A Systematic Review of Common and Brain-Disease-Specific RNA Editing Alterations Providing Novel Insights into Neurological and Neurodegenerative Disease Manifestations
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Search Strategy
2.2. Eligibility Criteria
2.3. Statistical Analysis and Data Extraction
2.4. Registration for Studies
3. Results
4. Discussion
4.1. The Diagnostic/Prognostic Potential of RNA Editing in Neurological/Neurodegenerative and Psychiatric Disorders
4.2. Future Perspectives and Challenges in the RNA Editing Research Field
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Farajollahi, S.; Maas, S. Molecular diversity through RNA editing: A balancing act. Trends Genet. 2010, 26, 221–230. [Google Scholar] [CrossRef]
- Licht, K.; Jantsch, M.F. Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J. Cell Biol. 2016, 213, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Huntley, M.A.; Lou, M.; Goldstein, L.D.; Lawrence, M.; Dijkgraaf, G.J.P.; Kaminker, J.S.; Gentleman, R. Complex regulation of ADAR-mediated RNA-editing across tissues. BMC Genom. 2016, 17, 61. [Google Scholar] [CrossRef] [PubMed]
- Daniel, C.; Lagergren, J.; Öhman, M. RNA editing of non-coding RNA and its role in gene regulation. Biochimie 2015, 117, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Deffit, S.N.; Hundley, H.A. To edit or not to edit: Regulation of ADAR editing specificity and efficiency. Wiley Interdiscip. Rev. RNA 2016, 7, 113–127. [Google Scholar] [CrossRef]
- Blanc, V.; Davidson, N.O. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev. Syst. Biol. Med. 2010, 2, 594–602. [Google Scholar] [CrossRef]
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Kannisto, E.D.; Enriquez, S.M.; Gollnick, P.; Baysal, B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 2015, 6, 6881. [Google Scholar] [CrossRef]
- Sharma, S.; Wang, J.; Alqassim, E.; Portwood, S.; Gomez, E.C.; Maguire, O.; Basse, P.H.; Wang, E.S.; Segal, B.H.; Baysal, B.E. Mitochondrial hypoxic stress induces widespread RNA editing by APOBEC3G in natural killer cells. Genome Biol. 2019, 20, 37. [Google Scholar] [CrossRef]
- Nevo-Caspi, Y.; Amariglio, N.; Rechavi, G.; Paret, G. A-to-I RNA Editing is Induced upon Hypoxia. Shock 2011, 35, 585–589. [Google Scholar] [CrossRef]
- Wedekind, J.E.; Dance, G.S.; Sowden, M.; Smith, H.C. Messenger RNA editing in mammals: New members of the APOBEC family seeking roles in the family business. Trends Genet. 2003, 19, 207–216. [Google Scholar] [CrossRef]
- Roberts, S.A.; Lawrence, M.S.; Klimczak, L.J.; Grimm, S.A.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 970–976. [Google Scholar] [CrossRef] [PubMed]
- Rice, G.I.; Kasher, P.R.; Forte, G.M.A.; Mannion, N.M.; Greenwood, S.M.; Szynkiewicz, M.; Dickerson, J.E.; Bhaskar, S.S.; Zampini, M.; Briggs, T.A.; et al. Mutations in ADAR1 cause AGS with Type 1 IFN signature. Nat. Genet. 2012, 44, 1243–1248. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Nellaker, C.; McIntyre, R.E.; Buendia-Buendia, J.E.; Bumpstead, S.; Ponting, C.P.; Flint, J.; Durbin, R.; Keane, T.M.; Adams, D.J. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 2012, 13, 26. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Manzari, C.; Mastropasqua, F.; Aiello, I.; D’Erchia, A.M.; Pesole, G. Profiling RNA editing in human tissues: Towards the inosinome Atlas. Sci. Rep. 2015, 5, 14941. [Google Scholar] [CrossRef]
- Behm, M.; Öhman, M. RNA Editing: A Contributor to Neuronal Dynamics in the Mammalian Brain. Trends Genet. 2016, 32, 165–175. [Google Scholar] [CrossRef]
- Ekdahl, Y.; Farahani, H.S.; Behm, M.; Lagergren, J.; Öhman, M. A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res. 2012, 22, 1477–1487. [Google Scholar] [CrossRef]
- Li, J.B.; Church, G.M. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat. Neurosci. 2013, 16, 1518–1522. [Google Scholar] [CrossRef]
- Sanjana, N.E.; Levanon, E.Y.; Hueske, E.A.; Ambrose, J.M.; Li, J.B. Activity-Dependent A-to-I RNA Editing in Rat Cortical Neurons. Genetics 2012, 192, 281–287. [Google Scholar] [CrossRef]
- Ball, S.M.; Atlason, P.T.; Shittu-Balogun, O.O.; Molnár, E. Assembly and intracellular distribution of kainate receptors is determined by RNA editing and subunit composition. J. Neurochem. 2010, 114, 1805–1818. [Google Scholar] [CrossRef]
- Daniel, C.; Wahlstedt, H.; Ohlson, J.; Björk, P.; Öhman, M. Adenosine-to-Inosine RNA Editing Affects Trafficking of the γ-Aminobutyric Acid Type A (GABAA) Receptor. J. Biol. Chem. 2011, 286, 2031–2040. [Google Scholar] [CrossRef]
- Huang, H.; Tan, B.Z.; Shen, Y.; Tao, J.; Jiang, F.; Sung, Y.Y.; Ng, C.K.; Raida, M.; Köhr, G.; Higuchi, M.; et al. RNA Editing of the IQ Domain in Cav1.3 Channels Modulates Their Ca2+-Dependent Inactivation. Neuron 2012, 73, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Bazzazi, H.; Johny, M.B.; Adams, P.J.; Soong, T.W.; Yue, D.T. Continuously Tunable Ca2+ Regulation of RNA-Edited CaV1.3 Channels. Cell Rep. 2013, 5, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Irimia, M.; Denuc, A.; Ferrán, J.L.; Pernaute, B.; Puelles, L.; Roy, S.W.; Garcia-Fernàndez, J.; Marfany, G. Evolutionarily conserved A-to-I editing increases protein stability of the alternative splicing factorNova1. RNA Biol. 2012, 9, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.B.; Dredge, B.K.; Stefani, G.; Zhong, R.; Buckanovich, R.J.; Okano, H.J.; Yang, Y.Y.; Darnell, R.B. Nova-1 Regulates Neuron-Specific Alternative Splicing and Is Essential for Neuronal Viability. Neuron 2000, 25, 359–371. [Google Scholar] [CrossRef]
- Singh, M. Dysregulated A to I RNA editing and non-coding RNAs in neurodegeneration. Front. Genet. 2013, 3, 326. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 88, 105906. [Google Scholar] [CrossRef]
- Grigorenko, E.V.; Bell, W.L.; Glazier, S.; Pons, T.; Deadwyler, S. Editing status at the Q/R site of the GluR2 and GluR6 glutamate receptor subunits in the surgically excised hippocampus of patients with refractory epilepsy. Neuroreport 1998, 9, 2219–2224. [Google Scholar] [CrossRef]
- Kortenbruck, G.; Berger, E.; Speckmann, E.-J.; Musshoff, U. RNA Editing at the Q/R Site for the Glutamate Receptor Subunits GLUR2, GLUR5, and GLUR6 in Hippocampus and Temporal Cortex from Epileptic Patients. Neurobiol. Dis. 2001, 8, 459–468. [Google Scholar] [CrossRef]
- Vollmar, W.; Gloger, J.; Berger, E.; Kortenbruck, G.; Köhling, R.; Speckmann, E.-J.; Musshoff, U. RNA editing (R/G site) and flip–flop splicing of the AMPA receptor subunit GluR2 in nervous tissue of epilepsy patients. Neurobiol. Dis. 2004, 15, 371–379. [Google Scholar] [CrossRef]
- Krestel, H.; Raffel, S.; von Lehe, M.; Jagella, C.; Moskau-Hartmann, S.; Becker, A.; Elger, C.E.; Seeburg, P.H.; Nirkko, A. Differences between RNA and DNA due to RNA editing in temporal lobe epilepsy. Neurobiol. Dis. 2013, 56, 66–73. [Google Scholar] [CrossRef]
- Srivastava, P.K.; Bagnati, M.; Delahaye-Duriez, A.; Ko, J.-H.; Rotival, M.; Langley, S.R.; Shkura, K.; Mazzuferi, M.; Danis, B.; Van Eyll, J.; et al. Genome-wide analysis of differential RNA editing in epilepsy. Genome Res. 2017, 27, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Akbarian, S.; Smith, M.A.; Jones, E.G. Editing for an AMPA receptor subunit RNA in prefrontal cortex and striatum in Alzheimer’s disease, Huntington’s disease and schizophrenia. Brain Res. 1995, 699, 297–304. [Google Scholar] [CrossRef]
- Sodhi, M.S.; Burnet, P.W.J.; Makoff, A.J.; Kerwin, R.W.; Harrison, P.J. RNA editing of the 5-HT2C receptor is reduced in schizophrenia. Mol. Psychiatry 2001, 6, 373–379. [Google Scholar] [CrossRef][Green Version]
- Dracheva, S.; Elhakem, S.L.; Marcus, S.M.; Siever, L.J.; McGurk, S.R.; Haroutunian, V. RNA editing and alternative splicing of human serotonin 2C receptor in schizophrenia. J. Neurochem. 2003, 87, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Breen, M.S.; Dobbyn, A.; Li, Q.; Roussos, P.; Hoffman, G.E.; Stahl, E.; Chess, A.; Sklar, P.; Li, J.B.; Devlin, B.; et al. Global landscape and genetic regulation of RNA editing in cortical samples from individuals with schizophrenia. Nat. Neurosci. 2019, 22, 1402–1412. [Google Scholar] [CrossRef]
- Niswender, C.M.; Herrick-Davis, K.; Dilley, G.E.; Meltzer, H.Y.; Overholser, J.C.; Stockmeier, C.A.; Emeson, R.B.; Sanders-Bush, E. RNA Editing of the Human Serotonin 5-HT2C Receptor Alterations in Suicide and Implications for Serotonergic Pharmacotherapy. Neuropsychopharmacology 2001, 24, 478–491. [Google Scholar] [CrossRef]
- Lyddon, R.; Dwork, A.J.; Keddache, M.; Siever, L.J.; Dracheva, S. Serotonin 2c receptor RNA editing in major depression and suicide. World J. Biol. Psychiatry 2013, 14, 590–601. [Google Scholar] [CrossRef]
- Di Narzo, A.F.; Kozlenkov, A.; Roussos, P.; Hao, K.; Hurd, Y.; Lewis, D.A.; Sibille, E.; Siever, L.J.; Koonin, E.; Dracheva, S. A unique gene expression signature associated with serotonin 2C receptor RNA editing in the prefrontal cortex and altered in suicide. Hum. Mol. Genet. 2014, 23, 4801–4813. [Google Scholar] [CrossRef]
- Weissmann, D.; Van Der Laan, S.; Underwood, M.D.; Salvetat, N.; Cavarec, L.; Vincent, L.; Molina, F.; Mann, J.J.; Arango, V.; Pujol, J.F. Region-specific alterations of A-to-I RNA editing of serotonin 2c receptor in the cortex of suicides with major depression. Transl. Psychiatry 2016, 6, e878. [Google Scholar] [CrossRef]
- Gurevich, I.; Tamir, H.; Arango, V.; Dwork, A.J.; Mann, J.J.; Schmauss, C. Altered Editing of Serotonin 2C Receptor Pre-mRNA in the Prefrontal Cortex of Depressed Suicide Victims. Neuron 2002, 34, 349–356. [Google Scholar] [CrossRef]
- Iwamoto, K.; Kato, T. RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neurosci. Lett. 2003, 346, 169–172. [Google Scholar] [CrossRef]
- Chimienti, F.; Cavarec, L.; Vincent, L.; Salvetat, N.; Arango, V.; Underwood, M.D.; Mann, J.J.; Pujol, J.-F.; Weissmann, D. Brain region-specific alterations of RNA editing in PDE8A mRNA in suicide decedents. Transl. Psychiatry 2019, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Dick, A.L.W.; Khermesh, K.; Paul, E.; Stamp, F.; Levanon, E.Y.; Chen, A. Adenosine-to-Inosine RNA Editing within Corticolimbic Brain Regions Is Regulated in Response to Chronic Social Defeat Stress in Mice. Front. Psychiatry 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Eran, A.; Li, J.B.; Vatalaro, K.; McCarthy, J.; Rahimov, F.; Collins, C.; Markianos, K.; Margulies, D.M.; Brown, E.N.; Calvo, S.E.; et al. Comparative RNA editing in autistic and neurotypical cerebella. Mol. Psychiatry 2013, 18, 1041–1048. [Google Scholar] [CrossRef][Green Version]
- Tran, S.S.; Jun, H.-I.; Bahn, J.H.; Azghadi, A.; Ramaswami, G.; Van Nostrand, E.L.; Nguyen, T.B.; Hsiao, Y.-H.E.; Lee, C.; Pratt, G.A.; et al. Widespread RNA editing dysregulation in brains from autistic individuals. Nat. Neurosci. 2019, 22, 25–36. [Google Scholar] [CrossRef]
- Takuma, H.; Kwak, S.; Yoshizawa, T.; Kanazawa, I. Reduction of GluR2 RNA editing, a molecular change that increases calcium influx through AMPA receptors, selective in the spinal ventral gray of patients with amyotrophic lateral sclerosis. Ann. Neurol. 1999, 46, 806–815. [Google Scholar] [CrossRef]
- Kawahara, Y.; Ito, K.; Sun, H.; Aizawa, H.; Kanazawa, I.; Kwak, S. RNA editing and death of motor neurons. Nature 2004, 427, 801. [Google Scholar] [CrossRef]
- Flomen, R.; Makoff, A. Increased RNA editing in EAAT2 pre-mRNA from amyotrophic lateral sclerosis patients: Involvement of a cryptic polyadenylation site. Neurosci. Lett. 2011, 497, 139–143. [Google Scholar] [CrossRef]
- Hideyama, T.; Yamashita, T.; Aizawa, H.; Tsuji, S.; Kakita, A.; Takahashi, H.; Kwak, S. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol. Dis. 2012, 45, 1121–1128. [Google Scholar] [CrossRef]
- D’Erchia, A.M.; Gallo, A.; Manzari, C.; Raho, S.; Horner, D.S.; Chiara, M.; Valletti, A.; Aiello, I.; Mastropasqua, F.; Ciaccia, L.; et al. Massive transcriptome sequencing of human spinal cord tissues provides new insights into motor neuron degeneration in ALS. Sci. Rep. 2017, 7, 10046. [Google Scholar] [CrossRef]
- Moore, S.; Alsop, E.; Lorenzini, I.; Starr, A.; Rabichow, B.E.; Mendez, E.; Levy, J.L.; Burciu, C.; Reiman, R.; Chew, J.; et al. ADAR2 mislocalization and widespread RNA editing aberrations in C9orf72-mediated ALS/FTD. Acta Neuropathol. 2019, 138, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Gaisler-Salomon, I.; Kravitz, E.; Feiler, Y.; Safran, M.; Biegon, A.; Amariglio, N.; Rechavi, G. Hippocampus-specific deficiency in RNA editing of GluA2 in Alzheimer’s disease. Neurobiol. Aging 2014, 35, 1785–1791. [Google Scholar] [CrossRef] [PubMed]
- Khermesh, K.; D’Erchia, A.M.; Barak, M.; Annese, A.; Wachtel, C.; Levanon, E.Y.; Picardi, E.; Eisenberg, E. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease. RNA 2016, 22, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Annese, A.; Manzari, C.; Lionetti, C.; Picardi, E.; Horner, D.S.; Chiara, M.; Caratozzolo, M.F.; Tullo, A.; Fosso, B.; Pesole, G.; et al. Whole transcriptome profiling of Late-Onset Alzheimer’s Disease patients provides insights into the molecular changes involved in the disease. Sci. Rep. 2018, 8, 4282. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Dammer, E.B.; Felsky, D.; Duong, D.M.; Klein, H.-U.; White, C.C.; Zhou, M.; Logsdon, B.A.; McCabe, C.; Xu, J.; et al. Atlas of RNA editing events affecting protein expression in aged and Alzheimer’s disease human brain tissue. Nat. Commun. 2021, 12, 7035. [Google Scholar] [CrossRef]
- Kiesel, P.; Bodemer, W.; Gibson, T.; Zischler, H.; Kaup, F.-J. Prion infected rhesus monkeys to study differential transcription of Alu DNA elements and editing of Alu transcripts in neuronal cells and blood cells. J. Med. Primatol. 2012, 41, 176–182. [Google Scholar] [CrossRef]
- Kanata, E.; Llorens, F.; Dafou, D.; Dimitriadis, A.; Thüne, K.; Xanthopoulos, K.; Bekas, N.; Espinosa, J.C.; Schmitz, M.; Marín-Moreno, A.; et al. RNA editing alterations define manifestation of prion diseases. Proc. Natl. Acad. Sci. USA 2019, 116, 19727–19735. [Google Scholar] [CrossRef]
- Higuchi, M.; Maas, S.; Single, F.N.; Hartner, J.C.; Rozov, A.; Burnashev, N.; Feldmeyer, D.; Sprengel, R.; Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 2000, 406, 78–81. [Google Scholar] [CrossRef]
- Hosaka, T.; Tsuji, H.; Kwak, S. RNA Editing: A New Therapeutic Target in Amyotrophic Lateral Sclerosis and Other Neurological Diseases. Int. J. Mol. Sci. 2021, 22, 10958. [Google Scholar] [CrossRef]
- Yamashita, T.; Hideyama, T.; Hachiga, K.; Teramoto, S.; Takano, J.; Iwata, N.; Saido, T.C.; Kwak, S. A role for calpain-dependent cleavage of TDP-43 in amyotrophic lateral sclerosis pathology. Nat. Commun. 2012, 3, 1307. [Google Scholar] [CrossRef]
- Yamashita, T.; Kwak, S. The molecular link between inefficient GluA2 Q/R site-RNA editing and TDP-43 pathology in motor neurons of sporadic amyotrophic lateral sclerosis patients. Brain Res. 2014, 1584, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Aizawa, H.; Sawada, J.; Hideyama, T.; Yamashita, T.; Katayama, T.; Hasebe, N.; Kimura, T.; Yahara, O.; Kwak, S. TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol. 2010, 120, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Brusa, R.; Zimmermann, F.; Koh, D.S.; Feldmeyer, D.; Gass, P.; Seeburg, P.H.; Sprengel, R. Early-Onset Epilepsy and Postnatal Lethality Associated with an Editing-Deficient GluR-B Allele in Mice. Science 1995, 270, 1677–1680. [Google Scholar] [CrossRef] [PubMed]
- Streit, A.K.; Derst, C.; Wegner, S.; Heinemann, U.; Zahn, R.K.; Decher, N. RNA editing of Kv1.1 channels may account for reduced ictogenic potential of 4-aminopyridine in chronic epileptic rats. Epilepsia 2011, 52, 645–648. [Google Scholar] [CrossRef]
- Bernard, A.; Ferhat, L.; Dessi, F.; Charton, G.; Represa, A.; Ben-Ari, Y.; Khrestchatisky, M. Q/R editing of the rat GluR5 and GluR6 kainate receptors in vivo and in vitro: Evidence for independent developmental, pathological and cellular regulation. Eur. J. Neurosci. 1999, 11, 604–616. [Google Scholar] [CrossRef]
- Ferrick-Kiddie, E.A.; Rosenthal, J.J.C.; Ayers, G.D.; Emeson, R.B. Mutations underlying Episodic Ataxia type-1 antagonize Kv1.1 RNA editing. Sci. Rep. 2017, 7, 41095. [Google Scholar] [CrossRef]
- Ramaswami, G.; Li, J.B. RADAR: A rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014, 42, D109–D113. [Google Scholar] [CrossRef]
- Kiran, A.; Baranov, P.V. DARNED: A DAtabase of RNa EDiting in humans. Bioinformatics 2010, 26, 1772–1776. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; D’Erchia, A.M.; Lo Giudice, C.; Pesole, G. REDIportal: A comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017, 45, D750–D757. [Google Scholar] [CrossRef]
- Hwang, T.; Park, C.-K.; Leung, A.K.; Gao, Y.; Hyde, T.M.; Kleinman, J.E.; Rajpurohit, A.; Tao, R.; Shin, J.H.; Weinberger, D.R. Dynamic regulation of RNA editing in human brain development and disease. Nat. Neurosci. 2016, 19, 1093–1099. [Google Scholar] [CrossRef]
- Cole, D.C.; Chung, Y.; Gagnidze, K.; Hajdarovic, K.H.; Rayon-Estrada, V.; Harjanto, D.; Bigio, B.; Gal-Toth, J.; Milner, T.A.; McEwen, B.S.; et al. Loss of APOBEC1 RNA-editing function in microglia exacerbates age-related CNS pathophysiology. Proc. Natl. Acad. Sci. USA 2017, 114, 13272–13277. [Google Scholar] [CrossRef]
- Rayon-Estrada, V.; Harjanto, D.; Hamilton, C.E.; Berchiche, Y.A.; Gantman, E.C.; Sakmar, T.P.; Bulloch, K.; Gagnidze, K.; Harroch, S.; McEwen, B.S.; et al. Epitranscriptomic profiling across cell types reveals associations between APOBEC1-mediated RNA editing, gene expression outcomes, and cellular function. Proc. Natl. Acad. Sci. USA 2017, 114, 13296–13301. [Google Scholar] [CrossRef] [PubMed]
- Harjanto, D.; Papamarkou, T.; Oates, C.J.; Rayon-Estrada, V.; Papavasiliou, F.N.; Papavasiliou, A. RNA editing generates cellular subsets with diverse sequence within populations. Nat. Commun. 2016, 7, 12145. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, B.R.; Hamilton, C.E.; Mwangi, M.M.; Dewell, S.; Papavasiliou, F.N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 2011, 18, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Deininger, P.L. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, W.T.; Warren, S.T. A Decade of Molecular Studies of Fragile X Syndrome. Annu. Rev. Neurosci. 2002, 25, 315–338. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Gudsnuk, K.; Kuo, S.-H.; Cotrina, M.L.; Rosoklija, G.; Sosunov, A.; Sonders, M.S.; Kanter, E.; Castagna, C.; Yamamoto, A.; et al. Loss of mTOR-Dependent Macroautophagy Causes Autistic-like Synaptic Pruning Deficits. Neuron 2014, 83, 1131–1143. [Google Scholar] [CrossRef] [PubMed]
- Won, H.; Mah, W.; Kim, E. Autism spectrum disorder causes, mechanisms, and treatments: Focus on neuronal synapses. Front. Mol. Neurosci. 2013, 6, 19. [Google Scholar] [CrossRef]
- Starr, A.; Sattler, R. Synaptic dysfunction and altered excitability in C9ORF72 ALS/FTD. Brain Res. 2018, 1693, 98–108. [Google Scholar] [CrossRef]
- Schneider, J.L.; Miller, A.M.; Woesner, M.E. Autophagy and Schizophrenia: A Closer Look at How Dysregulation of Neuronal Cell Homeostasis Influences the Pathogenesis of Schizophrenia. Einstein J. Biol. Med. 2016, 31, 34–39. [Google Scholar] [CrossRef]
- Perera, R.M.; Zoncu, R. The Lysosome as a Regulatory Hub. Annu. Rev. Cell Dev. Biol. 2016, 32, 223–253. [Google Scholar] [CrossRef] [PubMed]
- Silvestris, D.A.; Picardi, E.; Cesarini, V.; Fosso, B.; Mangraviti, N.; Massimi, L.; Martini, M.; Pesole, G.; Locatelli, F.; Gallo, A. Dynamic inosinome profiles reveal novel patient stratification and gender-specific differences in glioblastoma. Genome Biol. 2019, 20, 33. [Google Scholar] [CrossRef] [PubMed]
- Giacopuzzi, E.; Gennarelli, M.; Sacco, C.; Filippini, A.; Mingardi, J.; Magri, C.; Barbon, A. Genome-wide analysis of consistently RNA edited sites in human blood reveals interactions with mRNA processing genes and suggests correlations with cell types and biological variables. BMC Genom. 2018, 19, 963. [Google Scholar] [CrossRef] [PubMed]
- Vlachogiannis, N.; Gatsiou, A.; Silvestris, D.A.; Stamatelopoulos, K.; Tektonidou, M.G.; Gallo, A.; Sfikakis, P.P.; Stellos, K. Increased adenosine-to-inosine RNA editing in rheumatoid arthritis. J. Autoimmun. 2020, 106, 102329. [Google Scholar] [CrossRef]
- van der Laan, S.; Salvetat, N.; Weissmann, D.; Molina, F. Emerging RNA editing biomarkers will foster drug development. Drug Discov. Today 2017, 22, 1056–1063. [Google Scholar] [CrossRef]
- Salvetat, N.; Chimienti, F.; Cayzac, C.; Dubuc, B.; Checa-Robles, F.; Dupre, P.; Mereuze, S.; Patel, V.; Genty, C.; Lang, J.-P.; et al. Phosphodiesterase 8A to discriminate in blood samples depressed patients and suicide attempters from healthy controls based on A-to-I RNA editing modifications. Transl. Psychiatry 2021, 11, 255. [Google Scholar] [CrossRef]
- Hosaka, T.; Yamashita, T.; Tamaoka, A.; Kwak, S. Extracellular RNAs as Biomarkers of Sporadic Amyotrophic Lateral Sclerosis and Other Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 3148. [Google Scholar] [CrossRef]
- Gardner, O.K.; Wang, L.; Van Booven, D.; Whitehead, P.L.; Hamilton-Nelson, K.L.; Adams, L.D.; Starks, T.D.; Hofmann, N.K.; Vance, J.M.; Cuccaro, M.L.; et al. RNA editing alterations in a multi-ethnic Alzheimer disease cohort converge on immune and endocytic molecular pathways. Hum. Mol. Genet. 2019, 28, 3053–3061. [Google Scholar] [CrossRef]
- Diroma, M.A.; Ciaccia, L.; Pesole, G.; Picardi, E. Elucidating the editome: Bioinformatics approaches for RNA editing detection. Brief. Bioinform. 2019, 20, 436–447. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Chen, K.; Wylie, T.; Larson, D.E.; McLellan, M.D.; Mardis, E.R.; Weinstock, G.M.; Wilson, R.K.; Ding, L. VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 2009, 25, 2283–2285. [Google Scholar] [CrossRef]
- Picardi, E.; Pesole, G. REDItools: High-throughput RNA editing detection made easy. Bioinformatics 2013, 29, 1813–1814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiao, X. Genome sequence–independent identification of RNA editing sites. Nat. Methods 2015, 12, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Lu, Y.; Yan, S.; Xing, Q.; Tian, W. SPRINT: An SNP-free toolkit for identifying RNA editing sites. Bioinformatics 2017, 33, 3538–3548. [Google Scholar] [CrossRef] [PubMed]
- Piechotta, M.; Wyler, E.; Ohler, U.; Landthaler, M.; Dieterich, C. JACUSA: Site-specific identification of RNA editing events from replicate sequencing data. BMC Bioinform. 2017, 18, 7. [Google Scholar] [CrossRef]
- John, D.; Weirick, T.; Dimmeler, S.; Uchida, S. RNAEditor: Easy detection of RNA editing events and the introduction of editing islands. Brief. Bioinform. 2017, 18, 993–1001. [Google Scholar] [CrossRef]
- Picardi, E.; Horner, D.S.; Pesole, G. Single-cell transcriptomics reveals specific RNA editing signatures in the human brain. RNA 2017, 23, 860–865. [Google Scholar] [CrossRef]
- Picardi, E.; D’Erchia, A.M.; Gallo, A.; Montalvo, A.; Pesole, G. Uncovering RNA Editing Sites in Long Non-Coding RNAs. Front. Bioeng. Biotechnol. 2014, 49, 12. [Google Scholar] [CrossRef]
- Zheng, Y.; Jinyan, L.; Song, R.; Wang, S.; Li, T.; Zhang, X.; Chen, K.; Li, T.; Li, J. Accurate detection for a wide range of mutation and editing sites of microRNAs from small RNA high-throughput sequencing profiles. Nucleic Acids Res. 2016, 44, e123. [Google Scholar] [CrossRef]
- Wu, S.; Yang, M.; Kim, P.; Zhou, X. ADeditome provides the genomic landscape of A-to-I RNA editing in Alzheimer’s disease. Brief. Bioinform. 2021, 22, bbaa384. [Google Scholar] [CrossRef]
- Schaffer, A.A.; Kopel, E.; Hendel, A.; Picardi, E.; Levanon, E.Y.; Eisenberg, E. The cell line A-to-I RNA editing catalogue. Nucleic Acids Res. 2020, 48, 5849–5858. [Google Scholar] [CrossRef]
- Merkle, T.; Merz, S.; Reautschnig, P.; Blaha, A.; Li, Q.; Vogel, P.; Wettengel, J.; Li, J.B.; Stafforst, T. Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat. Biotechnol. 2019, 37, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Yi, Z.; Zhu, S.; Wang, C.; Cao, Z.; Zhou, Z.; Yuan, P.; Yu, Y.; Tian, F.; Liu, Z.; et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 2019, 37, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Rauch, S.; He, E.; Srienc, M.; Zhou, H.; Zhang, Z.; Dickinson, B.C. Programmable RNA-Guided RNA Effector Proteins Built from Human Parts. Cell 2019, 178, 122–134.e12. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Franklin, B.; Koob, J.; Kellner, M.J.; Ladha, A.; Joung, J.; Kirchgatterer, P.; Cox, D.B.T.; Zhang, F. A cytosine deaminase for programmable single-base RNA editing. Science 2019, 365, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Lv, J.; Li, Y.; Mao, S.; Li, Z.; Jing, Z.; Sun, Y.; Zhang, X.; Shen, S.; Wang, X.; et al. Programmable C-to-U RNA editing using the human APOBEC 3A deaminase. EMBO J. 2020, 39, e104741. [Google Scholar] [CrossRef] [PubMed]
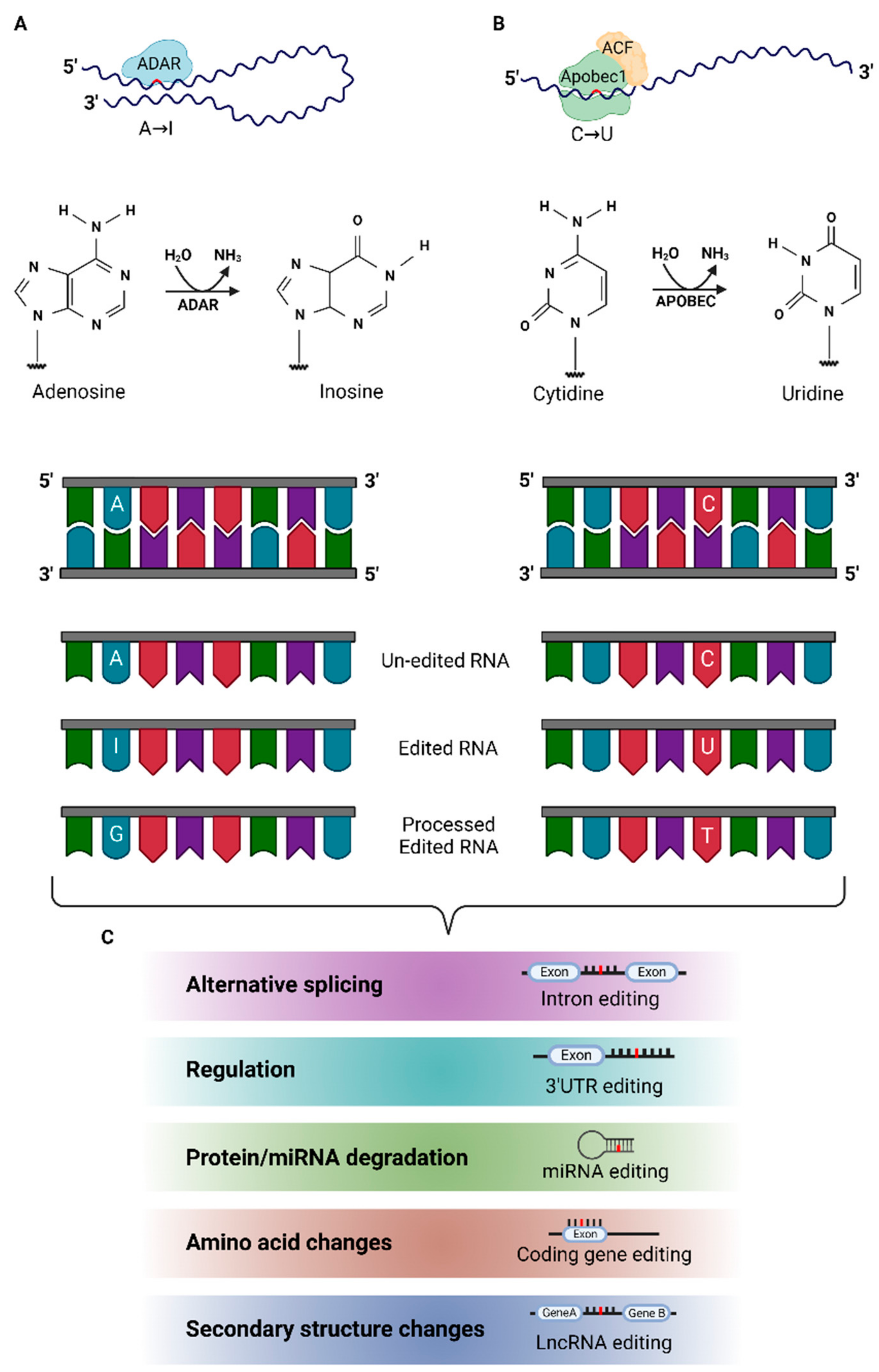

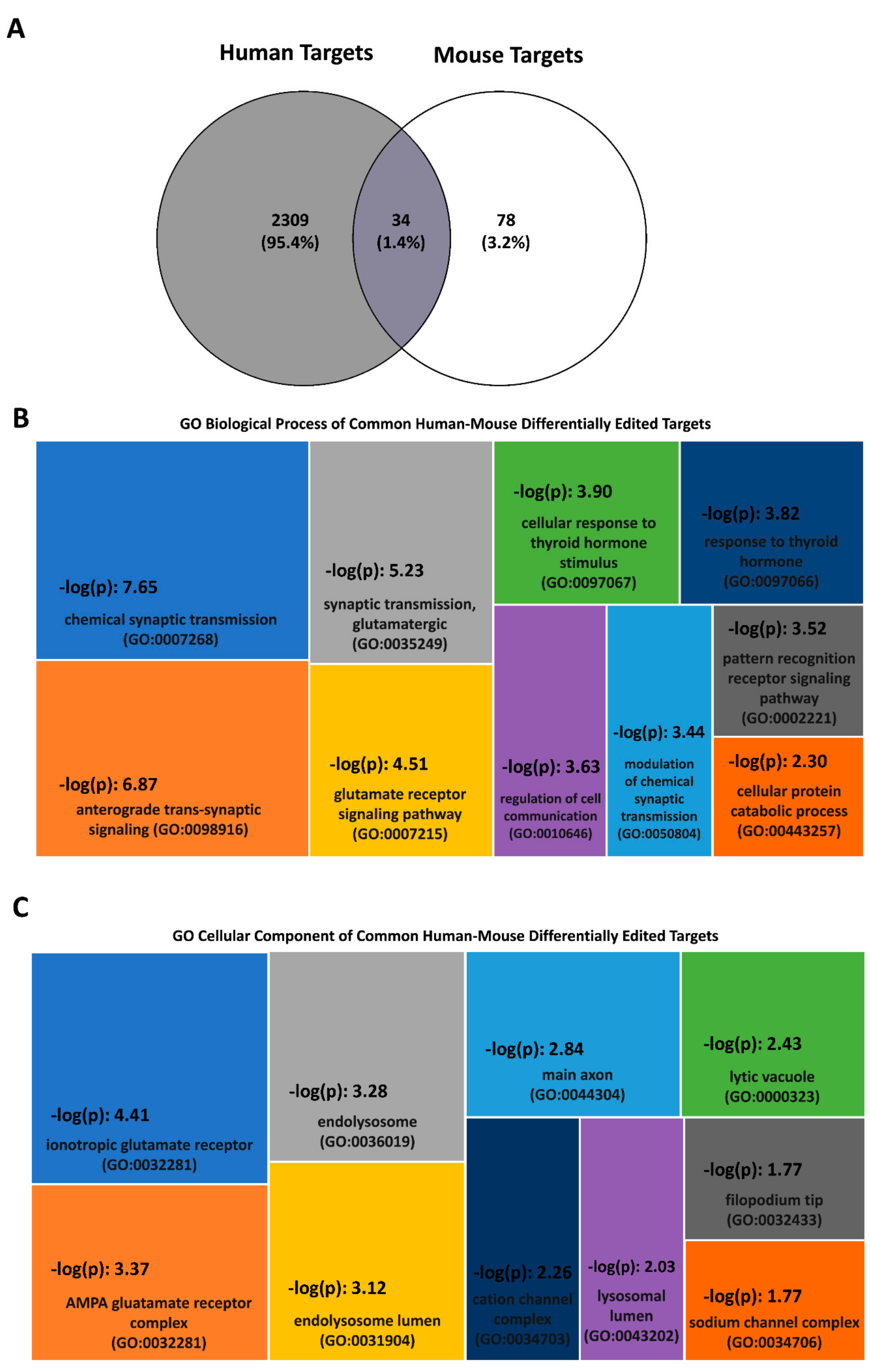
| Disorder | Species/Brain Region | Study Type/Target(s) | Methodology/Validation Method | DE Targets/Trend Relative to Controls | Remarks | Ref. |
|---|---|---|---|---|---|---|
| Epilepsy | Human/HPC | Focused/Grik2 | RFLPs | Grik2: ↑ Q/R site | Suggested as a compensatory mechanism | [27] |
| Human/TC | Focused/Grik1 and Grik2 | RFLPs | Grik1: ↑ Q/R site Grik2: ↑ Q/R site | Suggested as a compensatory mechanism | [28] | |
| Human/HPC | Focused/Gria2 | RFLPs | Gria2: ↑ R/G site | Suggested as contributor in disease pathogenesis | [29] | |
| Human/HPC | Focused/Gria2-4 Grik1-2, KCNA1, 5-HT2C | Sanger sequencing | KCNA1: ↓ I/V site | Inversely associated with disease duration | [30] | |
| Mouse/HPC | Transcriptome-wide/whole transcriptome | NGS/Sanger sequencing | 19 DE targets experimentally validated, ↑ Grik2, Ctsb, Rpa, Sparc, Slc1a2, Ovca2, ↓ Ncl, Wipi2, Klhl24, Hspa4l, Slc37a3, Cyfip2 | DE targets involved in disease related pathways | [31] | |
| SCZ | Human/PFC | Focused/Gria2 | RFLPs | Gria2: ↓ Q/R site | Possible contributor to disease pathogenesis | [32] |
| Human/FC | Focused/5-HT2C | Cloning and Sanger sequencing | 5-HT2C: ↓ site B | ↑ Unedited isoform | [33] | |
| Human/PFC | Focused/5-HT2C | Cloning and Sanger sequencing | 5-HT2C: ns site differences | Trend: ↑ unedited isoform | [34] | |
| Human/ACC/DLPFC | Transcriptome-wide/whole transcriptome | NGS meta-analysis/independent (validation) cohort in silico analysis | >100 DE sites per region, DE overlap between brain regions (n = 29) | ↑ Global editing, DE targets involved in disease associated pathways | [35] | |
| Suicide | Human/PFC | Focused/5-HT2C | Primer extension | 5-HT2C: site A | ↑ Site A | [36] |
| Human/PFC | Focused/5-HT2C | Targeted NGS/cloning and Sanger sequencing | 5-HT2C isoforms due to editing | ↑ ABCD isoform (hypoactive) | [37] | |
| Human/PFC | Focused/5-HT2C | Targeted NGS/validation cohort analysis | 5-HT2C edited isoforms, ↑ ABCD isoform | ABCD isoform associated with gene expression alterations | [38] | |
| Human/ACC/DLPF | Focused/5-HT2C | CE-SSCP | 5-HT2C edited isoforms, ACC: ↑ A, ABDE, ↓ D/DLPFC: ↑ AB | Region-specific differential representation | [39] | |
| Depression and suicide | Human/PFC | Focused/5-HT2C | Cloning and Sanger sequencing | 5-HT2C: ↑ site C’ (Ε), ↓ site D | [40] | |
| Human/PFC | Focused/5-HT2C | Primer extension and DHPLC/Sanger sequencing | 5-HT2C: sites A and D ns | Trend, Depr.: ↑ D, Suicide: ↑ A | [41] | |
| Human/ACC/DLPFC | Focused/PDE8A | CE-SSCP | PDE8A edited isoforms, ACC: ↑ ABCEF, ABC, ABEFG, BFG ↓ B, ABDE/DLPFC: ↑ ABEFG, BCEG, ↓ ABF, BEG | Region specific differential representation | [42] | |
| Human/whole blood | Focused/PDE8A | Targeted NGS | PDE8A: ↓ sites B, C, E and D, F ns ↓ Isoforms B, BC, BD, BE, BF | Similar patterns with the brain of suicide decedents | [42] | |
| CSDS | Mouse/PFC/BLA | Focused/recoding in neuronal function related transcripts (551 sites) | Targeted NGS (mmPCR_seq) | PFC: ↑ Commd2, Rsad1, Iqgap1, Klf16, Nova1 ↓ Wipi2, Zfp81, Rn45s, Rwdd2b, Dagla, BLA: ↑ Htr2c (C, D site), Gabra3, Tcp11l1, Qpctl ↓ Zfp324, Copa, Gria4, Fubp3, Nova1 | Region-specific DE | [43] |
| Autism | Human/CB | Focused/synaptic transcripts (10 targets) | Pyrosequencing/validation: Padlock probes and NGS (5 targets) | Gria 4: ↑ R/G site, Grik2 and 5-HT2C edited isoforms differential representation | Gria4 editing associated with differential splicing isoform usage | [44] |
| Human/TC, FC, CB | Transcriptome-wide/whole transcriptome | NGS/2nd cohort meta-analysis/cloning and Sanger sequencing | ↑ Ctsb, Neat1 ↓ Gsk3b, Nova1, Grik1, FAM213A, Dennd3 | ↓ Global editing | [45] |
| Disorder | Species/Brain Region | Study Type/Target(s) | Methodology/ Validation Method | DE Targets/Trend Relative to Controls | Remarks | Ref. |
|---|---|---|---|---|---|---|
| ALS | Human/SC | Focused/Gria2 | RFLPs/Sanger sequencing | Gria2: ↓ Q/R site | [46] | |
| Human/neurons $ | Focused/Gria2 | RFLPs | Gria2: ↓ Q/R site | No editing changes in Purkinje cells | [47] | |
| Human/SC and motor cortex | Focused/EAAT2 (astroglial glutamate transporter) | Cloning and Sanger sequencing | EAAT2: ↑ intron7 | Alternative polyadenylation and intron 7 retention transcripts (in vitro functional evidence) | [48] | |
| Human/SC neurons $ | Focused/Gria2 | RFLPs | Gria2: ↓ Q/R site | [49] | ||
| Human/SC | Transcriptome-wide/focus on database listed A-I editing sites | NGS | Gria2: ↓ Q/R site ns trend | Low sample number, n = 5–6/group | [50] | |
| ALS (C9orf72) | Human/SC, motor cortex, FC, CB | Transcriptome-wide/whole transcriptome | NGS/ADAR1 and/or ADAR2 deficient hiPSC-MNs cells and cells with aberrant ADAR2 localization | 1526 DE transcripts | No changes in global editing, region-specific hypo- and hyper-edited patterns | [51] |
| HD | Human/striatum | Focused/Gria2 | RFLPs | Gria2: ↓ Q/R site | [32] | |
| AD | Human/PFC | Focused/Gria2 | RFLPs | Gria2: ↓ Q/R site | [32] | |
| Human/HPC | Focused/Gria2 | Sanger sequencing/primer extension | Gria2: ↓ Q/R site | [52] | ||
| Human/HPC, temporal and frontal lobe | Focused/recoding in synaptic transcripts (72 targets, 118 sites) | Targeted NGS (mmPCR_seq) | ↓ 5-HT2C receptor isoforms, HPC: ↓ Cacna1d, Ddx58, Fbxl6, Fis1, Flj43663, Gria3, Gria4, Igfbp7, Kcna1, Meg3, Narf, Nova1, Ptpn14, Unc80 ↑ Copa Temporal lobe: ↓ Ccni, Fbxl6, Flj43663, Gria2, Gria4, Grik1, Grik2, Meg3, Mfn1, Tme63b, Unc80 ↑ Narf/Frontal lobe: ↓ Mfn1, Grik2, Meg3, Gria2, Unc80, Ddx58 | ↓ Recoding | [53] | |
| Human/HPC | Transcriptome-wide | NGS | 11 DE targets, ↓ Gria2, Gria3, Gria4, Grik1, Grik2 ↑ Blcap, Copa, Vn1r1, Znf235, Znf397, Znf582 | ↓ Recoding | [54] | |
| Human/ACC/DLPFC/PCC/aPFC/pSTG/IFGo/FFG/CB/TC | Transcriptome-wide/focus on database listed A-I editing sites | NGS | ↓ Editing in SYT11, MCUR1, SOD2, ORAI2, HSDL2, PFKP, and GPRC5B | DLPFC Samples: ↓ ADAR1 ↑ ADAR3 expression in AD cases | [55] | |
| Prion diseases | sCJD and vCJD Rhesus monkeys/CB | Focused/Alu | Cloning and Sanger sequencing | ↓ Alu editing | Strain specific differences | [56] |
| sCJD Mouse/Cortex | Transcriptome-wide | NGS and Sanger sequencing | 3 DE targets experimentally validated, Mouse pre-clinical: ↓ Sidt2, ↑ Fkrp/Mouse clinical: ↑ Rragd | ↓ Global editing, Human cross-validation: ↓ Paqr8, ↑ Ctss, Rrgad | [57] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karagianni, K.; Pettas, S.; Christoforidou, G.; Kanata, E.; Bekas, N.; Xanthopoulos, K.; Dafou, D.; Sklaviadis, T. A Systematic Review of Common and Brain-Disease-Specific RNA Editing Alterations Providing Novel Insights into Neurological and Neurodegenerative Disease Manifestations. Biomolecules 2022, 12, 465. https://doi.org/10.3390/biom12030465
Karagianni K, Pettas S, Christoforidou G, Kanata E, Bekas N, Xanthopoulos K, Dafou D, Sklaviadis T. A Systematic Review of Common and Brain-Disease-Specific RNA Editing Alterations Providing Novel Insights into Neurological and Neurodegenerative Disease Manifestations. Biomolecules. 2022; 12(3):465. https://doi.org/10.3390/biom12030465
Chicago/Turabian StyleKaragianni, Korina, Spyros Pettas, Georgia Christoforidou, Eirini Kanata, Nikolaos Bekas, Konstantinos Xanthopoulos, Dimitra Dafou, and Theodoros Sklaviadis. 2022. "A Systematic Review of Common and Brain-Disease-Specific RNA Editing Alterations Providing Novel Insights into Neurological and Neurodegenerative Disease Manifestations" Biomolecules 12, no. 3: 465. https://doi.org/10.3390/biom12030465
APA StyleKaragianni, K., Pettas, S., Christoforidou, G., Kanata, E., Bekas, N., Xanthopoulos, K., Dafou, D., & Sklaviadis, T. (2022). A Systematic Review of Common and Brain-Disease-Specific RNA Editing Alterations Providing Novel Insights into Neurological and Neurodegenerative Disease Manifestations. Biomolecules, 12(3), 465. https://doi.org/10.3390/biom12030465







