Hypoxia Alters the Proteome Profile and Enhances the Angiogenic Potential of Dental Pulp Stem Cell-Derived Exosomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Isolation, Culture, and Identification
2.2. Exosome Isolation and Identification
2.3. Exosome Uptake
2.4. Cell Proliferation Assay
2.5. Cell Migration Assay
2.6. Tube Formation Assay
2.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.8. Western Blot Analysis
2.9. Protein Extraction, Digestion and iTRAQ Labeling
2.10. Peptide Fractionation, High-Performance Liquid Chromatography (HPLC), and Mass Spectrometry Analysis
2.11. Bioinformatics Analysis
2.12. Immunofluorescent Analysis
2.13. Immunohistochemical Staining
2.14. LOXL2 Knockdown in HUVECs
2.15. Statistical Analysis
3. Results
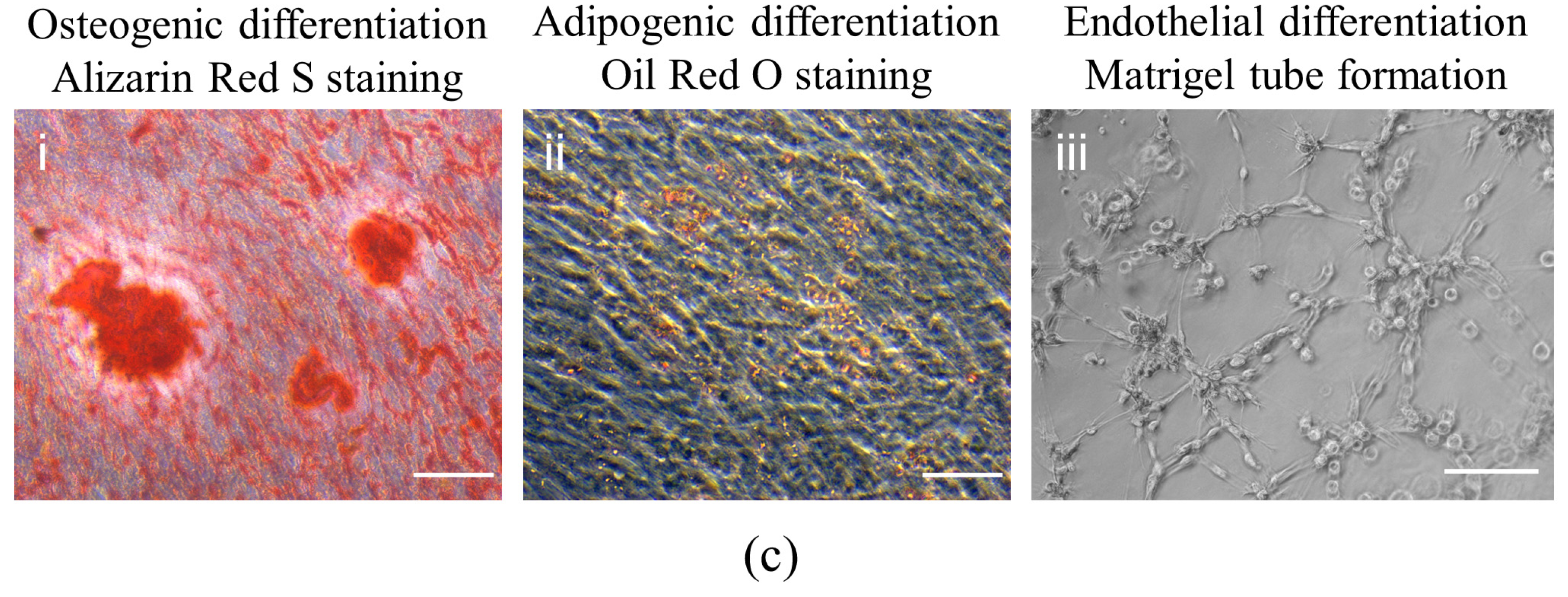
3.1. Isolation and Characterization of DPSCs
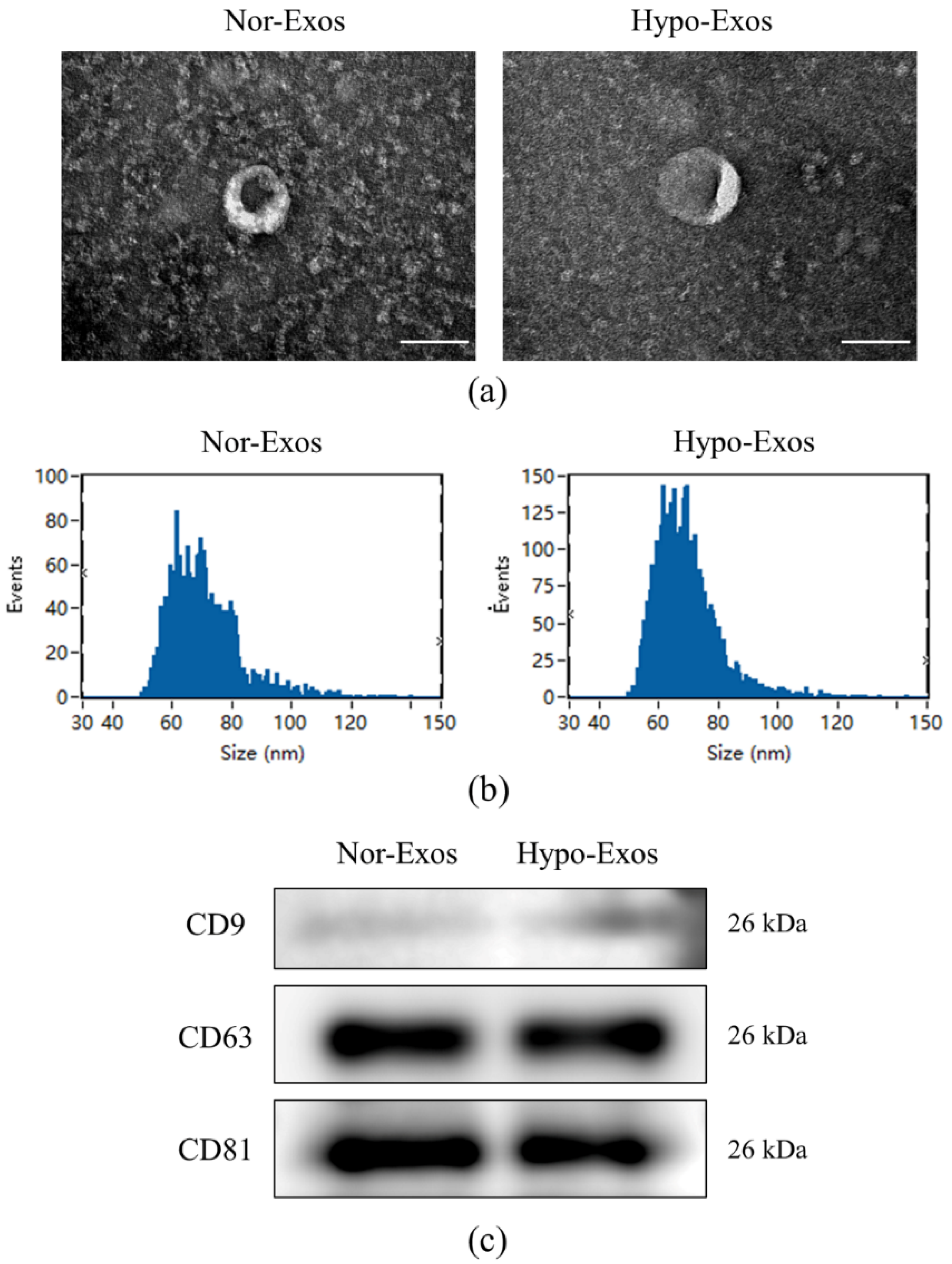
3.2. Isolation and Characteristics of Nor-Exos and Hypo-Exos
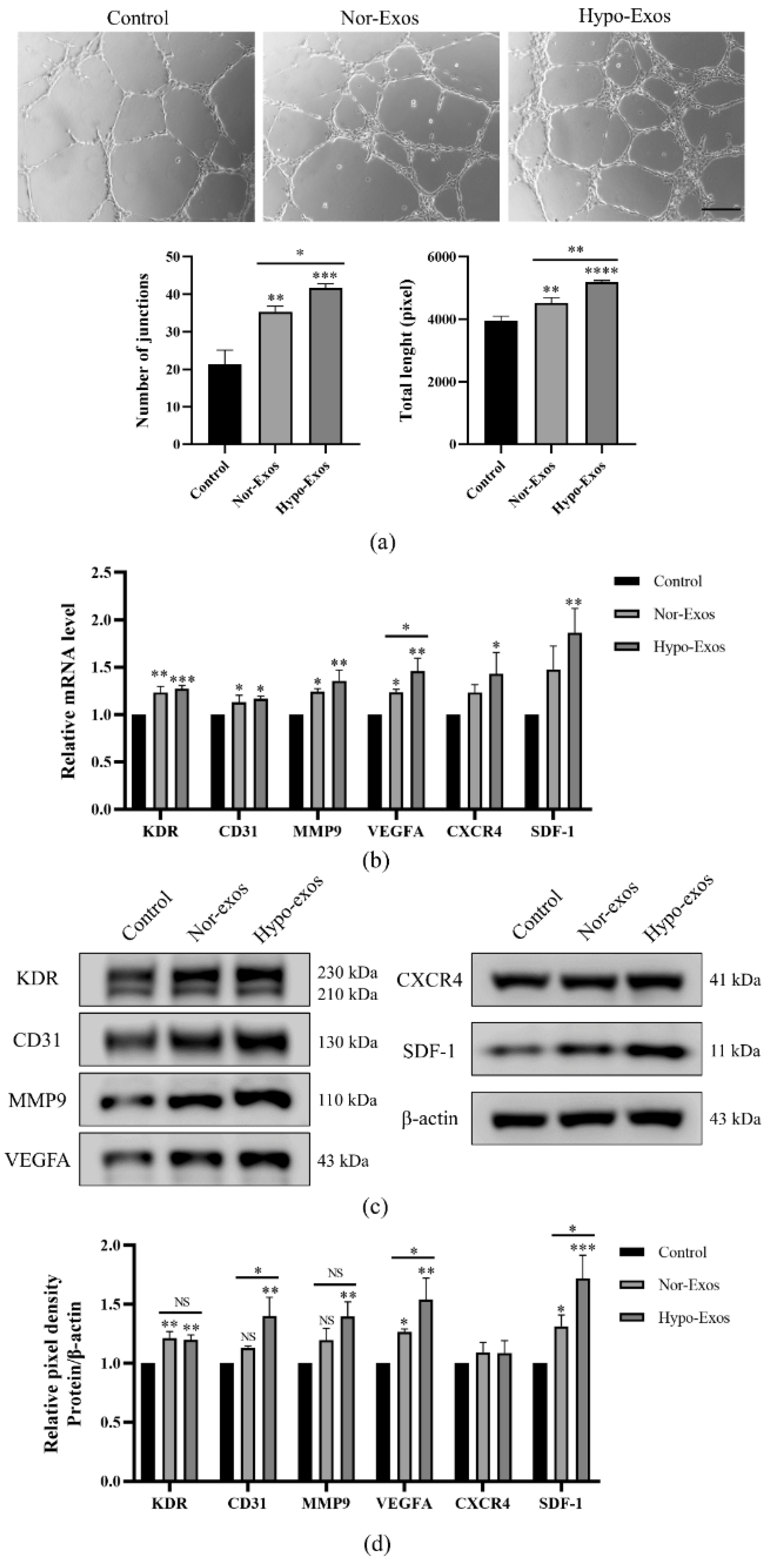
3.3. Hypo-Exos Enhance the Angiogenic Capacity of HUVECs In Vitro
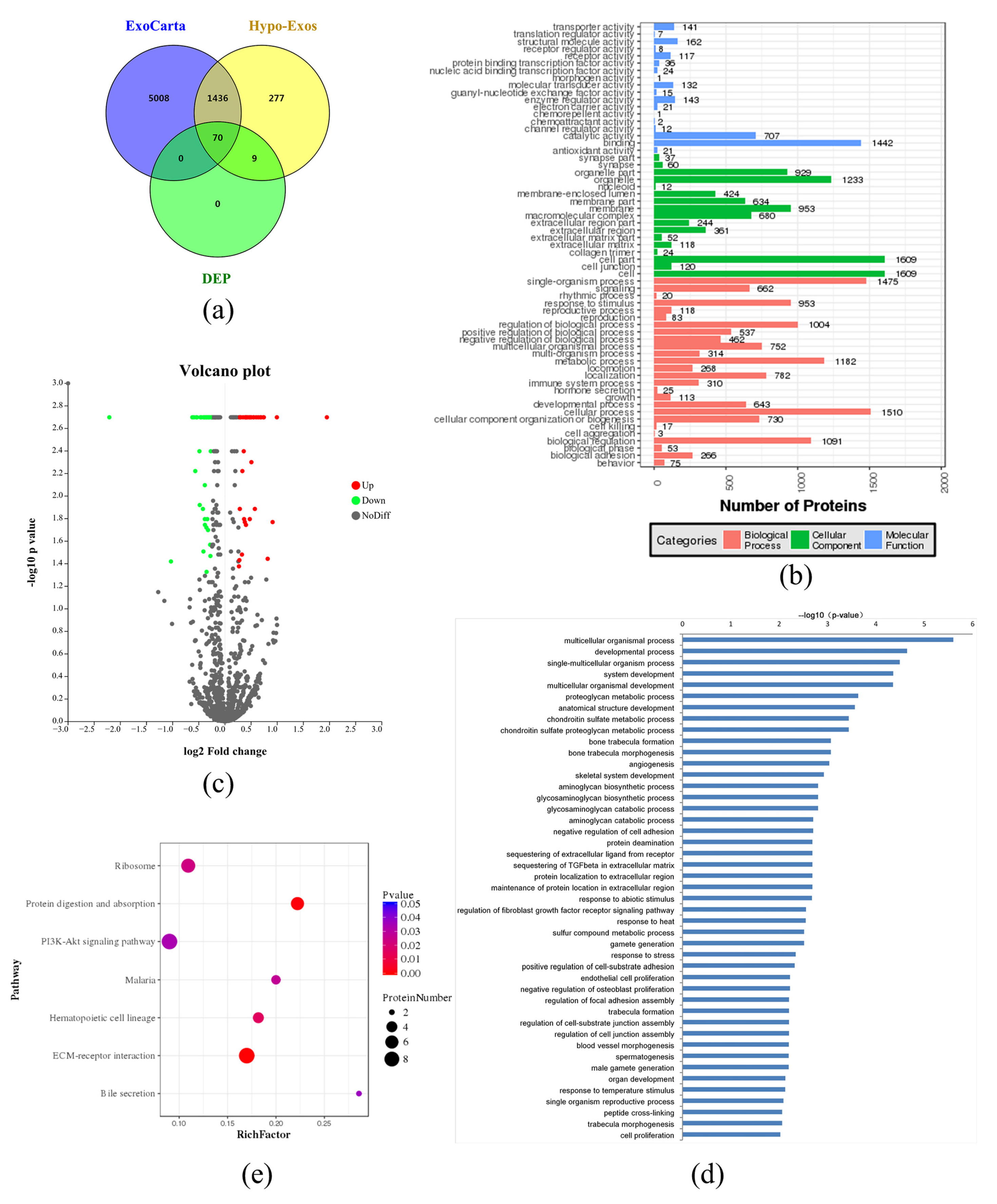
3.4. Exosomal Protein Identification via iTRAQ-Based Proteomics Analysis
3.5. LOXL2 May Be One of the Key Molecules in Hypo-Exos-Mediated Angiogenesis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- França, C.M.; Riggers, R.; Muschler, J.L.; Widbiller, M.; Lococo, P.M.; Diogenes, A.; Bertassoni, L.E. 3D-Imaging of whole neuronal and vascular networks of the human dental pulp via CLARITY and light sheet microscopy. Sci. Rep. 2019, 9, 10860. [Google Scholar] [CrossRef] [Green Version]
- Maqsood, M.; Kang, M.; Wu, X.; Chen, J.; Teng, L.; Qiu, L. Adult mesenchymal stem cells and their exosomes: Sources, characteristics, and application in regenerative medicine. Life Sci. 2020, 256, 118002. [Google Scholar] [CrossRef]
- Guo, H.; Li, B.; Wu, M.; Zhao, W.; He, X.; Sui, B.; Dong, Z.; Wang, L.; Shi, S.; Huang, X.; et al. Odontogenesis-related developmental microenvironment facilitates deciduous dental pulp stem cell aggregates to revitalize an avulsed tooth. Biomaterials 2021, 279, 121223. [Google Scholar] [CrossRef]
- Zhang, S.; Thiebes, A.L.; Kreimendahl, F.; Ruetten, S.; Buhl, E.M.; Wolf, M.; Jockenhoevel, S.; Apel, C. Extracellular vesicles-loaded fibrin gel supports rapid neovascularization for dental pulp regeneration. Int. J. Mol. Sci. 2020, 21, 4226. [Google Scholar] [CrossRef]
- Xian, X.; Gong, Q.; Li, C.; Guo, B.; Jiang, H. Exosomes with highly angiogenic potential for possible use in pulp gegeneration. J. Endod. 2018, 44, 751–758. [Google Scholar] [CrossRef]
- Wu, M.; Liu, X.; Li, Z.; Huang, X.; Guo, H.; Guo, X.; Yang, X.; Li, B.; Xuan, K.; Jin, Y. SHED aggregate exosomes shuttled miR-26a promote angiogenesis in pulp regeneration via TGF-β/SMAD2/3 signalling. Cell Prolif. 2021, 54, e13074. [Google Scholar] [CrossRef]
- Merckx, G.; Hosseinkhani, B.; Kuypers, S.; Deville, S.; Irobi, J.; Nelissen, I.; Michiels, L.; Lambrichts, I.; Bronckaers, A. Angiogenic effects of human dental pulp and bone marrow-derived mesenchymal stromal cells and their extracellular vesicles. Cells 2020, 9, 312. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Qiu, W.; Pan, Y.; Li, J.; Chen, Z.; Zhang, K.; Luo, Y.; Wu, B.; Xu, W. Exosomes from LPS-stimulated hDPSCs activated the angiogenic potential of HUVECs in vitro. Stem Cells Int. 2021, 2021, 6685307. [Google Scholar] [CrossRef]
- Li, J.; Ju, Y.; Liu, S.; Fu, Y.; Zhao, S. Exosomes derived from lipopolysaccharide-preconditioned human dental pulp stem cells regulate Schwann cell migration and differentiation. Connect. Tissue Res. 2021, 62, 277–286. [Google Scholar] [CrossRef]
- Chen, W.J.; Xie, J.; Lin, X.; Ou, M.H.; Zhou, J.; Wei, X.L.; Chen, W.X. The role of small extracellular vesicles derived from lipopolysaccharide-preconditioned human dental pulp stem cells in dental pulp regeneration. J. Endod. 2021, 47, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, X.; Yin, Y.; He, X.T.; An, Y.; Tian, B.M.; Hong, Y.L.; Wu, L.A.; Chen, F.M. The proangiogenic effects of extracellular vesicles secreted by dental pulp stem cells derived from periodontally compromised teeth. Stem Cell. Res. Ther. 2020, 11, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Li, X.; Wu, R.X.; He, X.T.; An, Y.; Xu, X.Y.; Sun, H.H.; Wu, L.A.; Chen, F.M. Periodontitis-compromised dental pulp stem cells secrete extracellular vesicles carrying miRNA-378a promote local angiogenesis by targeting Sufu to activate the Hedgehog/Gli1 signalling. Cell Prolif. 2021, 54, e13026. [Google Scholar] [CrossRef]
- Rombouts, C.; Giraud, T.; Jeanneau, C.; About, I. Pulp vascularization during tooth development, regeneration, and therapy. J. Dent. Res. 2017, 96, 137–144. [Google Scholar] [CrossRef]
- Aranha, A.M.; Zhang, Z.; Neiva, K.G.; Costa, C.A.; Hebling, J.; Nor, J.E. Hypoxia enhances the angiogenic potential of human dental pulp cells. J. Endod. 2010, 36, 1633–1637. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Yan, Q.; Liang, P.; Zhou, P.; Zhang, Y.; Ji, P. iTRAQ-based proteomic analysis exploring the influence of hypoxia on the proteome of dental pulp stem cells under 3D culture. Proteomics 2018, 18, 1700215. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-King, H.; García, N.A.; Ontoria-Oviedo, I.; Ciria, M.; Montero, J.A.; Sepúlveda, P. Hypoxia inducible factor-1α potentiates Jagged 1-mediated angiogenesis by mesenchymal stem cell-derived exosomes. Stem Cells 2017, 35, 1747–1759. [Google Scholar] [CrossRef] [Green Version]
- Dissanayaka, W.L.; Han, Y.; Zhang, L.; Zou, T.; Zhang, C. Bcl-2 Overexpression and hypoxia synergistically enhance angiogenic properties of dental pulp stem cells. Int. J. Mol. Sci. 2020, 21, 6159. [Google Scholar] [CrossRef]
- Xue, C.; Shen, Y.; Li, X.; Li, B.; Zhao, S.; Gu, J.; Chen, Y.; Ma, B.; Wei, J.; Han, Q.; et al. Exosomes derived from hypoxia-treated human adipose mesenchymal stem cells enhance angiogenesis through the PKA signaling pathway. Stem Cells Dev. 2018, 27, 456–465. [Google Scholar] [CrossRef]
- Han, Y.; Ren, J.; Bai, Y.; Pei, X.; Han, Y. Exosomes from hypoxia-treated human adipose-derived mesenchymal stem cells enhance angiogenesis through VEGF/VEGF-R. Int. J. Biochem. Cell Biol. 2019, 109, 59–68. [Google Scholar] [CrossRef]
- Liu, W.; Li, L.; Rong, Y.; Qian, D.; Chen, J.; Zhou, Z.; Luo, Y.; Jiang, D.; Cheng, L.; Zhao, S.; et al. Hypoxic mesenchymal stem cell-derived exosomes promote bone fracture healing by the transfer of miR-126. Acta Biomater. 2020, 103, 196–212. [Google Scholar] [CrossRef]
- Gao, W.; He, R.; Ren, J.; Zhang, W.; Wang, K.; Zhu, L.; Liang, T. Exosomal HMGB1 derived from hypoxia-conditioned bone marrow mesenchymal stem cells increases angiogenesis via the JNK/HIF-1α pathway. FEBS Open Bio. 2021, 11, 1364–1373. [Google Scholar] [CrossRef] [PubMed]
- Yuan, N.; Ge, Z.; Ji, W.; Li, J. Exosomes secreted from hypoxia-preconditioned mesenchymal stem cells prevent steroid-induced osteonecrosis of the femoral head by promoting angiogenesis in rats. BioMed Res. Int. 2021, 2021, 6655225. [Google Scholar] [CrossRef] [PubMed]
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Song, D.; Cao, X.; Wu, R.; Liu, B.; Ye, W.; Wu, J.; Yue, X. Comparative proteomic analysis of milk-derived exosomes in human and bovine colostrum and mature milk samples by iTRAQ-coupled LC-MS/MS. Food Res. Int. 2017, 92, 17–25. [Google Scholar] [CrossRef]
- Deng, Z.; Wang, J.; Xiao, Y.; Li, F.; Niu, L.; Liu, X.; Meng, L.; Zheng, H. Ultrasound-mediated augmented exosome release from astrocytes alleviates amyloid-β-induced neurotoxicity. Theranostics 2021, 11, 4351–4362. [Google Scholar] [CrossRef]
- Schietke, R.; Warnecke, C.; Wacker, I.; Schodel, J.; Mole, D.R.; Campean, V.; Amann, K.; Goppelt-Struebe, M.; Behrens, J.; Eckardt, K.U.; et al. The lysyl oxidases LOX and LOXL2 are necessary and sufficient to repress E-cadherin in hypoxia: Insights into cellular transformation processes mediated by HIF-1. J. Biol. Chem. 2010, 285, 6658–6669. [Google Scholar] [CrossRef] [Green Version]
- De Jong, O.G.; Van der Waals, L.M.; Kools, F.R.W.; Verhaar, M.C.; Van Balkom, B.W.M. Lysyl oxidase-like 2 is a regulator of angiogenesis through modulation of endothelial-to-mesenchymal transition. J. Cell. Physiol. 2019, 234, 10260–10269. [Google Scholar] [CrossRef] [Green Version]
- Zaffryar-Eilot, S.; Marshall, D.; Voloshin, T.; Bar-Zion, A.; Spangler, R.; Kessler, O.; Ghermazien, H.; Brekhman, V.; Suss-Toby, E.; Adam, D.; et al. Lysyl oxidase-like-2 promotes tumour angiogenesis and is a potential therapeutic target in angiogenic tumours. Carcinogenesis 2013, 34, 2370–2379. [Google Scholar]
- Ahmed, N.E.; Murakami, M.; Kaneko, S.; Nakashima, M. The effects of hypoxia on the stemness properties of human dental pulp stem cells (DPSCs). Sci. Rep. 2016, 6, 35476. [Google Scholar] [CrossRef] [Green Version]
- Moore, K.; Lemischka, I. Stem cells and their Niches. Science 2006, 311, 1880–1885. [Google Scholar] [CrossRef] [Green Version]
- Remy, M.; Ferraro, F.; Le Salver, P.; Rey, S.; Genot, E.; Djavaheri-Mergny, M.; Thebaud, N.; Boiziau, C.; Boeuf, H. Isolation and culture of human stem cells from apical papilla under low oxygen concentration highlight original properties. Cells 2019, 8, 1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujio, M.; Xing, Z.; Sharabi, N.; Xue, Y.; Yamamoto, A.; Hibi, H.; Ueda, M.; Fristad, I.; Mustafa, K. Conditioned media from hypoxic-cultured human dental pulp cells promotes bone healing during distraction osteogenesis. J. Tissue Eng. Regen. Med. 2017, 11, 2116–2126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korbecki, J.; Kojder, K.; Kapczuk, P.; Kupnicka, P.; Gawrońska-Szklarz, B.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. The effect of hypoxia on the expression of CXC chemokines and CXC chemokine receptors-A review of literature. Int. J. Mol. Sci. 2021, 22, 843. [Google Scholar] [CrossRef] [PubMed]
- Phinney, D.G.; Pittenger, M.F. Concise Review: MSC-derived exosomes for cell-free therapy. Stem Cells 2017, 35, 851–858. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; He, Y.; Li, L.; Mao, W.; Chen, X.; Ni, H.; Dong, Y.; Lyu, F. Exosomal MMP2 derived from mature osteoblasts promotes angiogenesis of endothelial cells via VEGF/Erk1/2 signaling pathway. Exp. Cell Res. 2019, 383, 111541. [Google Scholar] [CrossRef]
- Rosini, S.; Pugh, N.; Bonna, A.M.; Hulmes, D.J.S.; Farndale, R.W.; Adams, J.C. Thrombospondin-1 promotes matrix homeostasis by interacting with collagen and lysyl oxidase precursors and collagen cross-linking sites. Sci. Signal. 2018, 11, eaar2566. [Google Scholar] [CrossRef] [Green Version]
- Cosgrove, D.; Dufek, B.; Meehan, D.T.; Delimont, D.; Hartnett, M.; Samuelson, G.; Gratton, M.A.; Phillips, G.; MacKenna, D.A.; Bain, G. Lysyl oxidase like-2 contributes to renal fibrosis in Col4α3/Alport mice. Kidney Int. 2018, 94, 303–314. [Google Scholar] [CrossRef]
- Yi, X.; Zhou, Y.; Chen, Y.; Feng, X.; Liu, C.; Jiang, D.S.; Geng, J.; Li, X.; Jiang, X.; Fang, Z.M. The expression patterns and roles of lysyl oxidases in aortic dissection. Front. Cardiovasc. Med. 2021, 8, 692856. [Google Scholar] [CrossRef]
- Wang, Z.; Shi, Y.; Ying, C.; Jiang, Y.; Hu, J. Hypoxia-induced PLOD1 overexpression contributes to the malignant phenotype of glioblastoma via NF-κB signaling. Oncogene 2021, 40, 1458–1475. [Google Scholar] [CrossRef]
- Ezzoukhry, Z.; Henriet, E.; Piquet, L.; Boyé, K.; Bioulac-Sage, P.; Balabaud, C.; Couchy, G.; Zucman-Rossi, J.; Moreau, V.; Saltel, F. TGF-β1 promotes linear invadosome formation in hepatocellular carcinoma cells, through DDR1 up-regulation and collagen I cross-linking. Eur. J. Cell Biol. 2016, 95, 503–512. [Google Scholar] [CrossRef]
- Sansilvestri-Morel, P.; Harouki-Crochemore, N.; Bertin, F.; Bertheux, H.; Vermeil de Conchard, G.; Diguet, N.; Desfosses, E.; Lecomte, M.; Gonzalez, A.; Diez, J.; et al. Deficiency of procollagen C-proteinase enhancer 1 in mice has no major impact on cardiac collagen and function under basal conditions. J. Cardiovasc. Pharmacol. 2021, 78, e703–e713. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.J.; Peters, D.G. Serial analysis of the vascular endothelial transcriptome under static and shear stress conditions. Physiol. Genom. 2008, 34, 185–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bignon, M.; Pichol-Thievend, C.; Hardouin, J.; Malbouyres, M.; Brechot, N.; Nasciutti, L.; Barret, A.; Teillon, J.; Guillon, E.; Etienne, E.; et al. Lysyl oxidase-like protein-2 regulates sprouting angiogenesis and type IV collagen assembly in the endothelial basement membrane. Blood 2011, 118, 3979–3989. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Zheng, W.; Li, H.; Wu, W.; Liu, X.; Sun, Z.; Hu, H.; Du, L.; Jia, Q.; Liu, Q. LOXL2 upregulates hypoxia-inducible factor-1α signaling through Snail-FBP1 axis in hepatocellular carcinoma cells. Oncol. Rep. 2020, 43, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, H.; Zhu, L.; Zhang, X.; Liu, D.; Li, Q.; Ni, B.; Hu, L.; Zhang, Z.; Zhang, Y.; et al. Reciprocal regulation of LOXL2 and HIF1α drives the Warburg effect to support pancreatic cancer aggressiveness. Cell Death Dis. 2021, 12, 1106. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Doyle, L.M.; Wang, M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Gong, M.; Yu, B.; Wang, J.; Wang, Y.; Liu, M.; Paul, C.; Millard, R.W.; Xiao, D.S.; Ashraf, M.; Xu, M. Mesenchymal stem cells release exosomes that transfer miRNAs to endothelial cells and promote angiogenesis. Oncotarget 2017, 8, 45200–45212. [Google Scholar] [CrossRef] [Green Version]



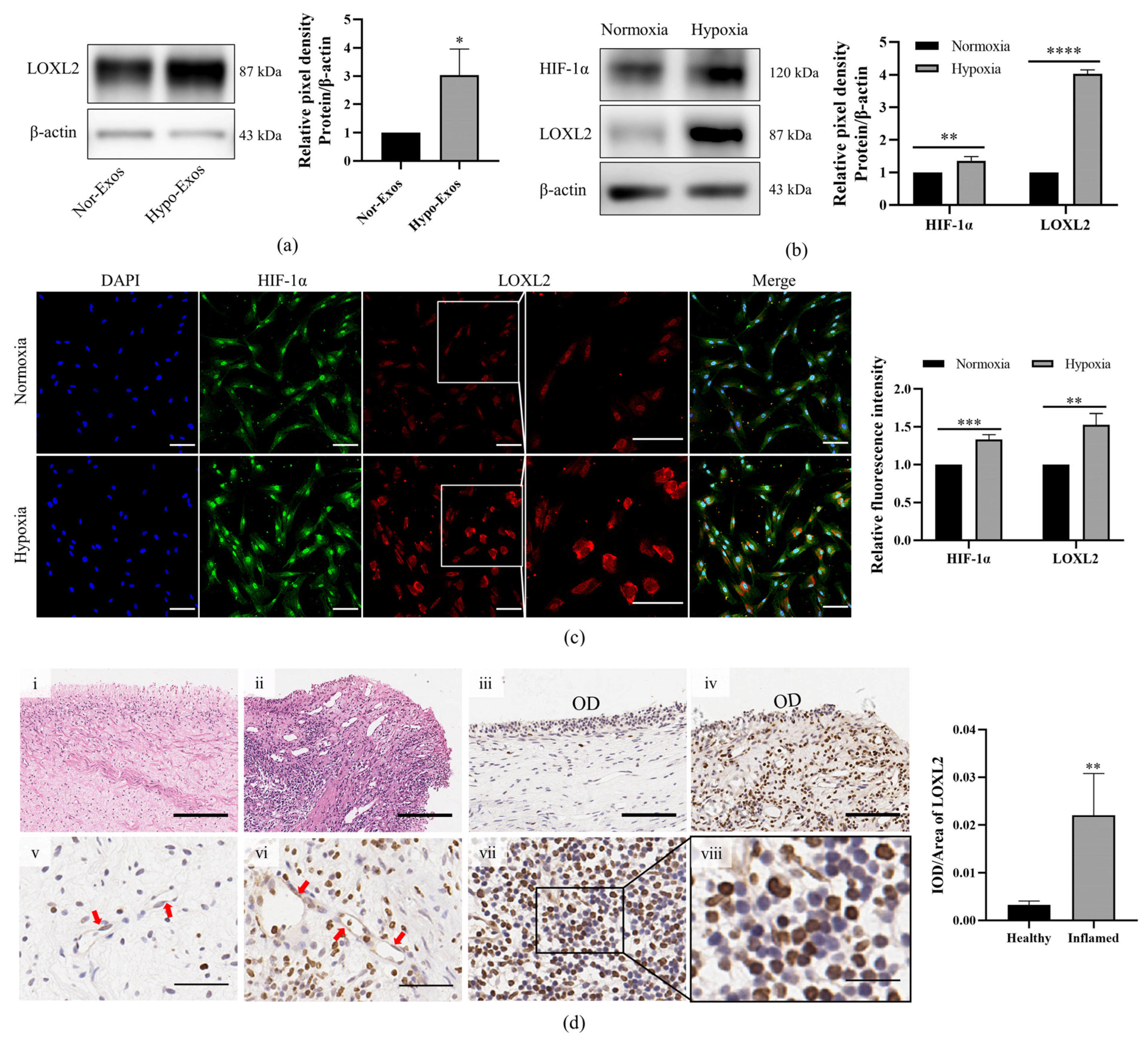
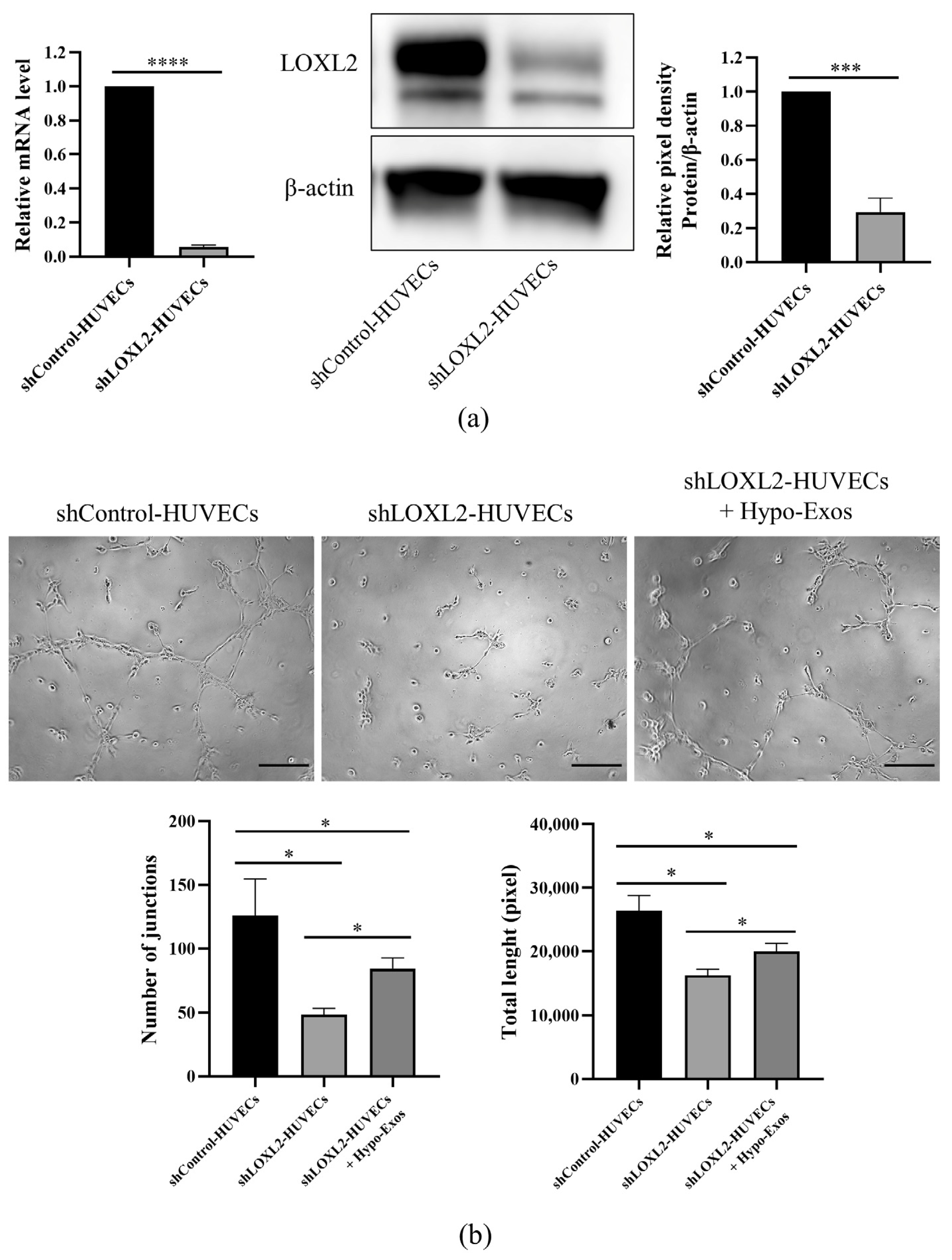
| Protein ID | Protein Name | Abbreviation | Ratio | p-Value | Expression |
|---|---|---|---|---|---|
| Q4LDE5 | Sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1 | SVEP1 | 1.666 | 0.002 | up |
| Q99715 | Collagen alpha-1(XII) chain | COL12A1 | 1.675 | 0.002 | up |
| P07585 | Decorin | DCN | 1.554 | 0.002 | up |
| Q8N474 | Secreted frizzled-related protein 1 | SFRP1 | 1.283 | 0.004 | up |
| Q08397 | Lysyl oxidase homolog 1 | LOXL1 | 1.311 | 0.002 | up |
| P07996 | Thrombospondin-1 | THBS1 | 1.306 | 0.002 | up |
| P02743 | Serum amyloid p-component | APCS | 1.325 | 0.002 | up |
| P06748 | Nucleophosmin | NPM1 | 1.991 | 0.002 | up |
| P62753 | 40S ribosomal protein S6 | RPS6 | 1.218 | 0.013 | up |
| Q9Y383 | Putative RNA-binding protein Luc7-like 2 | LUC7L2 | 1.39 | 0.016 | up |
| P11166 | Solute carrier family 2, facilitated glucose transporter member 1 | SLC2A1 | 1.272 | 0.002 | up |
| Q9Y4K0 | Lysyl oxidase homolog 2 | LOXL2 | 1.615 | 0.002 | up |
| P31431 | Syndecan-4 | SDC4 | 1.487 | 0.013 | up |
| P62269 | 40S ribosomal protein S18 | RPS18 | 1.211 | 0.037 | up |
| P26373 | 60S ribosomal protein L13 | RPL13 | 1.302 | 0.017 | up |
| P12110 | Collagen alpha-2(VI) chain | COL6A2 | 1.371 | 0.002 | up |
| Q8IUX7 | Adipocyte enhancer-binding protein 1 | AEBP1 | 1.335 | 0.002 | up |
| Q99613 | Eukaryotic translation initiation factor 3 subunit C | EIF3C | 1.289 | 0.016 | up |
| P27635 | 60S ribosomal protein L10 | RPL10 | 1.321 | 0.018 | up |
| P00736 | Complement C1r subcomponent | C1R | 1.317 | 0.002 | up |
| Q15582 | Transforming growth factor-beta-induced protein ig-h3 | TGFBI | 1.459 | 0.002 | up |
| P35555 | Fibrillin-1 | FBN1 | 1.595 | 0.002 | up |
| P19338 | Nucleolin | NCL | 1.343 | 0.002 | up |
| Q2TAA8 | Translin-associated factor X-interacting protein 1 | TSNAXIP1 | 3.862 | 0.002 | up |
| P61247 | 40S ribosomal protein S3a | RPS3A | 1.26 | 0.006 | up |
| P02545 | Prelamin-A/C | LMNA | 1.204 | 0.042 | up |
| P35442 | Thrombospondin-2 | THBS2 | 1.316 | 0.002 | up |
| Q02809 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | PLOD1 | 1.216 | 0.002 | up |
| P08253 | 72 kDa type IV collagenase | MMP2 | 1.399 | 0.002 | up |
| Q92620 | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 | DHX38 | 1.762 | 0.036 | up |
| P06576 | ATP synthase subunit beta, mitochondrial | ATP5B | 1.221 | 0.002 | up |
| P35556 | Fibrillin-2 | FBN2 | 1.513 | 0.002 | up |
| P21810 | Biglycan | BGN | 1.881 | 0.017 | up |
| Q96KK5 | Histone H2A type 1-H | HIST1H2AH | 1.253 | 0.033 | up |
| P62851 | 40S ribosomal protein S25 | RPS25 | 1.42 | 0.005 | up |
| P62241 | 40S ribosomal protein S8 | RPS8 | 1.392 | 0.002 | up |
| P12111 | Collagen alpha-3(VI) chain | COL6A3 | 1.465 | 0.002 | up |
| P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein | HSPG2 | 1.234 | 0.002 | up |
| P12109 | Collagen alpha-1(VI) chain | COL6A1 | 1.418 | 0.002 | up |
| P35908 | Keratin, type II cytoskeletal 2 epidermal | KRT2 | 0.725 | 0.002 | down |
| P46013 | Proliferation marker protein Ki-67 | MKI67 | 0.489 | 0.038 | down |
| P31689 | DnaJ homolog subfamily A member 1 | DNAJA1 | 0.822 | 0.027 | down |
| P08473 | Neprilysin | MME | 0.757 | 0.002 | down |
| P09382 | Galectin-1 | LGALS1 | 0.797 | 0.002 | down |
| P01591 | Immunoglobulin J chain | JCHAIN | 0.713 | 0.004 | down |
| P0DOX8 | Immunoglobulin lambda-1 light chain | IGL1 | 0.768 | 0.002 | down |
| P02647 | Apolipoprotein A-I | APOA1 | 0.812 | 0.002 | down |
| P13645 | Keratin, type I cytoskeletal 10 | KRT10 | 0.663 | 0.002 | down |
| P20742 | Pregnancy zone protein | PZP | 0.767 | 0.018 | down |
| P13647 | Keratin, type II cytoskeletal 5 | KRT5 | 0.784 | 0.002 | down |
| O14786 | Neuropilin-1 | NRP1 | 0.807 | 0.002 | down |
| P00441 | Superoxide dismutase [Cu-Zn] | SOD1 | 0.825 | 0.034 | down |
| P24821 | Tenascin | TNC | 0.755 | 0.002 | down |
| Q9Y625 | Glypican-6 | GPC6 | 0.766 | 0.008 | down |
| P29144 | Tripeptidyl-peptidase 2 | TPP2 | 0.675 | 0.006 | down |
| Q14432 | cGMP-inhibited 3′,5′-cyclic phosphodiesterase A | PDE3A | 0.216 | 0.002 | down |
| P04264 | Keratin, type II cytoskeletal 1 | KRT1 | 0.684 | 0.002 | down |
| P13987 | CD59 glycoprotein | CD59 | 0.781 | 0.002 | down |
| Q14344 | Guanine nucleotide-binding protein subunit alpha-13 | GNA13 | 0.799 | 0.02 | down |
| P69891 | Hemoglobin subunit gamma-1 | HBG1 | 0.76 | 0.002 | down |
| Q8N339 | Metallothionein-1M | MT1M | 0.716 | 0.012 | down |
| Q9Y639 | Neuroplastin | NPTN | 0.812 | 0.002 | down |
| O43866 | CD5 antigen-like | CD5L | 0.782 | 0.019 | down |
| Q9ULC3 | Ras-related protein Rab-23 | RAB23 | 0.792 | 0.016 | down |
| Q99536 | Synaptic vesicle membrane protein VAT-1 homolog | VAT1 | 0.829 | 0.004 | down |
| Q15113 | Procollagen C-endopeptidase enhancer 1 | PCOLCE | 0.649 | 0.002 | down |
| Q06830 | Peroxiredoxin-1 | PRDX1 | 0.766 | 0.016 | down |
| P10599 | Thioredoxin | TXN | 0.723 | 0.002 | down |
| Q8N883 | Zinc finger protein 614 | ZNF614 | 0.807 | 0.002 | down |
| Q8IWU5 | Extracellular sulfatase Sulf-2 | SULF2 | 0.752 | 0.031 | down |
| Q13449 | Limbic system-associated membrane protein | LSAMP | 0.784 | 0.047 | down |
| P04220 | Ig mu heavy chain disease protein | IGHM | 0.691 | 0.002 | down |
| P35527 | Keratin, type I cytoskeletal 9 | KRT9 | 0.733 | 0.002 | down |
| O43854 | EGF-like repeat and discoidin I-like domain-containing protein 3 | EDIL3 | 0.783 | 0.002 | down |
| P01023 | Alpha-2-macroglobulin | A2M | 0.779 | 0.002 | down |
| P54709 | Sodium/potassium-transporting ATPase subunit beta-3 | ATP1B3 | 0.747 | 0.013 | down |
| P26006 | Integrin alpha-3 | ITGA3 | 0.81 | 0.002 | down |
| P60033 | CD81 antigen | CD81 | 0.758 | 0.002 | down |
| P11142 | Heat shock cognate 71 kDa protein | HSPA8 | 0.828 | 0.002 | down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, B.; Xian, X.; Lin, X.; Huang, L.; Liang, A.; Jiang, H.; Gong, Q. Hypoxia Alters the Proteome Profile and Enhances the Angiogenic Potential of Dental Pulp Stem Cell-Derived Exosomes. Biomolecules 2022, 12, 575. https://doi.org/10.3390/biom12040575
Li B, Xian X, Lin X, Huang L, Liang A, Jiang H, Gong Q. Hypoxia Alters the Proteome Profile and Enhances the Angiogenic Potential of Dental Pulp Stem Cell-Derived Exosomes. Biomolecules. 2022; 12(4):575. https://doi.org/10.3390/biom12040575
Chicago/Turabian StyleLi, Baoyu, Xuehong Xian, Xinwei Lin, Luo Huang, Ailin Liang, Hongwei Jiang, and Qimei Gong. 2022. "Hypoxia Alters the Proteome Profile and Enhances the Angiogenic Potential of Dental Pulp Stem Cell-Derived Exosomes" Biomolecules 12, no. 4: 575. https://doi.org/10.3390/biom12040575
APA StyleLi, B., Xian, X., Lin, X., Huang, L., Liang, A., Jiang, H., & Gong, Q. (2022). Hypoxia Alters the Proteome Profile and Enhances the Angiogenic Potential of Dental Pulp Stem Cell-Derived Exosomes. Biomolecules, 12(4), 575. https://doi.org/10.3390/biom12040575





