Interplay between Alba and Cren7 Regulates Chromatin Compaction in Sulfolobus solfataricus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sulfolobus Solfataricus Strain and Growth Conditions
2.2. Micrococcal Nuclease Digestion
2.3. On-Substrate Lysis
2.4. Expression and Purification of Recombinant Architectural Proteins
2.5. Formation of Alba Heterodimers
2.6. In Vitro Chromatin Reconstitution
2.7. Atomic Force Microscopy
2.8. RNA Extraction and Quantitative PCR
3. Results
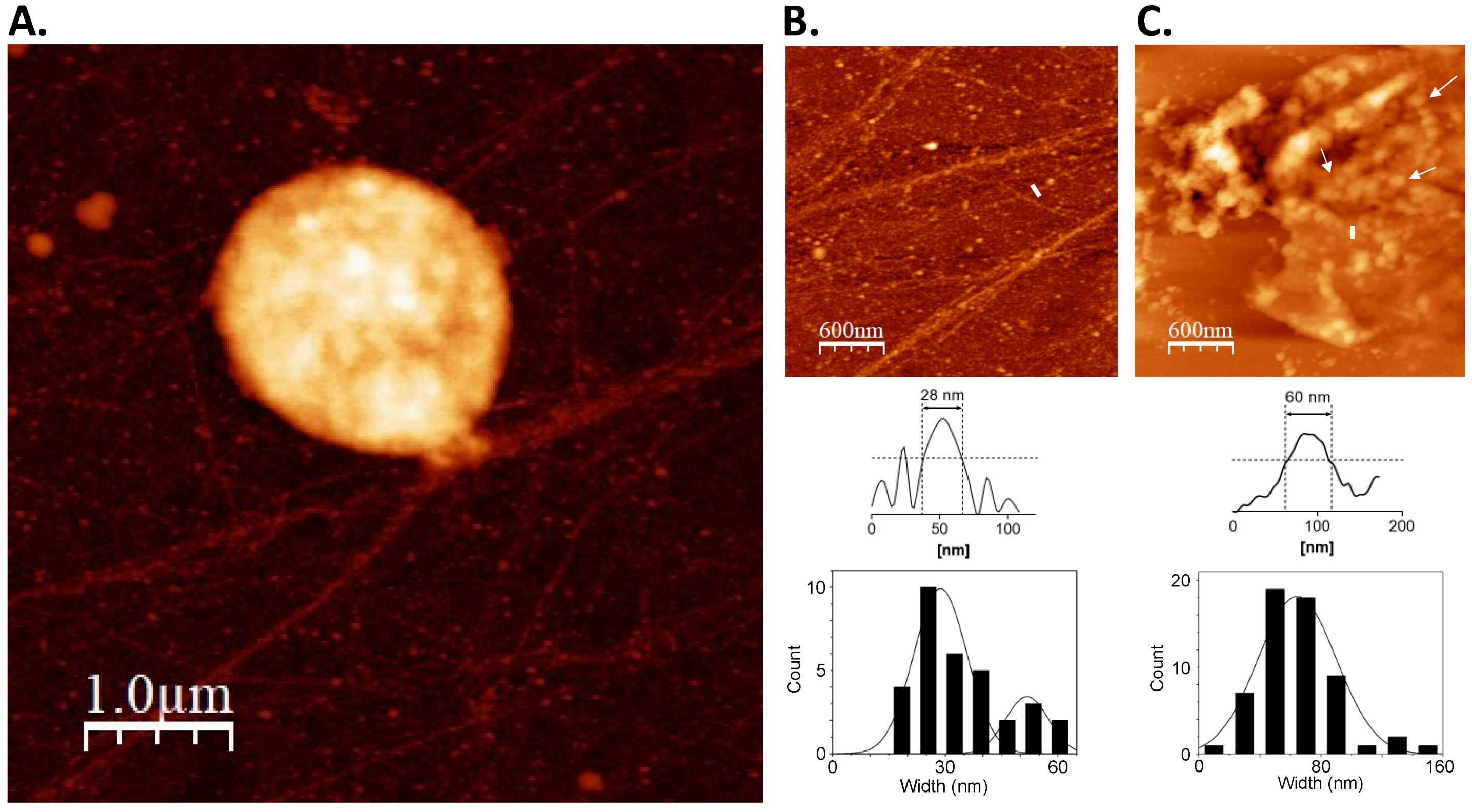
3.1. On-Substrate Lysis Reveals Fundamental Structure of the Nucleoid
3.2. Alba Mediates the Formation of Regularly Structured Complexes while Cren7 Forms Loose Nucleoprotein Structures with Long DNA
3.3. Interplay between Architectural Proteins Can Alter Chromatin Structures In Vitro
3.4. Expression Level of Architectural Protein Regulators Changes with Growth Phase
4. Discussion
4.1. Architectural Landscape of the S. solfataricus Chromatin
4.2. Alba-Mediated Chromatin Structures
4.3. Chromatin Dynamics in S. solfataricus
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luijsterburg, M.S.; White, M.F.; van Driel, R.; Dame, R.T. The Major Architects of Chromatin: Architectural Proteins in Bacteria, Archaea and Eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 393–418. [Google Scholar] [CrossRef]
- Sandman, K.; Reeve, J.N. Archaeal Chromatin Proteins: Different Structures but Common Function? Curr. Opin. Microbiol. 2005, 8, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Guo, L.; Huang, L. Archaeal Chromatin Proteins. Sci. China Life Sci. 2012, 55, 377–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, M.F.; Bell, S.D. Holding It Together: Chromatin in the Archaea. Trends Genet. 2002, 18, 621–626. [Google Scholar] [CrossRef]
- Luger, K.; Dechassa, M.L.; Tremethick, D.J. New Insights into Nucleosome and Chromatin Structure: An Ordered State or a Disordered Affair? Nat. Rev. Mol. Cell Biol. 2012, 13, 436–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peeters, E.; Driessen, R.P.C.; Werner, F.; Dame, R.T. The Interplay between Nucleoid Organization and Transcription in Archaeal Genomes. Nat. Rev. Microbiol. 2015, 13, 333–341. [Google Scholar] [CrossRef]
- Dame, R.T. The Role of Nucleoid-Associated Proteins in the Organization and Compaction of Bacterial Chromatin: Organization and Compaction of Bacterial Chromatin. Mol. Microbiol. 2005, 56, 858–870. [Google Scholar] [CrossRef]
- Dame, R.T.; Rashid, F.-Z.M.; Grainger, D.C. Chromosome Organization in Bacteria: Mechanistic Insights into Genome Structure and Function. Nat. Rev. Genet. 2020, 21, 227–242. [Google Scholar] [CrossRef]
- Reeve, J.N.; Sandman, K.; Daniels, C.J. Archaeal Histones, Nucleosomes, and Transcription Initiation. Cell 1997, 89, 999–1002. [Google Scholar] [CrossRef] [Green Version]
- Sandman, K.; Krzycki, J.A.; Dobrinski, B.; Lurz, R.; Reeve, J.N. HMf, a DNA-Binding Protein Isolated from the Hyperthermophilic Archaeon Methanothermus Fervidus, Is Most Closely Related to Histones. Proc. Natl. Acad. Sci. USA 1990, 87, 5788–5791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henneman, B.; van Emmerik, C.; van Ingen, H.; Dame, R.T. Structure and Function of Archaeal Histones. PLoS Genet. 2018, 14, e1007582. [Google Scholar] [CrossRef] [PubMed]
- Grote, M.; Dijk, J.; Reinhardt, R. Ribosomal and DNA Binding Proteins of the Thermoacidophilic Archaebacterium Sulfolobus Acidocaldarius. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 1986, 873, 405–413. [Google Scholar] [CrossRef]
- Le Cam, E.; Culard, F.; Larquet, E.; Delain, E.; Cognet, J.A. DNA Bending Induced by the Archaebacterial Histone-like Protein MC1. J. Mol. Biol. 1999, 285, 1011–1021. [Google Scholar] [CrossRef]
- Guo, L.; Feng, Y.; Zhang, Z.; Yao, H.; Luo, Y.; Wang, J.; Huang, L. Biochemical and Structural Characterization of Cren7, a Novel Chromatin Protein Conserved among Crenarchaea. Nucleic Acids Res. 2008, 36, 1129–1137. [Google Scholar] [CrossRef]
- Jelinska, C.; Conroy, M.J.; Craven, C.J.; Hounslow, A.M.; Bullough, P.A.; Waltho, J.P.; Taylor, G.L.; White, M.F. Obligate Heterodimerization of the Archaeal Alba2 Protein with Alba1 Provides a Mechanism for Control of DNA Packaging. Structure 2005, 13, 963–971. [Google Scholar] [CrossRef]
- Wardleworth, B.N.; Russell, R.J.M.; Bell, S.D.; Taylor, G.L.; White, M.F. Structure of Alba: An Archaeal Chromatin Protein Modulated by Acetylation. EMBO J. 2002, 21, 4654–4662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, S.D.; Botting, C.H.; Wardleworth, B.N.; Jackson, S.P.; White, M.F. The Interaction of Alba, a Conserved Archaeal Chromatin Protein, with Sir2 and Its Regulation by Acetylation. Science 2002, 296, 148–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, H.; Guo, R.; Wen, Y.; Liu, D.; Huang, L. An Abundant DNA Binding Protein from the Hyperthermophilic Archaeon Sulfolobus Shibatae Affects DNA Supercoiling in a Temperature-Dependent Fashion. J. Bacteriol. 2000, 182, 3929–3933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, M.B. Studies with Cyanidium Caldarium, an Anomalously Pigmented Chlorophyte. Arch. Mikrobiol. 1959, 32, 270–277. [Google Scholar] [CrossRef]
- Brock, T.D.; Brock, K.M.; Belly, R.T.; Weiss, R.L. Sulfolobus: A New Genus of Sulfur-Oxidizing Bacteria Living at Low PH and High Temperature. Arch. Mikrobiol. 1972, 84, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yoshimura, S.H.; Hizume, K.; Ohniwa, R.L.; Ishihama, A.; Takeyasu, K. Fundamental Structural Units of the Escherichia Coli Nucleoid Revealed by Atomic Force Microscopy. Nucleic Acids Res. 2004, 32, 1982–1992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurer, S.; Fritz, J.; Muskhelishvili, G. A Systematic in Vitro Study of Nucleoprotein Complexes Formed by Bacterial Nucleoid-Associated Proteins Revealing Novel Types of DNA Organization. J. Mol. Biol. 2009, 387, 1261–1276. [Google Scholar] [CrossRef] [PubMed]
- Horcas, I.; Fernández, R.; Gómez-Rodríguez, J.M.; Colchero, J.; Gómez-Herrero, J.; Baro, A.M. WSXM: A Software for Scanning Probe Microscopy and a Tool for Nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef] [PubMed]
- Telford, D.J.; Stewart, B.W. Micrococcal Nuclease: Its Specificity and Use for Chromatin Analysis. Int. J. Biochem. 1989, 21, 127–137. [Google Scholar] [CrossRef]
- Shimamura, A.; Tremethick, D.; Worcel, A. Characterization of the Repressed 5S DNA Minichromosomes Assembled in Vitro with a High-Speed Supernatant of Xenopus Laevis Oocytes. Mol. Cell Biol. 1988, 8, 4257–4269. [Google Scholar] [CrossRef] [Green Version]
- Ammar, R.; Torti, D.; Tsui, K.; Gebbia, M.; Durbic, T.; Bader, G.D.; Giaever, G.; Nislow, C. Chromatin Is an Ancient Innovation Conserved between Archaea and Eukarya. eLife 2012, 1, e00078. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, H.; Prieto, E.I.; Nambu, T.; Mashimo, C.; Kashiwagi, K.; Okinaga, T.; Atomi, H.; Takeyasu, K. Different Proteins Mediate Step-Wise Chromosome Architectures in Thermoplasma Acidophilum and Pyrobaculum Calidifontis. Front. Microbiol. 2020, 11, 1247. [Google Scholar] [CrossRef] [PubMed]
- Črnigoj, M.; Podlesek, Z.; Zorko, M.; Jerala, R.; Anderluh, G.; Ulrih, N.P. Interactions of Archaeal Chromatin Proteins Alba1 and Alba2 with Nucleic Acids. PLoS ONE 2013, 8, e58237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurens, N.; Driessen, R.P.C.; Heller, I.; Vorselen, D.; Noom, M.C.; Hol, F.J.H.; White, M.F.; Dame, R.T.; Wuite, G.J.L. Alba Shapes the Archaeal Genome Using a Delicate Balance of Bridging and Stiffening the DNA. Nat. Commun. 2012, 3, 1328. [Google Scholar] [CrossRef] [PubMed]
- Driessen, R.P.C.; Meng, H.; Suresh, G.; Shahapure, R.; Lanzani, G.; Priyakumar, U.D.; White, M.F.; Schiessel, H.; van Noort, J.; Dame, R.T. Crenarchaeal Chromatin Proteins Cren7 and Sul7 Compact DNA by Inducing Rigid Bends. Nucleic Acids Res. 2013, 41, 196–205. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, M.; Chen, Y.; Wang, L.; Liu, Q.; Dong, Y.; Gong, Y.; Huang, L. Architectural Roles of Cren7 in Folding Crenarchaeal Chromatin Filament. Mol. Microbiol. 2019, 111, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Marsh, V.L.; Peak-Chew, S.Y.; Bell, S.D. Sir2 and the Acetyltransferase, Pat, Regulate the Archaeal Chromatin Protein, Alba. J. Biol. Chem. 2005, 280, 21122–21128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popławski, A.; Bernander, R. Nucleoid Structure and Distribution in Thermophilic Archaea. J. Bacteriol. 1997, 179, 7625–7630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hizume, K.; Nakai, T.; Araki, S.; Prieto, E.; Yoshikawa, K.; Takeyasu, K. Removal of Histone Tails from Nucleosome Dissects the Physical Mechanisms of Salt-Induced Aggregation, Linker Histone H1-Induced Compaction, and 30-Nm Fiber Formation of the Nucleosome Array. Ultramicroscopy 2009, 109, 868–873. [Google Scholar] [CrossRef] [Green Version]
- Hocher, A.; Rojec, M.; Swadling, J.B.; Esin, A.; Warnecke, T. The DNA-Binding Protein HTa from Thermoplasma Acidophilum Is an Archaeal Histone Analog. eLife 2019, 8, e52542. [Google Scholar] [CrossRef]
- Pereira, S.L.; Grayling, R.A.; Lurz, R.; Reeve, J.N. Archaeal Nucleosomes. Proc. Natl. Acad. Sci. USA 1997, 94, 12633–12637. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.C.; Qian, Z.; Adhya, S.L. Architecture of the Escherichia Coli Nucleoid. PLoS Genet. 2019, 15, e1008456. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, H.; Shin, M.; Oda, T.; Matsumi, R.; Ohniwa, R.L.; Itoh, T.; Shirahige, K.; Imanaka, T.; Atomi, H.; Yoshimura, S.H.; et al. Histone and TK0471/TrmBL2 Form a Novel Heterogeneous Genome Architecture in the Hyperthermophilic Archaeon Thermococcus Kodakarensis. Mol. Biol. Cell 2011, 22, 386–398. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, H.; Harwood, J.C.; Moore, K.M.; Paszkiewicz, K.; Durley, S.C.; Fukushima, H.; Atomi, H.; Takeyasu, K.; Kent, N.A. An Alternative Beads-on-a-String Chromatin Architecture in Thermococcus Kodakarensis. EMBO Rep. 2013, 14, 711–717. [Google Scholar] [CrossRef] [Green Version]
- Hizume, K.; Yoshimura, S.H.; Maruyama, H.; Kim, J.; Wada, H.; Takeyasu, K. Chromatin Reconstitution: Development of a Salt-Dialysis Method Monitored by Nano-Technology. Arch. Histol. Cytol. 2002, 65, 405–413. [Google Scholar] [CrossRef] [Green Version]
- Goyal, M.; Banerjee, C.; Nag, S.; Bandyopadhyay, U. The Alba Protein Family: Structure and Function. Biochim. Biophys. Acta 2016, 1864, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Takemata, N.; Samson, R.Y.; Bell, S.D. Physical and Functional Compartmentalization of Archaeal Chromosomes. Cell 2019, 179, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Gissot, M.; Walker, R.; Delhaye, S.; Alayi, T.D.; Huot, L.; Hot, D.; Callebaut, I.; Schaeffer-Reiss, C.; Dorsselaer, A.V.; Tomavo, S. Toxoplasma Gondii Alba Proteins Are Involved in Translational Control of Gene Expression. J. Mol. Biol. 2013, 425, 1287–1301. [Google Scholar] [CrossRef]
- Guo, R.; Xue, H.; Huang, L. Ssh10b, a Conserved Thermophilic Archaeal Protein, Binds RNA in Vivo. Mol. Microbiol. 2003, 50, 1605–1615. [Google Scholar] [CrossRef] [Green Version]
- Aravind, L.; Iyer, L.M.; Anantharaman, V. The Two Faces of Alba: The Evolutionary Connection between Proteins Participating in Chromatin Structure and RNA Metabolism. Genome Biol. 2003, 4, R64. [Google Scholar] [CrossRef] [Green Version]
- Rigden, D.J.; Galperin, M.Y. Sequence Analysis of GerM and SpoVS, Uncharacterized Bacterial “sporulation” Proteins with Widespread Phylogenetic Distribution. Bioinformatics 2008, 24, 1793–1797. [Google Scholar] [CrossRef] [PubMed]
- Hołówka, J.; Zakrzewska-Czerwińska, J. Nucleoid Associated Proteins: The Small Organizers That Help to Cope With Stress. Front. Microbiol. 2020, 11, 590. [Google Scholar] [CrossRef]
- Bowman, G.D.; Poirier, M.G. Post-Translational Modifications of Histones That Influence Nucleosome Dynamics. Chem. Rev. 2015, 115, 2274–2295. [Google Scholar] [CrossRef] [Green Version]
- Vorontsov, E.A.; Rensen, E.; Prangishvili, D.; Krupovic, M.; Chamot-Rooke, J. Abundant Lysine Methylation and N-Terminal Acetylation in Sulfolobus Islandicus Revealed by Bottom-Up and Top-Down Proteomics. Mol. Cell. Proteom. 2016, 15, 3388–3404. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Wang, Q.; Liu, T.; Peng, N.; Huang, L. Insights into the Post-Translational Modifications of Archaeal Sis10b (Alba): Lysine-16 Is Methylated, Not Acetylated, and This Does Not Regulate Transcription or Growth. Mol. Microbiol. 2018, 109, 192–208. [Google Scholar] [CrossRef]
- Dilweg, I.W.; Dame, R.T. Post-Translational Modification of Nucleoid-Associated Proteins: An Extra Layer of Functional Modulation in Bacteria? Biochem. Soc. Trans. 2018, 46, 1381–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laursen, S.P.; Bowerman, S.; Luger, K. Archaea: The Final Frontier of Chromatin. J. Mol. Biol. 2021, 433, 166791. [Google Scholar] [CrossRef] [PubMed]








| Target Gene | Forward Sequence (5′ → 3′) | Reverse Sequence (3′ → 5′) |
|---|---|---|
| glyA | AGGACGCAGACTTTGAACCT | AAACTCTAAACACAGCCGCA |
| albA | ACTCTCGATTGCCTTCCGTC | GGAAGAGCTATTAGCAAAGCCG |
| albA2 | CAGGTAGGGAAATTTCTAAAGCAG | CGCCTTCTATCTCTAACTTCACTT |
| creN7 | ACATGGCAGCTAGTTTCACC | TCTCTACCGAGGAACCTCCC |
| Sir2 | AGGTGTGATTGTGGTGGGAT | CAACGGGGTCTCCTCCATAT |
| Pat | ACAGAACACTAGGGATAGGGAC | GCGGAGAAAGTTGCTAGGTT |
| Setup | Number of Measured Objects | Contour Length | End-to-End Distance | ||
|---|---|---|---|---|---|
| Mean (µm) | SD | Mean (µm) | SD | ||
| Naked DNA | 25 | 1.03 | 0.14 | 0.364 | 0.15 |
| 6:1 Alba1-DNA | 25 | 0.954 | 0.20 | 0.156 | 0.12 |
| 24:1 Alba1-DNA | 25 | 1.20 | 0.26 | 0.654 | 0.20 |
| 0.3:1 Cren7-DNA | 20 | 0.948 | 0.11 | 0.464 | 0.20 |
| 0.6:1 Cren7-DNA | 20 | 0.899 | 0.11 | 0.297 | 0.15 |
| 1:1 Cren7-DNA | 20 | 0.704 | 0.12 | 0.373 | 0.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cajili, M.K.M.; Prieto, E.I. Interplay between Alba and Cren7 Regulates Chromatin Compaction in Sulfolobus solfataricus. Biomolecules 2022, 12, 481. https://doi.org/10.3390/biom12040481
Cajili MKM, Prieto EI. Interplay between Alba and Cren7 Regulates Chromatin Compaction in Sulfolobus solfataricus. Biomolecules. 2022; 12(4):481. https://doi.org/10.3390/biom12040481
Chicago/Turabian StyleCajili, Marc Kenneth M., and Eloise I. Prieto. 2022. "Interplay between Alba and Cren7 Regulates Chromatin Compaction in Sulfolobus solfataricus" Biomolecules 12, no. 4: 481. https://doi.org/10.3390/biom12040481





