Molecular Recognition of Proteins through Quantitative Force Maps at Single Molecule Level
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Labeling and Covalent Immobilization on Mica
2.2. HRP–Biotin Enzymatic Assays
2.3. Topography and Adhesion AFM Mapping Analysis
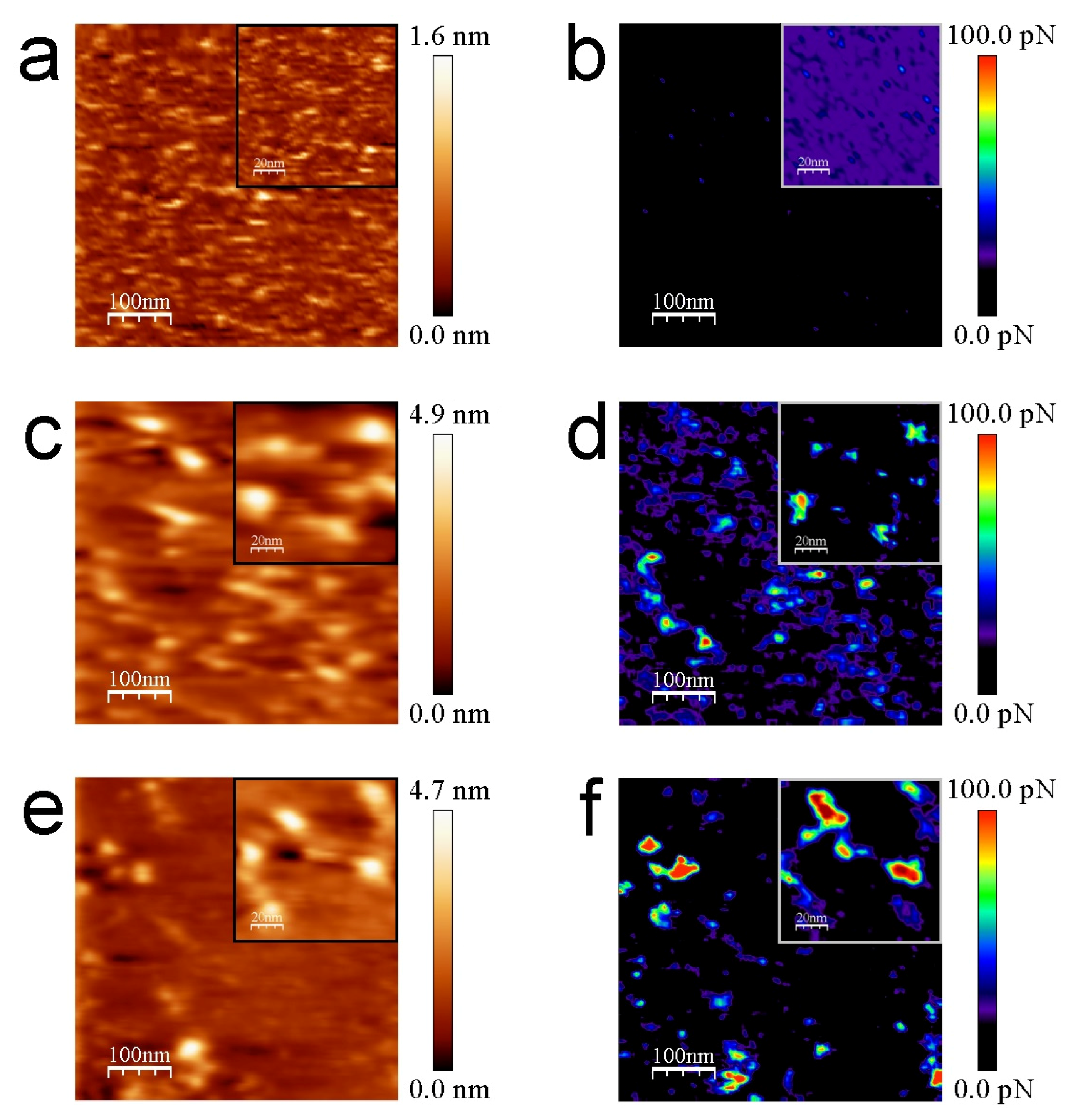
3. Results
3.1. HRP–Biotin Enzymatic Assays
3.2. AFM Measurements
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Binnig, G.; Rohrer, H. Scanning tunneling microscopy-from birth to adolescence. Rev. Mod. Phys. 1987, 59, 615. [Google Scholar] [CrossRef]
- Gross, L.; Moll, N.; Mohn, F.; Curioni, A.; Meyer, G.; Hanke, F.; Persson, M. High Resolution Molecular Orbital Imaging Using a ρ-Waste STM tip. Phys. Rev. Lett. 2011, 107, 086101. [Google Scholar] [CrossRef] [PubMed]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic Force Microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Gaub, H.; Newton, R.; Pfreundschuh, M.; Gerber, C.; Müller, D.J. Atomic force microscopy-based characterization and design of biointerfaces. Nat. Rev. Mater. 2017, 2, 17008. [Google Scholar] [CrossRef]
- Sugimoto, Y.; Pou, P.; Abe, M.; Jelinek, P.; Perez, R.; Morita, S.; Custance, O. Chemical Identification of individual surface atoms by atomic force microscopy. Nature 2007, 446, 64–67. [Google Scholar] [CrossRef]
- Müller, D.J.; Dufrêne, Y.F. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat. Nanotechnol. 2008, 3, 261–269. [Google Scholar] [CrossRef]
- Müller, D.J.; Dumitru, A.C.; Lo Giudice, C.; Gaub, H.E.; Hinterdorfer, P.; Hummer, G.; De Yoreo, J.J.; Alsteens, D. Atomic Force Microscopy-Based Force Spectroscopy and Multiparametric Imaging of Biomolecular and Cellular Systems. Chem. Rev. 2020, 121, 11701–11725. [Google Scholar] [CrossRef]
- Clausen- Schaumann, H.; Seitz, M.; Krautbauer, R.; Gaub, H.E. Force spectroscopy with single bio-molecules. Curr. Opin. Chem. Biol. 2000, 4, 524–530. [Google Scholar] [CrossRef]
- Hinterdorfer, P.; Dufrêne, Y.F. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods 2006, 3, 347–355. [Google Scholar] [CrossRef]
- Evans, E.; Ritchie, K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997, 72, 1541–1555. [Google Scholar] [CrossRef]
- Evans, E. Probing the Relation between Force-Lifetime-and Chemistry in Single Molecular Bonds. Annu. Rev. Biophys. Biomol. Struct. 2011, 30, 105–128. [Google Scholar] [CrossRef] [PubMed]
- Tapia-Rojo, R.; Marcuello, C.; Lostao, A.; Gómez-Moreno, C.; Mazo, J.J.; Falo, F. A physical picture for mechanical dissociation of biological complexes: From forces to free-energies. Phys. Chem. Chem. Phys. 2017, 19, 4567–4575. [Google Scholar] [CrossRef] [PubMed]
- Dammer, U.; Hegner, M.; Anselmetti, D.; Wagner, P.; Dreier, H.; Huber, W.; Guntherodt, H.H. Specific antigen/antibody interactions measured by force microscopy. Biophys J. 1996, 70, 2437–2441. [Google Scholar] [CrossRef]
- Wang, C.; Hu, R.; Morrissey, J.J.; Kharasch, E.D.; Singamaneni, S. Single Molecule Force Spectroscopy to Compare Natural versus Artificial Antibody-Antigen Interaction. Small 2017, 13, 1604255. [Google Scholar] [CrossRef]
- Ratto, T.V.; Langry, K.C.; Rudd, R.E.; Balhom, R.L.; Allen, M.J.; McElfresh, M.W. Force Spectroscopy of the Double Tethered Concanavalin-A Mannose Bond. Biophysical J. 2004, 86, 2430–2437. [Google Scholar] [CrossRef]
- Li, F.; Redick, S.D.; Erickson, H.P.; Moy, V.T. Force Measurements of the α5β1 Integrin-Fibronectin Interaction. Biophys. J. 2003, 84, 1252–1262. [Google Scholar] [CrossRef]
- Sewald, N.; Wilking, S.D.; Eckel, R.; Albu, S.; Wollschläger, K.; Gaus, K.; Becker, A.; Bartels, F.W.; Ros, R.; Anselmetti, D.J. Probing DNA-peptide interaction forces at the single molecule level. Pept. Sci. 2006, 12, 836–842. [Google Scholar] [CrossRef]
- Marcuello, C.; Foulon, L.; Chabbert, B.; Aguié-Béguin, V.; Molinari, M. Atomic force microscopy reveals how relative humidity impacts the Young’s modulus of lignocellulosic polymers and their adhesion with cellulose nanocrystals at the nanoscale. Int. J. Biol. Macromol. 2020, 147, 1064–1075. [Google Scholar] [CrossRef]
- Marcuello, C.; de Miguel, R.; Martínez-Júlvez, M.; Gómez-Moreno, C.; Lostao, A. Mechanostability of the Single Electron-Transfer Complexes of Anabaena Ferredoxin-NADP+ Reductase. ChemPhysChem 2015, 16, 3161–3169. [Google Scholar] [CrossRef]
- Pérez-Dominguez, S.; Caballero-Mancebo, S.; Marcuello, C.; Martínez-Júlvez, M.; Medina, M.; Lostao, A. Nanomechanical Study of Enzyme: Coenzyme Complexes: Bipartite Sites in Plastidic Ferredoxin-NADP+ Reductase for the Interaction with NADP+. Antioxidants 2022, 11, 537. [Google Scholar] [CrossRef]
- Turner, A.P.F. Biosensors—Sense and Sensitivity. Science 2000, 290, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, M.; Dettmann, W.; Gaub, H.E. Atomic force microscope imaging contrast based on molecular recognition. Biophys. J. 1997, 72, 445–448. [Google Scholar] [CrossRef]
- Stroh, C.M.; Ebner, A.; Geretschlager, M.; Freudenthale, G.; Kienberger, F.; Kamruzzahan, A.S.M.; Smith-Gill, S.J.; Gruber, H.J.; Hinterdorfer, P. Simultaneous topography and recognition imaging using force microscopy. Biophys. J. 2004, 87, 1981–1990. [Google Scholar] [CrossRef] [PubMed]
- Chtcheglova, L.A.; Hinterdorfer, P. Simultaneous AFM topography and recognition imaging at the plasma membrane of mammalian cells. Semin. Cell Dev. Biol. 2018, 73, 45–56. [Google Scholar] [CrossRef]
- Hofer, M.; Adamsmaier, S.; van Zanten, T.S.; Chtcheglova, L.A.; Manzo, C.; Duman, M.; Mayer, B.; Ebner, A.; Moertelmaier, M.; Kada, G.; et al. Molecular recognition imaging using tuning-fork based transverse dynamic force microscopy. Ultramicroscopy 2010, 110, 605–611. [Google Scholar] [CrossRef]
- De Pablo, P.J.; Colchero, J.; Gómez-Herrero, J.; Baró, A.M. Jumping mode scanning force microscopy. Appl. Phys. Lett. 1998, 73, 3300. [Google Scholar] [CrossRef]
- Borisov, S.M.; Wolfbeis, O.S. Optical Biosensors. Chem. Rev. 2008, 108, 423–461. [Google Scholar] [CrossRef]
- Cooper, M.A. Optical biosensors in drug discovery. Nat. Rev. Drug Discov. 2002, 1, 515–528. [Google Scholar] [CrossRef]
- Snider, J.; Kotiyar, M.; Saraon, P.; Yao, Z.; Jurisica, I.; Stagijar, I. Fundamentals of protein interaction network mapping. Mol. Syst. Biol. 2015, 11, 848. [Google Scholar] [CrossRef]
- Sevecka, M.; MacBeath, G. State-based discovery: A multidimensional screen for small-molecule modulators of EGF signaling. Nat. Methods 2006, 3, 825–831. [Google Scholar] [CrossRef][Green Version]
- Andrei, S.A.; Sijbesma, E.; Hann, M.; Davisi, J.; O’Mahony, G.; Perry, M.W.D.; Karawajcyz, A.; Eickhoff, J.; Brunsveld, L.; Doveston, R.G.; et al. Stabilization of protein-protein interactions in drug discovery. Expert Opin. Drug Discov. 2017, 12, 925–940. [Google Scholar] [CrossRef] [PubMed]
- Marcuello, C.; de Miguel, R.; Gómez-Moreno, C.; Martínez-Júlvez, M.; Lostao, A. An efficient method for enzyme immobilization evidenced by atomic force microscopy. Protein Eng. Des. Sel. 2012, 25, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Josephy, P.D.; Eling, T.; Mason, R.P. The horseradish peroxidase-catalyzed oxidation of 3,5,3’,5´-tetramethylbenzidine. Free radicals and charge-transfer complex intermediates. J. Biol. Chem. 1982, 257, 3669–3675. [Google Scholar] [CrossRef]
- Meinander, K.; Jensen, T.N.; Simonsen, S.B.; Helveg, S.; Lauritsen, J.V. Quantification of tip-broadening in non-contact atomic force microscopy with carbon nanotube tips. Nanotechnology 2012, 23, 405705. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Jiang, Z.; Jin, W.; Prewett, P.D.; Jiang, K. Estimation of AFM Tip Shape and Status in Linewidth and Profile Measurement. J. Nanosci. Nanotechnol. 2011, 11, 11041–11044. [Google Scholar] [CrossRef] [PubMed]
- Lostao, A.; Peleato, M.L.; Gómez-Moreno, C.; Fillat, M.F. Oligomerization properties of FurA from the cyanobacterium Anabaena sp. PCC 7120: Direct visualization by in situ atomic force microscopy under different redox conditions. BBA-Proteins Proteom. 2010, 1804, 1723–1729. [Google Scholar] [CrossRef]
- Sotres, J.; Lostao, A.; Gómez-Moreno, C.; Baró, A.M. Jumping mode AFM imaging of biomolecules in the repulsive electrical double layer. Ultramicroscopy 2007, 107, 1207–1212. [Google Scholar] [CrossRef]
- Sotres, J.; Lostao, A.; Wildling, L.; Ebner, A.; Gómez-Moreno, C.; Gruber, H.J.; Hinterdorfer, P.; Baró, A.M. Unbinding Molecular Recognition Force Maps of Localized Single Receptor Molecules by Atomic Force Microscopy. ChemPhysChem 2008, 9, 590–599. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodríguez, J.M.; Colchero, J.; Gómez-Herrero, J.; Baró, A.M. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef]
- Valle-Delgado, J.J.; Molina-Bolivar, J.A.; Galisteo-González, F.; Gálvez-Ruiz, M.J.; Feiler, A.; Rutland, M.W. Interaction Forces between BSA layers Adsorbed on Silica Surfaces Measured with an Atomic Force Microscope. J. Phys. Chem. B 2004, 108, 5365–5371. [Google Scholar] [CrossRef]
- Green, N.M.; Joynson, M.A. A preliminary crystallographic investigation of avidin. Biochem. J. 1970, 118, 71–72. [Google Scholar] [CrossRef] [PubMed]
- Pähler, A.; Hendrickson, W.A.; Kolks, M.A.; Argaraña, C.E.; Cantor, C.R. Characterization and crystallization of core streptavidin. J. Biol. Chem. 1987, 262, 13933–13937. [Google Scholar] [CrossRef]
- Weber, P.C.; Cox, M.J.; Salemme, F.R.; Ohlendorf, D.H. Crystallographic data for Streptomyces avidinii streptavidin. J. Biol. Chem. 1987, 262, 12728–12729. [Google Scholar] [CrossRef]
- Neish, C.S.; Martin, I.L.; Henderson, R.M.; Edwardson, J.M. Direct visualization of ligand-protein interactions using atomic force microscopy. Br. J. Pharmacol. 2002, 135, 1943–1950. [Google Scholar] [CrossRef]
- Wang, Y.; Ran, S.; Yang, G. Single molecular investigation of DNA looping and aggregation by restriction endonuclease BspMI. Sci. Rep. 2014, 4, 5897. [Google Scholar] [CrossRef]
- Gad, M.; Mizutani, W.; Machida, M.; Ishikawa, M. Method for stretching DNA molecules on mica surface in one direction for AFM imaging. Nucleic Acids Symp. Ser. 2000, 44, 215–216. [Google Scholar] [CrossRef]
- Teulon, J.M.; Delcuze, Y.; Odorico, M.; Chen, S.W.; Parot, P.; Pellequer, J.L. Single and multiple bonds in (strept)avidin-biotin interactions. J. Mol. Recognit. 2011, 24, 490–502. [Google Scholar] [CrossRef]
- Almonte, L.; Lopez-Elvira, E.; Baró, A.M. Surface-Charge Differentiation of Streptavidin and Avidin by Atomic Force Microscopy-Force Spectroscopy. ChemPhysChem 2014, 15, 2768–2773. [Google Scholar] [CrossRef]
- Rico, F.; Russek, A.; González, L.; Grubmüller, H.; Scheuring, S. Heterogeneous and rate-dependent streptavidin-biotin unbinding revealed by high-speed force spectroscopy and atomistic simulations. PNAS 2019, 116, 6594–6601. [Google Scholar] [CrossRef]
- de Odrowaz-Piramowicz, M.; Czuba, P.; Targosz, M.; Burda, K.; Szymonski, M. Dynamic force measurements of avidin-biotin and streptavidin-biotin interactions using AFM. Acta Biochim. Pol. 2006, 53, 93–100. [Google Scholar] [CrossRef]
- Frauenfelder, H.; Sligar, S.G.; Wolynes, P.G. The energy landscapes and motions of proteins. Science 1991, 254, 1598–1603. [Google Scholar] [CrossRef] [PubMed]
- Rico, F.; Moy, V.T. Energy landscape roughness of the streptavidin-biotin interaction. J. Mol. Recognit. 2007, 20, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Köhler, M.; Karner, A.; Leitner, M.; Hytönen, V.P.; Kulomaa, M.; Hinterdorfer, P.; Ebner, A. pH-Dependent Deformations of the Energy Landscape of Avidin-like Proteins Investigated by Single Molecule Force Spectroscopy. Molecules 2014, 19, 12531–12546. [Google Scholar] [CrossRef] [PubMed]
- Walton, E.B.; Lee, S.; van Vliet, K.J. Extending Bell´s Model: How Force Transducer Stiffness Alters Measured Unbinding Forces and Kinetics of Molecular Complexes. Biophys. J. 2008, 94, 2621–2630. [Google Scholar] [CrossRef]
- Wilcheck, M.; Bayer, E.A.; Livhan, O. Essentials of biorecognition: The strept(avidin)-biotin system as a model for protein-protein and protein-ligand interactions. Immunol. Lett. 2006, 103, 27–32. [Google Scholar] [CrossRef]
- Bonanni, A.; Pividori, M.I.; Del Valle, M. Application of the avidin-biotin interaction to immobilize DNA in the development of electrochemical impedance genosensors. Anal. Bioanal. Chem. 2007, 389, 851–861. [Google Scholar] [CrossRef]
- Morpurgo, M.; Facchin, S.; Pignato, M.; Silvestri, D.; Casarin, E.; Realdon, N. Characterization of multifunctional nanosystems based on the avidin-nucleic acid interaction as signal enhancers of immune-detection. Anal. Chem. 2012, 54, 3433–3439. [Google Scholar] [CrossRef]
- Pignatto, M.; Realdon, N.; Morpurgo, M. Optimized Avidin Nucleic Acid Nanoassemblies by a Tailored PEGylation Strategy and Their Application as Molecular Amplifiers in Detection. Bioconjug. Chem. 2010, 21, 1254–1263. [Google Scholar] [CrossRef]
- Magro, M.; Faralli, A.; Baratella, D.; Bertipaglia, I.; Giannetti, S.; Salviulo, G.; Zboril, R.; Vianello, F. Avidin Functionalized Maghemite Nanoparticles and Their Application for Recombinant Human Biotinyl-SERCA Purification. Langmuir 2012, 28, 15392–15401. [Google Scholar] [CrossRef]





| Control with Mica without Protein | Control of HRP–Biotin without TMB | |||
|---|---|---|---|---|
| Without Protein | Avidin 2 μg | Streptavidin 2 μg | ||
| Absorbance 450 nm | 0.6367 | 0.0030 | 0.0024 | 0.0032 |
| Incubated Protein (µg) | Avidin Absorbance (450 nm) | Avidin Molecules/cm2 | Streptavidin Absorbance (450 nm) | Streptavidin Molecules/cm2 |
|---|---|---|---|---|
| 0.0 | 0 | 0 | 0 | 0 |
| 0.5 | 0.1458 | 1.26 · 1010 | 0.2051 | 7.91 · 1010 |
| 1.0 | 0.3267 | 2.83 · 1010 | 0.3308 | 1.27 · 1011 |
| 2.0 | 0.6422 | 5.56 · 1010 | 0.5938 | 2.28 · 1011 |
| 4.0 | 0.7635 | 6.61 · 1010 | 0.6838 | 2.63 · 1011 |
| 5.0 | 0.7943 | 6.87 · 1010 | -------- | -------- |
| 15.0 | 0.8495 | 7.35 · 1010 | 0.8895 | 3.42 · 1011 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcuello, C.; de Miguel, R.; Lostao, A. Molecular Recognition of Proteins through Quantitative Force Maps at Single Molecule Level. Biomolecules 2022, 12, 594. https://doi.org/10.3390/biom12040594
Marcuello C, de Miguel R, Lostao A. Molecular Recognition of Proteins through Quantitative Force Maps at Single Molecule Level. Biomolecules. 2022; 12(4):594. https://doi.org/10.3390/biom12040594
Chicago/Turabian StyleMarcuello, Carlos, Rocío de Miguel, and Anabel Lostao. 2022. "Molecular Recognition of Proteins through Quantitative Force Maps at Single Molecule Level" Biomolecules 12, no. 4: 594. https://doi.org/10.3390/biom12040594
APA StyleMarcuello, C., de Miguel, R., & Lostao, A. (2022). Molecular Recognition of Proteins through Quantitative Force Maps at Single Molecule Level. Biomolecules, 12(4), 594. https://doi.org/10.3390/biom12040594









