Mass Spectrometry-Based Proteome Profiling of Extracellular Vesicles Derived from the Cerebrospinal Fluid of Adult Rhesus Monkeys Exposed to Cocaine throughout Gestation
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Characterization of CSF-EVs
3.2. Proteomic Characterization of CSF-EVs by Mass Spectrometry (MS)
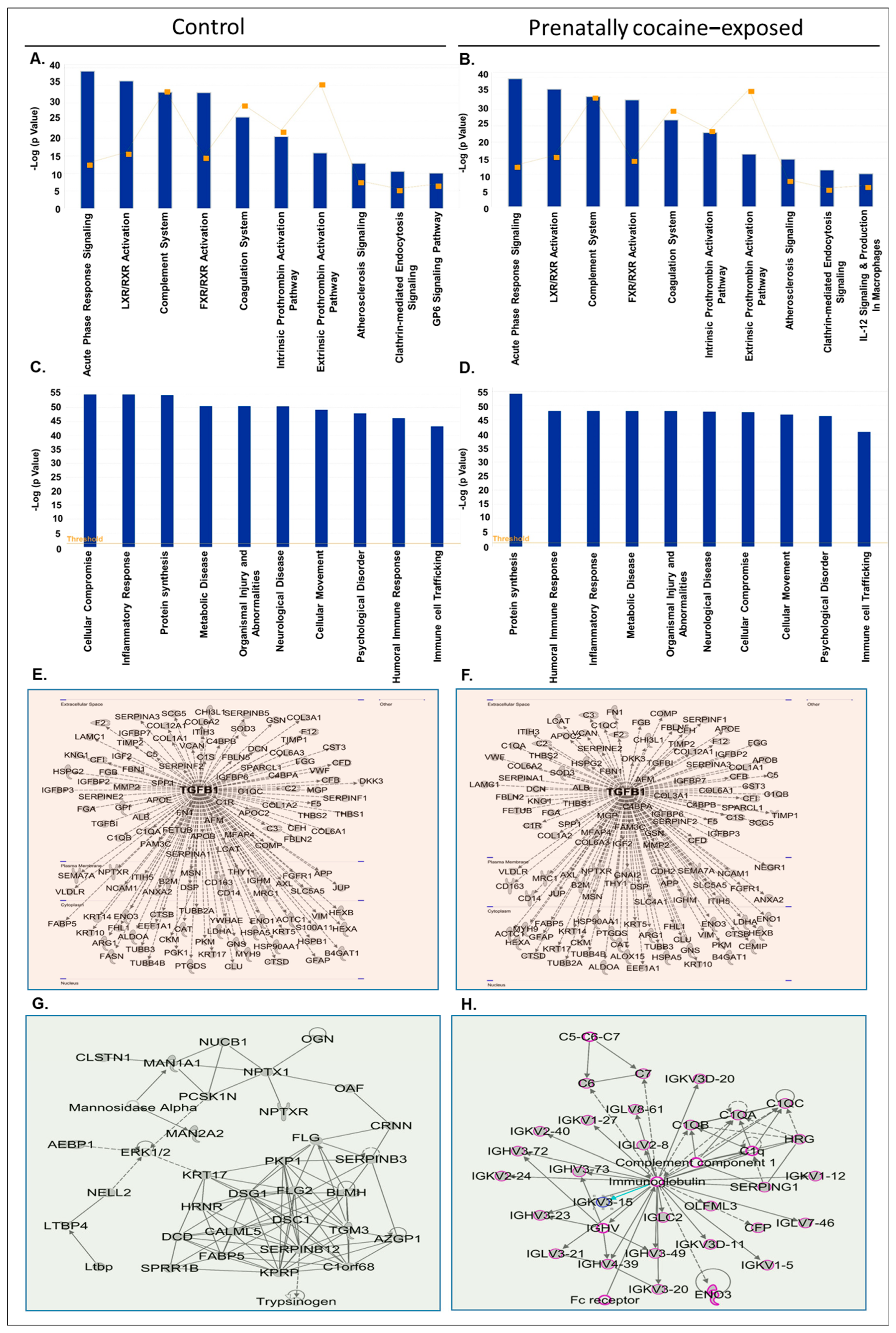
3.3. Ingenuity Pathway Analysis (IPA) of Proteins Identified in CSF-EVs
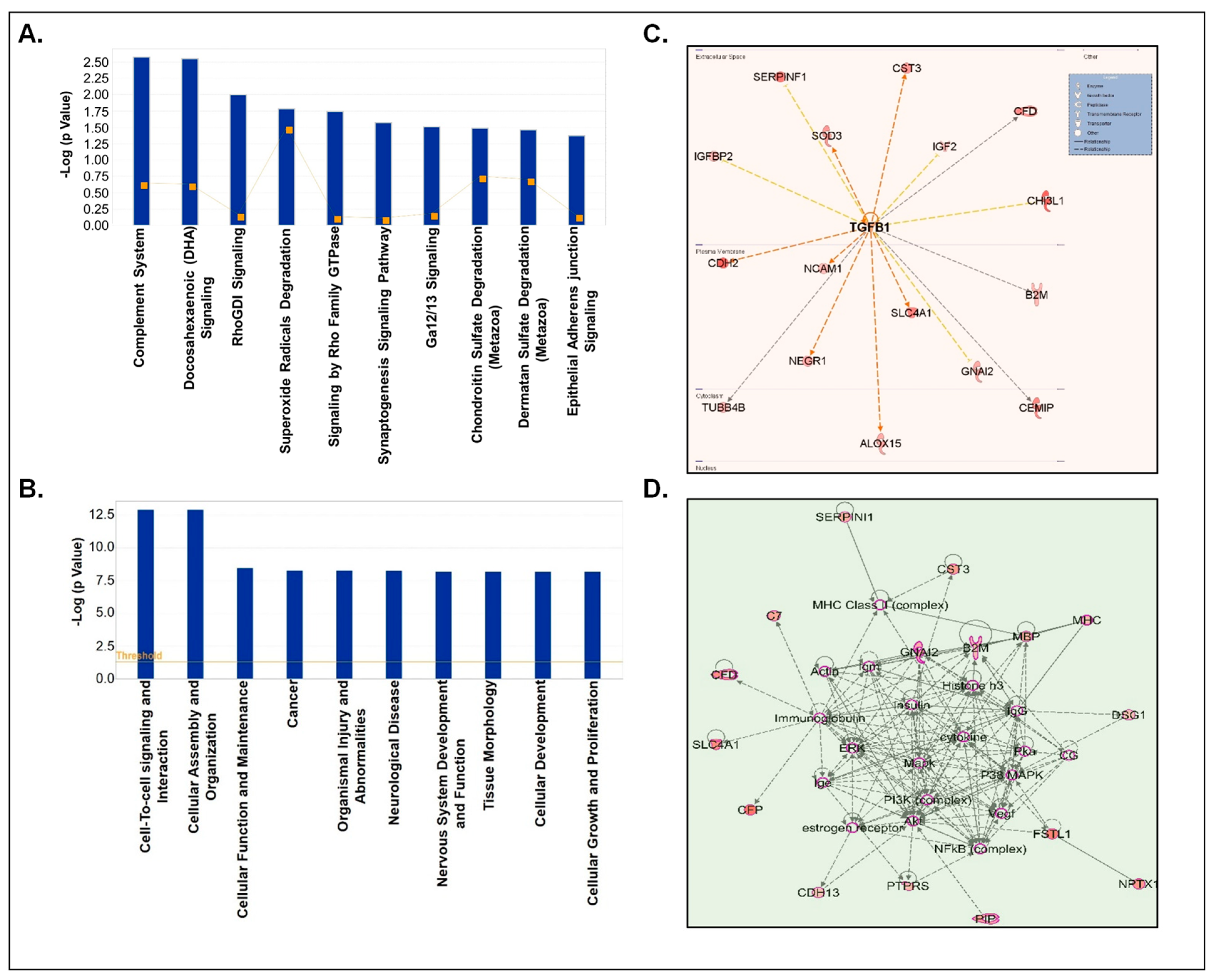
3.4. Processing of Differentially Loaded Proteins in CSF-EVs from Control and Prenatally Cocaine-Exposed Monkeys Using IPA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Substance Abuse and Mental Health Services Administration. Key Substance Use and Mental Health Indicators in the United States: Results from the 2019 National Survey on Drug Use and Health; PEP20-07-01-001; Center for Behavioral Health Statistics and Quality, Substance Abuse and Mental Health Services Administration: Rockville, MD, USA, 2020.
- Hedegaard, H.; Spencer, M.R.; Garnett, M.F. Increase in Drug Overdose Deaths Involving Cocaine: United States, 2009–2018; CDC: Atlanta, GA, USA, 2020; pp. 1–8.
- Kosofsky, B.E.; Wilkins, A.S.; Gressens, P.; Evrard, P. Transplacental Cocaine Exposure: A Mouse Model Demonstrating Neuroanatomic and Behavioral Abnormalities. J. Child Neurol. 1994, 9, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Wendell, A.D. Overview and epidemiology of substance abuse in pregnancy. Clin. Obstet. Gynecol. 2013, 56, 91–96. [Google Scholar] [CrossRef]
- Song, J.; Guan, X.W.; Ren, J.Q.; He, W. Developmental toxicity of cocaine exposure in mid-pregnancy mice. Acta Pharmacol. Sin. 2002, 23, 1029–1034. [Google Scholar]
- Schenker, S.; Yang, Y.; Johnson, R.F.; Downing, J.W.; Schenken, R.S.; Henderson, G.I.; King, T.S. The transfer of cocaine and its metabolites across the term human placenta. Clin. Pharmacol. Ther. 1993, 53, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Derauf, C.; Kekatpure, M.; Neyzi, N.; Lester, B.; Kosofsky, B. Neuroimaging of children following prenatal drug exposure. Semin. Cell Dev. Biol. 2009, 20, 441–454. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, L.R.; Gould, T.J. Multigenerational and transgenerational effects of paternal exposure to drugs of abuse on behavioral and neural function. Eur. J. Neurosci. 2018, 50, 2453–2466. [Google Scholar] [CrossRef]
- Hamilton, L.R.; Czoty, P.W.; Nader, M.A. Behavioral characterization of adult male and female rhesus monkeys exposed to cocaine throughout gestation. Psychopharmacology 2011, 213, 799–808. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, L.R.; Czoty, P.W.; Gage, H.D.; Nader, M.A. Characterization of the dopamine receptor system in adult rhesus monkeys exposed to cocaine throughout gestation. Psychopharmacology 2010, 210, 481–488. [Google Scholar] [CrossRef] [Green Version]
- Paule, M.G.; Gillam, M.P.; Binienda, Z.; Morris, P. Chronic Cocaine Exposure throughout Gestation in the Rhesus Monkey. Ann. N. Y. Acad. Sci. 1996, 801, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.S.S.; O’Connell, K.; Finnerty, T.K. Potential Role of Extracellular Vesicles in the Pathophysiology of Drug Addiction. Mol. Neurobiol. 2018, 55, 6906–6913. [Google Scholar] [CrossRef]
- Hollander, J.A.; Im, H.I.; Amelio, A.L.; Kocerha, J.; Bali, P.; Lu, Q.; Willoughby, D.; Wahlestedt, C.; Conkright, M.D.; Kenny, P.J. Striatal microRNA controls cocaine intake through CREB signalling. Nature 2010, 466, 197–202. [Google Scholar] [CrossRef] [Green Version]
- Quinn, R.K.; Brown, A.L.; Goldie, B.J.; Levi, E.M.; Dickson, P.W.; Smith, D.W.; Cairns, M.J.; Dayas, C.V. Distinct miRNA expression in dorsal striatal subregions is associated with risk for addiction in rats. Transl. Psychiatry 2015, 5, e503. [Google Scholar] [CrossRef]
- Shahjin, F.; Guda, R.S.; Schaal, V.L.; Odegaard, K.; Clark, A.; Gowen, A.; Xiao, P.; Lisco, S.J.; Pendyala, G.; Yelamanchili, S.V. Brain-Derived Extracellular Vesicle microRNA Signatures Associated with In Utero and Postnatal Oxycodone Exposure. Cells 2019, 9, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Li, C.; Zhou, Y.; Luo, C.; Ou, J.; Li, J.; Mo, Z. Expression of microRNAs in the serum exosomes of methamphetamine-dependent rats vs. ketamine-dependent rats. Exp. Ther. Med. 2018, 15, 3369–3375. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Kim, S.; Su, Y.; Sharma, M.; Kumar, P.; Singh, S.; Lee, J.; Furdui, C.M.; Singh, R.; Hsu, F.-C.; et al. Brain cell-derived exosomes in plasma serve as neurodegeneration biomarkers in male cynomolgus monkeys self-administrating oxycodone. EBioMedicine 2021, 63, 103192. [Google Scholar] [CrossRef] [PubMed]
- Morris, P.; Biniendaa, Z.; Gillam, M.P.; Harkey, M.R.; Zhou, C.; Henderson, G.L.; Paule, M.G. The effect of chronic cocaine exposure during pregnancy on maternal and infant outcomes in the rhesus monkey. Neurotoxicol. Teratol. 1996, 18, 147–154. [Google Scholar] [CrossRef]
- Morris, P.; Binienda, Z.; Gillam, M.P.; Klein, J.; McMartin, K.; Koren, G.; Duhart, H.M.; Slikker, W.; Paule, M.G. The effect of chronic cocaine exposure throughout pregnancy on maternal and infant outcomes in the rhesus monkey. Neurotoxicol. Teratol. 1997, 19, 47–57. [Google Scholar] [CrossRef]
- Pavelka, N.; Pelizzola, M.; Vizzardelli, C.; Capozzoli, M.; Splendiani, A.; Granucci, F.; Ricciardi-Castagnoli, P. A power law global error model for the identification of differentially expressed genes in microarray data. BMC Bioinform. 2004, 5, 203. [Google Scholar] [CrossRef] [Green Version]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Lambert, B.L.; Bauer, C.R. Developmental and behavioral consequences of prenatal cocaine exposure: A review. J. Perinatol. 2012, 32, 819–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arendt, R.E.; Minnes, S.; Singer, L.T. Fetal Cocaine Exposure: Neurologic Effects and Sensory-Motor Delays. Phys. Occup. Ther. Pediatr. 1996, 16, 129–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Kumar, S.; Crenshaw, B.J.; Williams, S.D.; Bell, C.R.; Matthews, Q.L.; Sims, B. Cocaine-Specific Effects on Exosome Biogenesis in Microglial Cells. Neurochem. Res. 2021, 46, 1006–1018. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, R.; Tamashiro-Orrego, A.; Promes, V.; Tu, L.; Shi, J.; Yang, Y. Cocaine Self-administration and Extinction Inversely Alter Neuron to Glia Exosomal Dynamics in the Nucleus Accumbens. Front. Cell Neurosci. 2020, 13, 581. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, S.; Nagy, C.; Ibrahim, P.; Théroux, J.-F.; Wakid, M.; Fiori, L.M.; Yang, J.; Rotzinger, S.; Foster, J.A.; Mechawar, N.; et al. Neuron-derived extracellular vesicles enriched from plasma show altered size and miRNA cargo as a function of antidepressant drug response. Mol. Psychiatry 2021, 26, 7417–7424. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.; Kim, H.; McGee, L.; Johnson, A.E.; Talwar, S.; Marugan, J.; Southall, N.; Hu, X.; Lal, M.; Mondal, D.; et al. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: A drug repurposing strategy for advanced cancer. Sci. Rep. 2018, 8, 8161. [Google Scholar] [CrossRef] [Green Version]
- Jensen, L.E.; Whitehead, A.S. Regulation of serum amyloid A protein expression during the acute-phase response. Biochem. J. 1998, 334 Pt 3, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Miida, T.; Yamada, T.; Seino, U.; Ito, M.; Fueki, Y.; Takahashi, A.; Kosuge, K.; Soda, S.; Hanyu, O.; Obayashi, K.; et al. Serum amyloid A (SAA)-induced remodeling of CSF-HDL. Biochim. Biophys. Acta (BBA) 2006, 1761, 424–433. [Google Scholar] [CrossRef]
- Barbierato, M.; Borri, M.; Facci, L.; Zusso, M.; Skaper, S.D.; Giusti, P. Expression and Differential Responsiveness of Central Nervous System Glial Cell Populations to the Acute Phase Protein Serum Amyloid A. Sci. Rep. 2017, 7, 12158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brionne, T.C.; Tesseur, I.; Masliah, E.; Wyss-Coray, T. Loss of TGF-beta 1 leads to increased neuronal cell death and microgliosis in mouse brain. Neuron 2003, 40, 1133–1145. [Google Scholar] [CrossRef] [Green Version]
- Doyle, K.P.; Cekanaviciute, E.; Mamer, L.E.; Buckwalter, M.S. TGFβ signaling in the brain increases with aging and signals to astrocytes and innate immune cells in the weeks after stroke. J. Neuroinflamm. 2010, 7, 62. [Google Scholar] [CrossRef] [Green Version]
- Bergau, N.; Maul, S.; Rujescu, D.; Simm, A.; Navarrete Santos, A. Reduction of Glycolysis Intermediate Concentrations in the Cerebrospinal Fluid of Alzheimer’s Disease Patients. Front. Neurosci. 2019, 13, 871. [Google Scholar] [CrossRef] [PubMed]
- Ökvist, A.; Johansson, S.; Kuzmin, A.; Bazov, I.; Merino-Martinez, R.; Ponomarev, I.; Mayfield, R.D.; Harris, R.A.; Sheedy, D.; Garrick, T.; et al. Neuroadaptations in Human Chronic Alcoholics: Dysregulation of the NF-κB System. PLoS ONE 2007, 2, e930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannon, M.J.; Johnson, M.M.; Michelhaugh, S.K.; Hartley, Z.J.; Halter, S.D.; David, J.A.; Kapatos, G.; Schmidt, C.J. A Molecular Profile of Cocaine Abuse Includes the Differential Expression of Genes that Regulate Transcription, Chromatin, and Dopamine Cell Phenotype. Neuropsychopharmacology 2014, 39, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- Guesnet, P.; Alessandri, J.M. Docosahexaenoic acid (DHA) and the developing central nervous system (CNS)—Implications for dietary recommendations. Biochimie 2011, 93, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Kovacic, P. Role of oxidative metabolites of cocaine in toxicity and addiction: Oxidative stress and electron transfer. Med. Hypotheses 2005, 64, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Cerretani, D.; Fineschi, V.; Bello, S.; Riezzo, I.; Turillazzi, E.; Neri, M. Role of Oxidative Stress in Cocaine-Induced Cardiotoxicity and Cocaine-Related Death. Curr. Med. Chem. 2012, 19, 5619–5623. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, K.-L.; Shukla, A.; Beroun, A.; Ishikawa, M.; Huang, X.; Wang, Y.; Wang, Y.Q.; Yang, Y.; Bastola, N.D.; et al. Cocaine Triggers Astrocyte-Mediated Synaptogenesis. Biol. Psychiatry 2021, 89, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhu, Y.-L.; Zhou, Y.-Y.; Liang, G.-F.; Wang, Y.-Y.; Hu, F.-H.; Xiao, Z.-D. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef] [Green Version]
- Fries, G.R.; Khan, S.; Stamatovich, S.; Dyukova, E.; Walss-Bass, C.; Lane, S.D.; Schmitz, J.M.; Wardle, M.C. Anhedonia in cocaine use disorder is associated with inflammatory gene expression. PLoS ONE 2018, 13, e0207231. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rather, H.A.; Mishra, S.; Su, Y.; Kumar, A.; Singh, S.; Misra, B.B.; Lee, J.; Furdui, C.M.; Hamilton, L.R.; Gould, R.W.; et al. Mass Spectrometry-Based Proteome Profiling of Extracellular Vesicles Derived from the Cerebrospinal Fluid of Adult Rhesus Monkeys Exposed to Cocaine throughout Gestation. Biomolecules 2022, 12, 510. https://doi.org/10.3390/biom12040510
Rather HA, Mishra S, Su Y, Kumar A, Singh S, Misra BB, Lee J, Furdui CM, Hamilton LR, Gould RW, et al. Mass Spectrometry-Based Proteome Profiling of Extracellular Vesicles Derived from the Cerebrospinal Fluid of Adult Rhesus Monkeys Exposed to Cocaine throughout Gestation. Biomolecules. 2022; 12(4):510. https://doi.org/10.3390/biom12040510
Chicago/Turabian StyleRather, Hilal A., Shalini Mishra, Yixin Su, Ashish Kumar, Sangeeta Singh, Biswapriya B. Misra, Jingyun Lee, Cristina M. Furdui, Lindsey R. Hamilton, Robert W. Gould, and et al. 2022. "Mass Spectrometry-Based Proteome Profiling of Extracellular Vesicles Derived from the Cerebrospinal Fluid of Adult Rhesus Monkeys Exposed to Cocaine throughout Gestation" Biomolecules 12, no. 4: 510. https://doi.org/10.3390/biom12040510
APA StyleRather, H. A., Mishra, S., Su, Y., Kumar, A., Singh, S., Misra, B. B., Lee, J., Furdui, C. M., Hamilton, L. R., Gould, R. W., Nader, S. H., Nader, M. A., & Deep, G. (2022). Mass Spectrometry-Based Proteome Profiling of Extracellular Vesicles Derived from the Cerebrospinal Fluid of Adult Rhesus Monkeys Exposed to Cocaine throughout Gestation. Biomolecules, 12(4), 510. https://doi.org/10.3390/biom12040510






