Cancer. Oncotarget 2016, 7, 87081–87090. [Google Scholar] [CrossRef]
- Wu, P.; Huang, R.; Xiong, Y.-L.; Wu, C. Protective Effects of Curcumin against Liver Fibrosis through Modulating DNA Methylation. Chin. J. Nat. Med. 2016, 14, 255–264. [Google Scholar] [CrossRef]
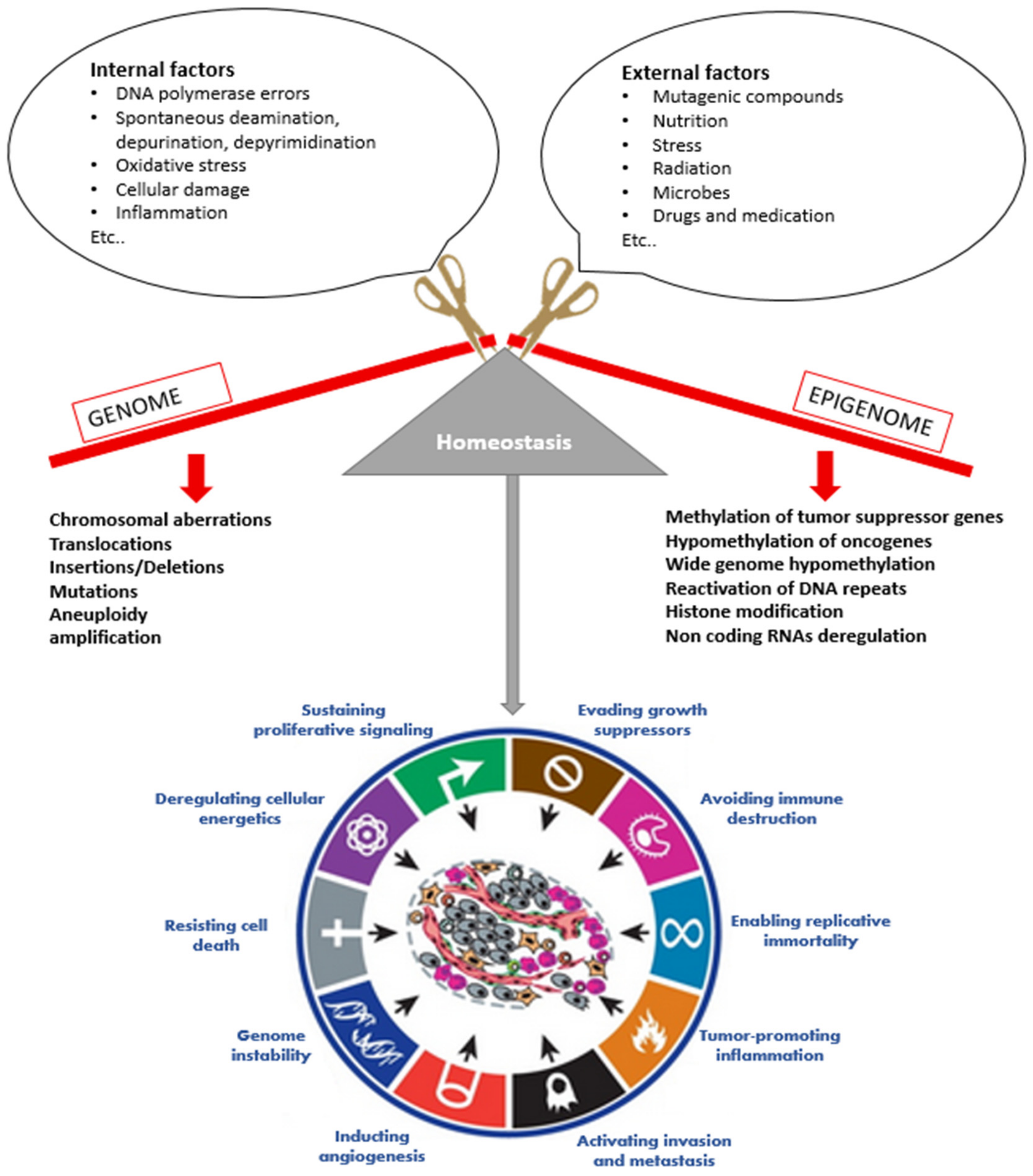

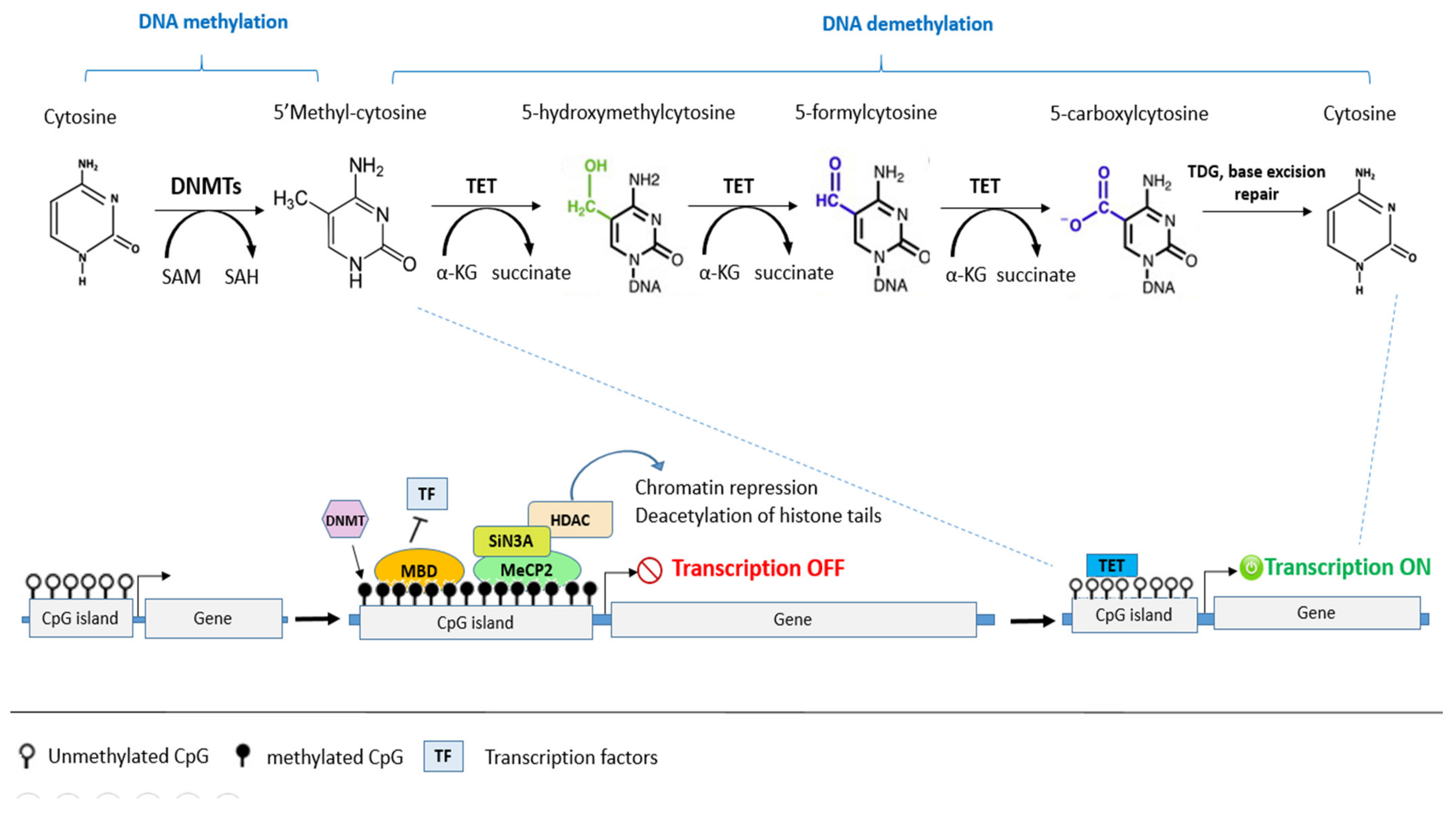
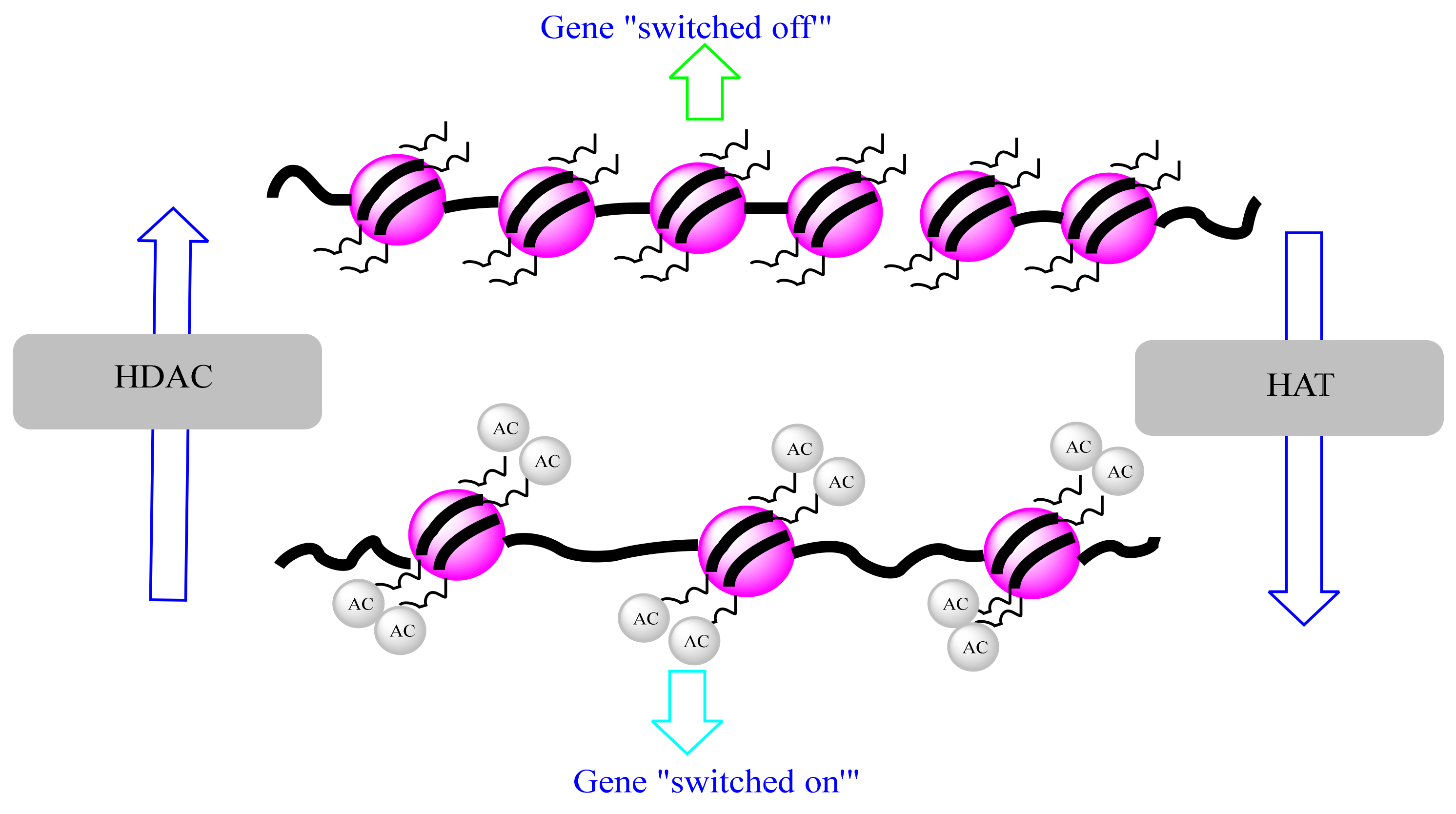
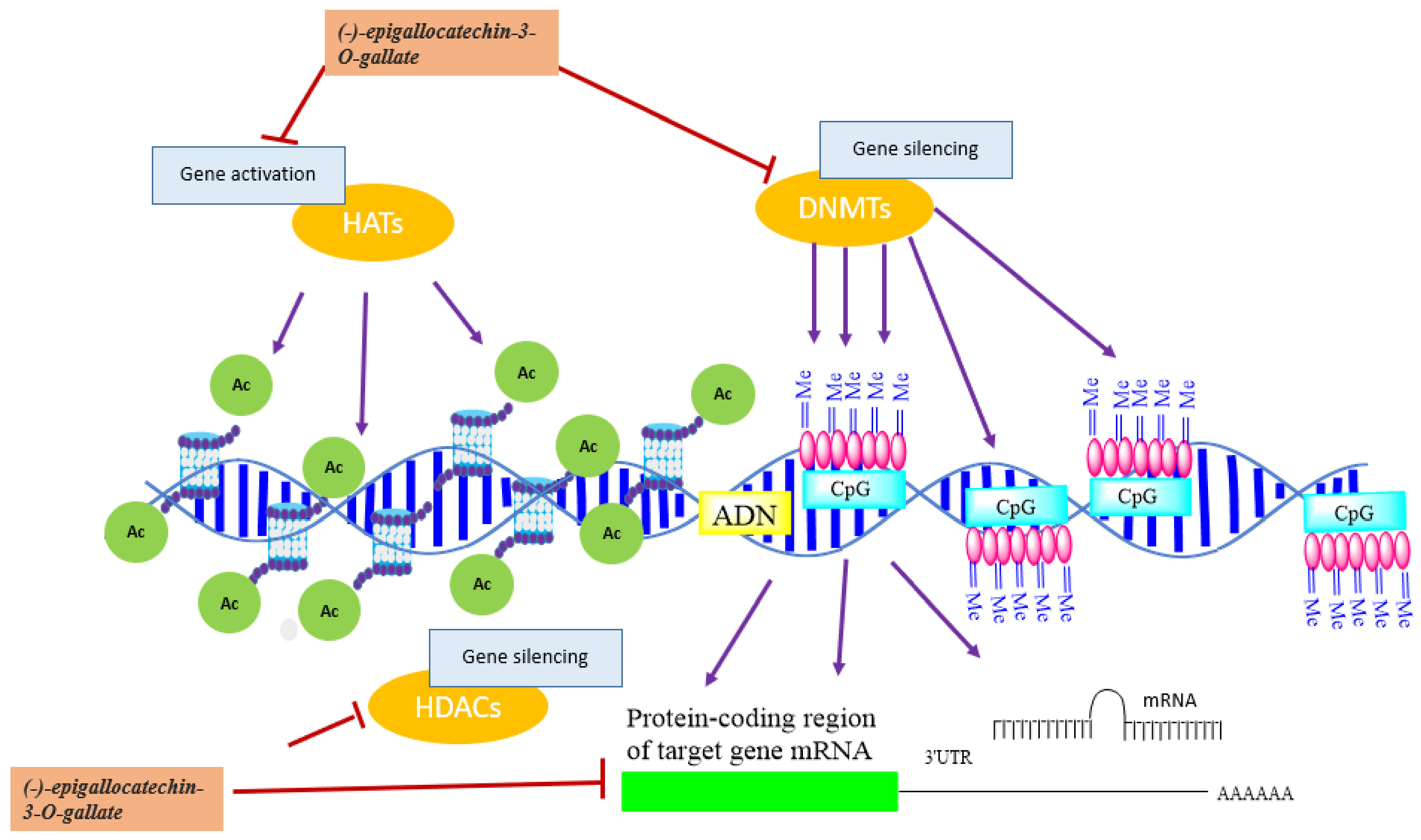

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouyahya, A.; Mechchate, H.; Oumeslakht, L.; Zeouk, I.; Aboulaghras, S.; Balahbib, A.; Zengin, G.; Kamal, M.A.; Gallo, M.; Montesano, D.; et al. The Role of Epigenetic Modifications in Human Cancers and the Use of Natural Compounds as Epidrugs: Mechanistic Pathways and Pharmacodynamic Actions. Biomolecules 2022, 12, 367. https://doi.org/10.3390/biom12030367
Bouyahya A, Mechchate H, Oumeslakht L, Zeouk I, Aboulaghras S, Balahbib A, Zengin G, Kamal MA, Gallo M, Montesano D, et al. The Role of Epigenetic Modifications in Human Cancers and the Use of Natural Compounds as Epidrugs: Mechanistic Pathways and Pharmacodynamic Actions. Biomolecules. 2022; 12(3):367. https://doi.org/10.3390/biom12030367
Chicago/Turabian StyleBouyahya, Abdelhakim, Hamza Mechchate, Loubna Oumeslakht, Ikrame Zeouk, Sara Aboulaghras, Abdelaali Balahbib, Gokhan Zengin, Mohammad Amjad Kamal, Monica Gallo, Domenico Montesano, and et al. 2022. "The Role of Epigenetic Modifications in Human Cancers and the Use of Natural Compounds as Epidrugs: Mechanistic Pathways and Pharmacodynamic Actions" Biomolecules 12, no. 3: 367. https://doi.org/10.3390/biom12030367
APA StyleBouyahya, A., Mechchate, H., Oumeslakht, L., Zeouk, I., Aboulaghras, S., Balahbib, A., Zengin, G., Kamal, M. A., Gallo, M., Montesano, D., & El Omari, N. (2022). The Role of Epigenetic Modifications in Human Cancers and the Use of Natural Compounds as Epidrugs: Mechanistic Pathways and Pharmacodynamic Actions. Biomolecules, 12(3), 367. https://doi.org/10.3390/biom12030367











