Nuclear and Cytoplasmatic Players in Mitochondria-Related CNS Disorders: Chromatin Modifications and Subcellular Trafficking
Abstract
1. Introduction
2. Chromatin Plasticity at A Glance
2.1. DNA Methylation/Hydroxymethylation and Their Readers
2.2. Histone-Modifying Enzymes
2.3. ATP-Dependent Chromatin Remodelers
3. Epigenetic Factors-Driven Neurodevelopmental Disorders with Mitochondrial Phenotype and Subcellular Trafficking Implications
3.1. Methylation Reading and Rett Syndrome
3.2. Rett-like Phenotypes Not Associated with MeCP2 Mutations
3.3. Histone Ubiquitination Writing and Mental Retardation X-Linked Syndromic Turner-Type
3.4. Histone Acetylation Writing and KANSL1/Koolene De Vries Syndrome
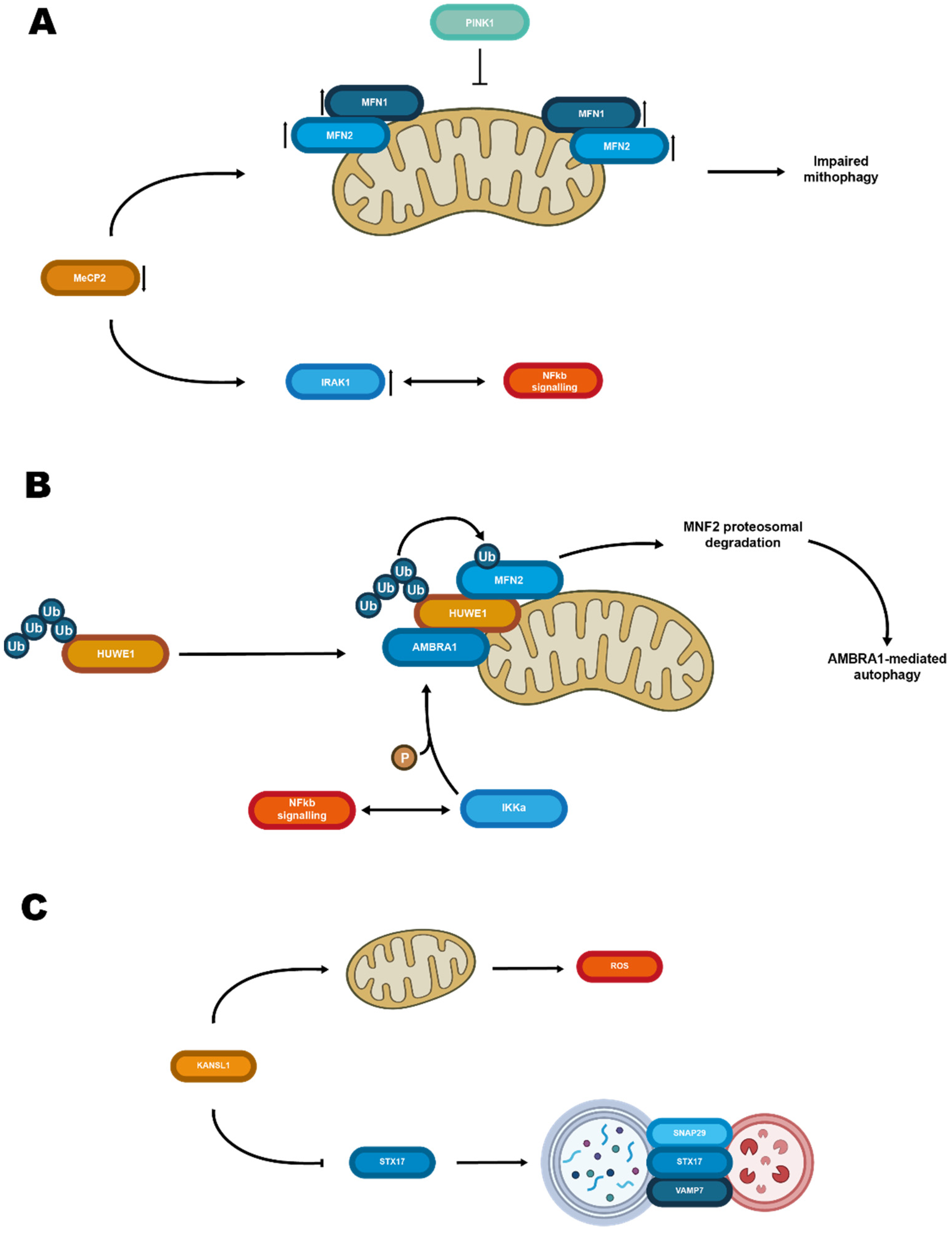
3.5. Some Epigenomic Outcomes and Mitochondria Meet on the Ubiquitin Ligase Route of Mitophagy: A Matter of Clearance
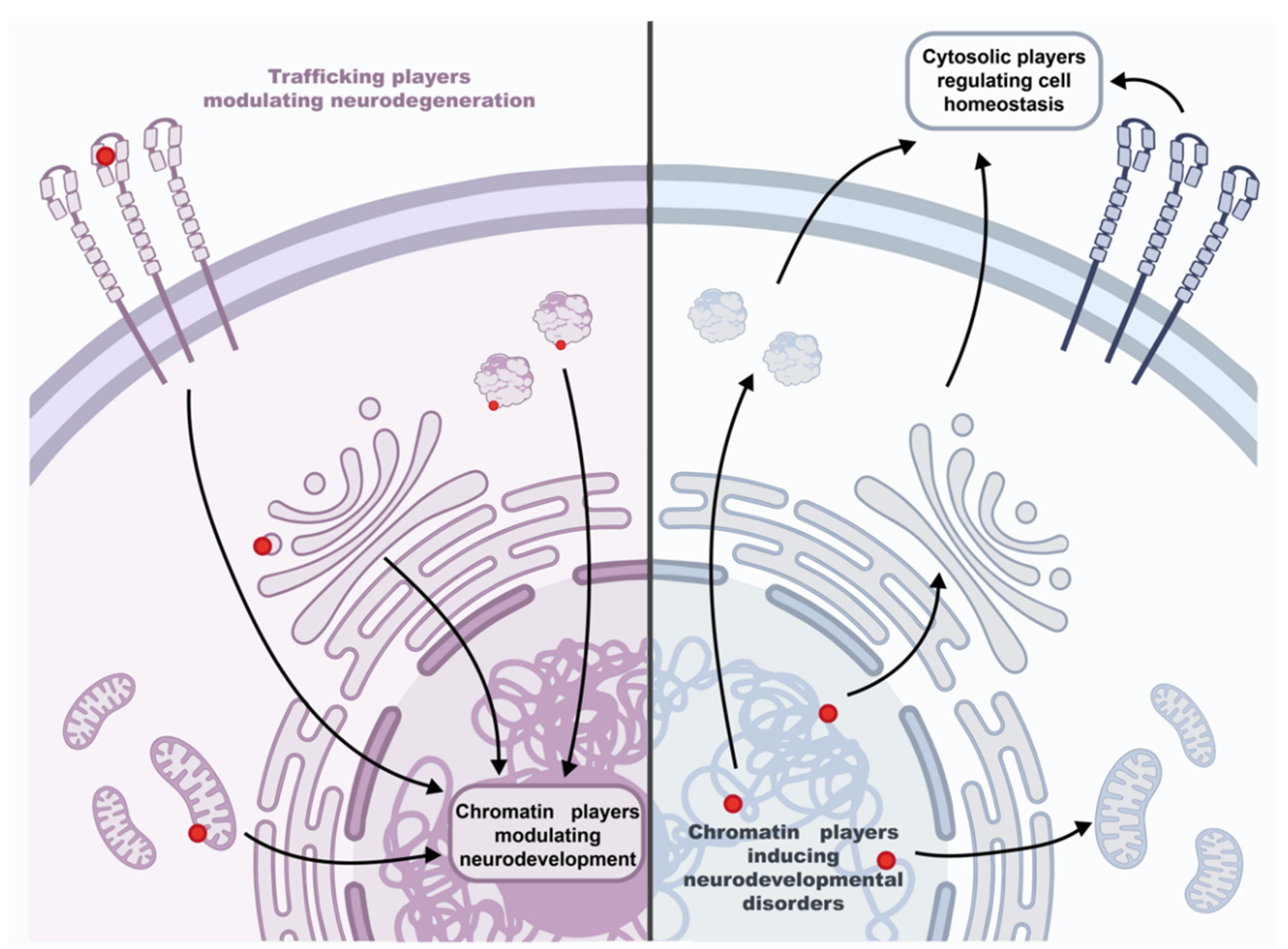
4. Traffic at A Glance
5. SNARE and Tethering Factors
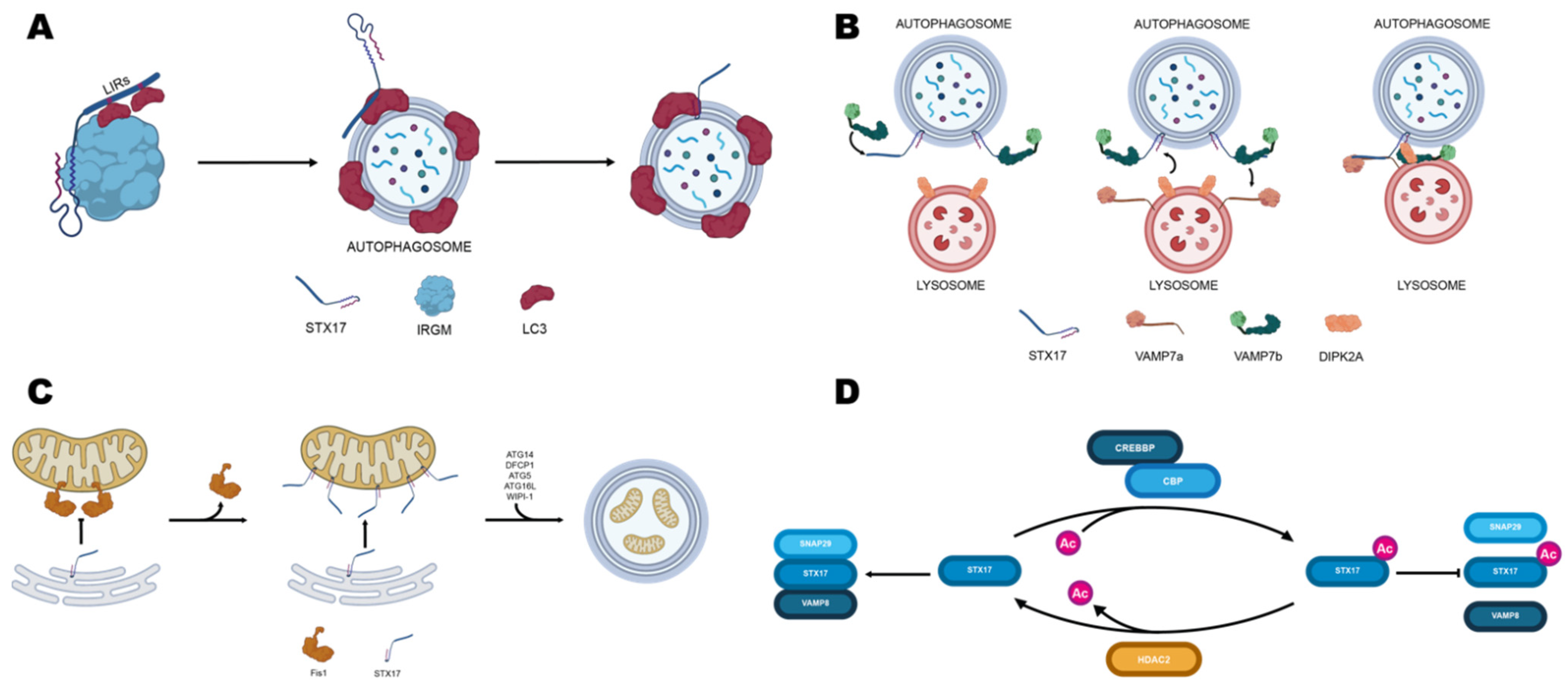
5.1. Syntaxin17
5.2. SNAP29
5.3. Tethering Factors
6. Rabs
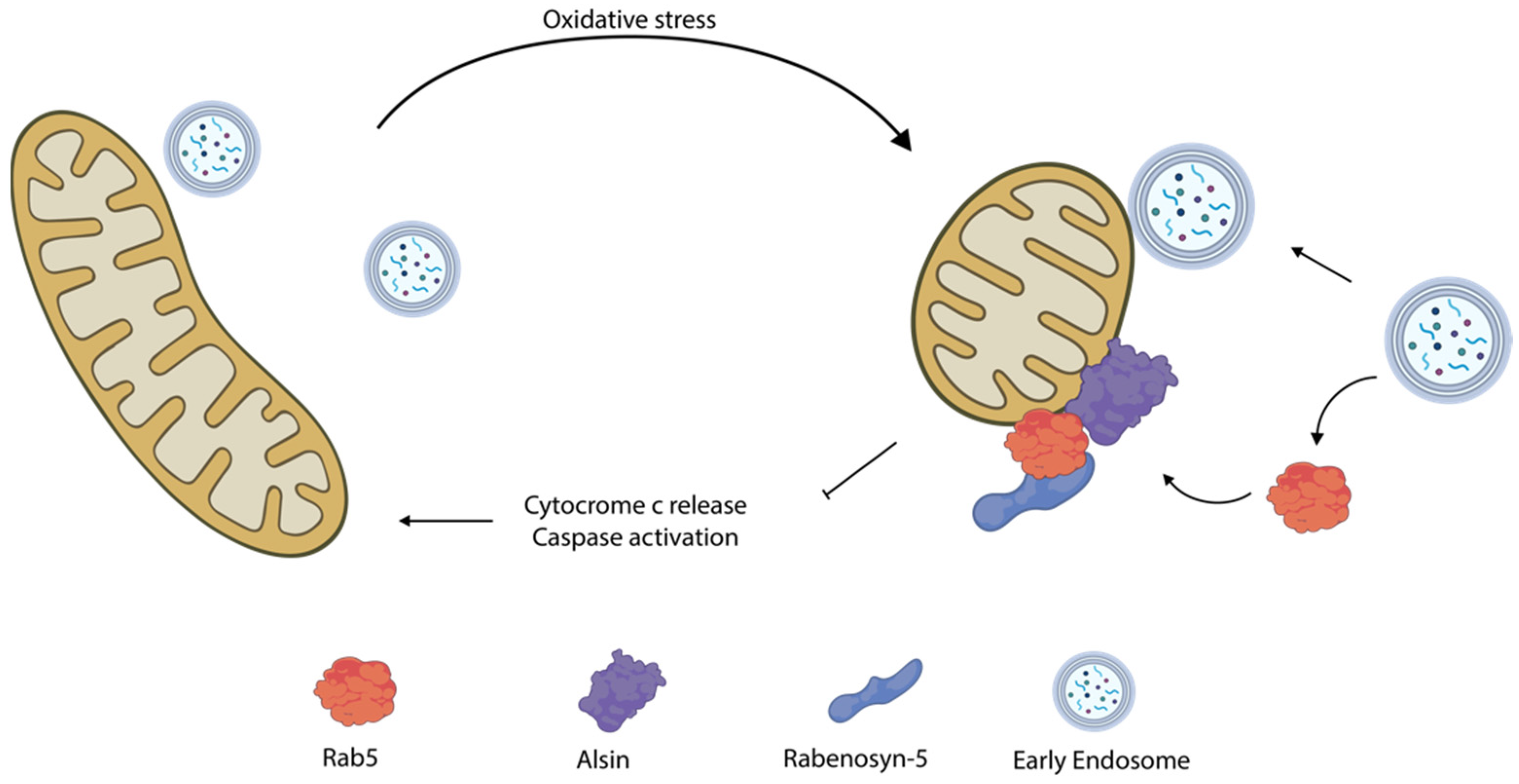
6.1. Rab5
6.2. Rab7
6.3. Rab8
6.4. Rab11
6.5. Rab32
7. Other Proteins
7.1. Mitofusins
7.2. Other Trafficking Factors
7.3. VAMP Associated Proteins
8. Post Translational Modifiers
8.1. HDACs
8.2. SUMOylation
8.3. Reversible Phosphorylation
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bader, V.; Winklhofer, K.F. Mitochondria at the interface between neurodegeneration and neuroinflammation. Semin. Cell Dev. Biol. 2020, 99, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C. Mitochondrial Dynamics and Its Involvement in Disease. Annu. Rev. Pathol. 2020, 15, 235–259. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C. Mitochondria: Dynamic organelles in disease, aging, and development. Cell 2006, 125, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Beitz, J.M. Parkinson’s disease: A review. Front. Biosci. 2014, 6, 65–74. [Google Scholar] [CrossRef]
- Raza, C.; Anjum, R.; Shakeel, N.U.A. Parkinson’s disease: Mechanisms, translational models and management strategies. Life Sci. 2019, 226, 77–90. [Google Scholar] [CrossRef]
- Chanou, A.; Hamperl, S. Single-Molecule Techniques to Study Chromatin. Front. Cell Dev. Biol. 2021, 9, 699771. [Google Scholar] [CrossRef]
- Smits, V.A.J.; Alonso-de Vega, I.; Warmerdam, D.O. Chromatin regulators and their impact on DNA repair and G2 checkpoint recovery. Cell Cycle 2020, 19, 2083–2093. [Google Scholar] [CrossRef]
- Cortés-Mendoza, J.; Díaz de León-Guerrero, S.; Pedraza-Alva, G.; Pérez-Martínez, L. Shaping synaptic plasticity: The role of activity-mediated epigenetic regulation on gene transcription. Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci. 2013, 31, 359–369. [Google Scholar] [CrossRef]
- Biswas, S.; Rao, C.M. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur. J. Pharmacol. 2018, 837, 8–24. [Google Scholar] [CrossRef]
- Gillette, T.G.; Hill, J.A. Readers, writers, and erasers: Chromatin as the whiteboard of heart disease. Circ. Res. 2015, 116, 1245–1253. [Google Scholar] [CrossRef]
- Vaughan, R.M.; Kupai, A.; Rothbart, S.B. Chromatin Regulation through Ubiquitin and Ubiquitin-like Histone Modifications. Trends Biochem. Sci. 2021, 46, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Villaseñor, R.; Baubec, T. Regulatory mechanisms governing chromatin organization and function. Curr. Opin. Cell Biol. 2021, 70, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Shen, H.-M.; Gopalakrishnan, V.; Gordon, N. Epigenetic Regulation of Autophagy Beyond the Cytoplasm: A Review. Front. Cell Dev. Biol. 2021, 9, 675599. [Google Scholar] [CrossRef] [PubMed]
- Basavarajappa, B.S.; Subbanna, S. Histone Methylation Regulation in Neurodegenerative Disorders. Int. J. Mol. Sci. 2021, 22, 4654. [Google Scholar] [CrossRef]
- Iwase, S.; Bérubé, N.G.; Zhou, Z.; Kasri, N.N.; Battaglioli, E.; Scandaglia, M.; Barco, A. Epigenetic Etiology of Intellectual Disability. J. Neurosci. 2017, 37, 10773–10782. [Google Scholar] [CrossRef]
- Rossignol, D.A.; Frye, R.E. Mitochondrial dysfunction in autism spectrum disorders: A systematic review and meta-analysis. Mol. Psychiatry 2012, 17, 290–314. [Google Scholar] [CrossRef]
- Valenti, D.; de Bari, L.; De Filippis, B.; Henrion-Caude, A.; Vacca, R.A. Mitochondrial dysfunction as a central actor in intellectual disability-related diseases: An overview of Down syndrome, autism, Fragile X and Rett syndrome. Neurosci. Biobehav. Rev. 2014, 46 Pt 2, 202–217. [Google Scholar] [CrossRef]
- Guo, J.U.; Su, Y.; Shin, J.H.; Shin, J.; Li, H.; Xie, B.; Zhong, C.; Hu, S.; Le, T.; Fan, G.; et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2014, 17, 215–222. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef]
- Lopes, A.F.C. Mitochondrial metabolism and DNA methylation: A review of the interaction between two genomes. Clin. Epigenetics 2020, 12, 182. [Google Scholar] [CrossRef]
- Della Ragione, F.; Vacca, M.; Fioriniello, S.; Pepe, G.; D’Esposito, M. MECP2, a multi-talented modulator of chromatin architecture. Brief. Funct. Genom. 2016, 15, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Fioriniello, S.; Marano, D.; Fiorillo, F.; D’Esposito, M.; Della Ragione, F. Epigenetic Factors That Control Pericentric Heterochromatin Organization in Mammals. Genes 2020, 11, 595. [Google Scholar] [CrossRef]
- Bellini, E.; Pavesi, G.; Barbiero, I.; Bergo, A.; Chandola, C.; Nawaz, M.S.; Rusconi, L.; Stefanelli, G.; Strollo, M.; Valente, M.M.; et al. MeCP2 post-translational modifications: A mechanism to control its involvement in synaptic plasticity and homeostasis? Front. Cell. Neurosci. 2014, 8, 236. [Google Scholar] [CrossRef] [PubMed]
- Tai, D.J.C.; Liu, Y.C.; Hsu, W.L.; Ma, Y.L.; Cheng, S.J.; Liu, S.Y.; Lee, E.H.Y. MeCP2 SUMOylation rescues Mecp2-mutant-induced behavioural deficits in a mouse model of Rett syndrome. Nat. Commun. 2016, 7, 10552. [Google Scholar] [CrossRef] [PubMed]
- Cardinaux, J.R.; Notis, J.C.; Zhang, Q.; Vo, N.; Craig, J.C.; Fass, D.M.; Brennan, R.G.; Goodman, R.H. Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol. Cell. Biol. 2000, 20, 1546–1552. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.A.J.; Shilatifard, A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 2020, 52, 1271–1281. [Google Scholar] [CrossRef]
- Dimitrova, E.; Turberfield, A.H.; Klose, R.J. Histone demethylases in chromatin biology and beyond. EMBO Rep. 2015, 16, 1620–1639. [Google Scholar] [CrossRef]
- Zocchi, L.; Sassone-Corsi, P. SIRT1-mediated deacetylation of MeCP2 contributes to BDNF expression. Epigenetics 2012, 7, 695–700. [Google Scholar] [CrossRef]
- Zhang, P.; Torres, K.; Liu, X.; Liu, C.-G.; Pollock, R.E. An Overview of Chromatin-Regulating Proteins in Cells. Curr. Protein Pept. Sci. 2016, 17, 401–410. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Marano, D.; Fioriniello, S.; Fiorillo, F.; Gibbons, R.J.; D’Esposito, M.; Della Ragione, F. ATRX Contributes to MeCP2-Mediated Pericentric Heterochromatin Organization during Neural Differentiation. Int. J. Mol. Sci. 2019, 20, 5371. [Google Scholar] [CrossRef] [PubMed]
- Adam, D. Mental health: On the spectrum. Nature 2013, 496, 416–418. [Google Scholar] [CrossRef] [PubMed]
- Leblond, C.S.; Le, T.-L.; Malesys, S.; Cliquet, F.; Tabet, A.-C.; Delorme, R.; Rolland, T.; Bourgeron, T. Operative list of genes associated with autism and neurodevelopmental disorders based on database review. Mol. Cell. Neurosci. 2021, 113, 103623. [Google Scholar] [CrossRef] [PubMed]
- Vacca, M.; Della Ragione, F.; Scalabrì, F.; D’Esposito, M. X inactivation and reactivation in X-linked diseases. Semin. Cell Dev. Biol. 2016, 56, 78–87. [Google Scholar] [CrossRef]
- Labadorf, A.; Hoss, A.G.; Lagomarsino, V.; Latourelle, J.C.; Hadzi, T.C.; Bregu, J.; MacDonald, M.E.; Gusella, J.F.; Chen, J.-F.; Akbarian, S.; et al. RNA Sequence Analysis of Human Huntington Disease Brain Reveals an Extensive Increase in Inflammatory and Developmental Gene Expression. PLoS ONE 2015, 10, e0143563. [Google Scholar] [CrossRef]
- Esposito, M.; Sherr, G.L. Epigenetic Modifications in Alzheimer’s Neuropathology and Therapeutics. Front. Neurosci. 2019, 13, 476. [Google Scholar] [CrossRef]
- Toker, L.; Tran, G.T.; Sundaresan, J.; Tysnes, O.-B.; Alves, G.; Haugarvoll, K.; Nido, G.S.; Dölle, C.; Tzoulis, C. Genome-wide histone acetylation analysis reveals altered transcriptional regulation in the Parkinson’s disease brain. Mol. Neurodegener. 2021, 16, 31. [Google Scholar] [CrossRef]
- Amir, R.E.; van den Veyver, I.B.; Wan, M.; Tran, C.Q.; Francke, U.; Zoghbi, H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999, 23, 185–188. [Google Scholar] [CrossRef]
- Vacca, M.; Filippini, F.; Budillon, A.; Rossi, V.; Della Ragione, F.; De Bonis, M.L.; Mercadante, G.; Manzati, E.; Gualandi, F.; Bigoni, S.; et al. MECP2 gene mutation analysis in the British and Italian Rett Syndrome patients: Hot spot map of the most recurrent mutations and bioinformatic analysis of a new MECP2 conserved region. Brain Dev. 2001, 23 (Suppl. S1), S246–S250. [Google Scholar] [CrossRef]
- Heilstedt, H.A.; Shahbazian, M.D.; Lee, B. Infantile hypotonia as a presentation of Rett syndrome. Am. J. Med. Genet. 2002, 111, 238–242. [Google Scholar] [CrossRef]
- De Felice, C.; Maffei, S.; Signorini, C.; Leoncini, S.; Lunghetti, S.; Valacchi, G.; D’Esposito, M.; Filosa, S.; Della Ragione, F.; Butera, G.; et al. Subclinical myocardial dysfunction in Rett syndrome. Eur. Hear. J. Cardiovasc. Imaging 2012, 13, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Cornford, M.E.; Philippart, M.; Jacobs, B.; Scheibel, A.B.; Vinters, H.V. Neuropathology of Rett syndrome: Case report with neuronal and mitochondrial abnormalities in the brain. J. Child Neurol. 1994, 9, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Eeg-Olofsson, O.; Al-Zuhair, A.G.; Teebi, A.S.; Daoud, A.S.; Zaki, M.; Besisso, M.S.; Al-Essa, M.M. Rett syndrome: A mitochondrial disease? J. Child Neurol. 1990, 5, 210–214. [Google Scholar] [CrossRef]
- Dotti, M.T.; Manneschi, L.; Malandrini, A.; De Stefano, N.; Caznerale, F.; Federico, A. Mitochondrial dysfunction in Rett syndrome. An ultrastructural and biochemical study. Brain Dev. 1993, 15, 103–106. [Google Scholar] [CrossRef]
- Filosa, S.; Pecorelli, A.; D’Esposito, M.; Valacchi, G.; Hajek, J. Exploring the possible link between MeCP2 and oxidative stress in Rett syndrome. Free Radic. Biol. Med. 2015, 88, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Müller, M. Disturbed redox homeostasis and oxidative stress: Potential players in the developmental regression in Rett syndrome. Neurosci. Biobehav. Rev. 2019, 98, 154–163. [Google Scholar] [CrossRef]
- Can, K.; Menzfeld, C.; Rinne, L.; Rehling, P.; Kügler, S.; Golubiani, G.; Dudek, J.; Müller, M. Neuronal Redox-Imbalance in Rett Syndrome Affects Mitochondria as Well as Cytosol, and Is Accompanied by Intensified Mitochondrial O2 Consumption and ROS Release. Front. Physiol. 2019, 10, 479. [Google Scholar] [CrossRef]
- Durand, T.; De Felice, C.; Signorini, C.; Oger, C.; Bultel-Poncé, V.; Guy, A.; Galano, J.-M.; Leoncini, S.; Ciccoli, L.; Pecorelli, A.; et al. F(2)-Dihomo-isoprostanes and brain white matter damage in stage 1 Rett syndrome. Biochimie 2013, 95, 86–90. [Google Scholar] [CrossRef]
- Villemagne, P.M.; Naidu, S.; Villemagne, V.L.; Yaster, M.; Wagner, H.N.J.; Harris, J.C.; Moser, H.W.; Johnston, M.V.; Dannals, R.F.; Wong, D.F. Brain glucose metabolism in Rett Syndrome. Pediatr. Neurol. 2002, 27, 117–122. [Google Scholar] [CrossRef]
- Viola, A.; Saywell, V.; Villard, L.; Cozzone, P.J.; Lutz, N.W. Metabolic fingerprints of altered brain growth, osmoregulation and neurotransmission in a Rett syndrome model. PLoS ONE 2007, 2, e157. [Google Scholar] [CrossRef]
- Haas, R.H.; Light, M.; Rice, M.; Barshop, B.A. Oxidative metabolism in Rett syndrome: 1. Clinical studies. Neuropediatrics 1995, 26, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Golubiani, G.; Lagani, V.; Solomonia, R.; Müller, M. Metabolomic Fingerprint of Mecp2-Deficient Mouse Cortex: Evidence for a Pronounced Multi-Facetted Metabolic Component in Rett Syndrome. Cells 2021, 10, 2494. [Google Scholar] [CrossRef] [PubMed]
- Vacca, M.; Tripathi, K.P.; Speranza, L.; Aiese Cigliano, R.; Scalabrì, F.; Marracino, F.; Madonna, M.; Sanseverino, W.; Perrone-Capano, C.; Guarracino, M.R.; et al. Effects of Mecp2 loss of function in embryonic cortical neurons: A bioinformatics strategy to sort out non-neuronal cells variability from transcriptome profiling. BMC Bioinform. 2016, 17 (Suppl. S2), 14. [Google Scholar] [CrossRef] [PubMed]
- Marano, D.; Fioriniello, S.; D’Esposito, M.; Della Ragione, F. Transcriptomic and Epigenomic Landscape in Rett Syndrome. Biomolecules 2021, 11, 967. [Google Scholar] [CrossRef]
- Aldosary, M.; Al-Bakheet, A.; Al-Dhalaan, H.; Almass, R.; Alsagob, M.; Al-Younes, B.; AlQuait, L.; Mustafa, O.M.; Bulbul, M.; Rahbeeni, Z.; et al. Rett Syndrome, a Neurodevelopmental Disorder, Whole-Transcriptome, and Mitochondrial Genome Multiomics Analyses Identify Novel Variations and Disease Pathways. OMICS 2020, 24, 160–171. [Google Scholar] [CrossRef]
- Kriaucionis, S.; Paterson, A.; Curtis, J.; Guy, J.; Macleod, N.; Bird, A. Gene expression analysis exposes mitochondrial abnormalities in a mouse model of Rett syndrome. Mol. Cell. Biol. 2006, 26, 5033–5042. [Google Scholar] [CrossRef]
- Forlani, G.; Giarda, E.; Ala, U.; Di Cunto, F.; Salani, M.; Tupler, R.; Kilstrup-Nielsen, C.; Landsberger, N. The MeCP2/YY1 interaction regulates ANT1 expression at 4q35: Novel hints for Rett syndrome pathogenesis. Hum. Mol. Genet. 2010, 19, 3114–3123. [Google Scholar] [CrossRef]
- Cervellati, C.; Sticozzi, C.; Romani, A.; Belmonte, G.; De Rasmo, D.; Signorile, A.; Cervellati, F.; Milanese, C.; Mastroberardino, P.G.; Pecorelli, A.; et al. Impaired enzymatic defensive activity, mitochondrial dysfunction and proteasome activation are involved in RTT cell oxidative damage. Biochim. Biophys. Acta 2015, 1852, 2066–2074. [Google Scholar] [CrossRef]
- Shulyakova, N.; Andreazza, A.C.; Mills, L.R.; Eubanks, J.H. Mitochondrial Dysfunction in the Pathogenesis of Rett Syndrome: Implications for Mitochondria-Targeted Therapies. Front. Cell. Neurosci. 2017, 11, 58. [Google Scholar] [CrossRef]
- Cicaloni, V.; Pecorelli, A.; Tinti, L.; Rossi, M.; Benedusi, M.; Cervellati, C.; Spiga, O.; Santucci, A.; Hayek, J.; Salvini, L.; et al. Proteomic profiling reveals mitochondrial alterations in Rett syndrome. Free Radic. Biol. Med. 2020, 155, 37–48. [Google Scholar] [CrossRef]
- Fehr, S.; Wilson, M.; Downs, J.; Williams, S.; Murgia, A.; Sartori, S.; Vecchi, M.; Ho, G.; Polli, R.; Psoni, S.; et al. The CDKL5 disorder is an independent clinical entity associated with early-onset encephalopathy. Eur. J. Hum. Genet. 2013, 21, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Nguyen, S.; McKeehan, K.; Wang, F.; McKeehan, W.L.; Liu, L. Microtubule-associated protein 1S (MAP1S) bridges autophagic components with microtubules and mitochondria to affect autophagosomal biogenesis and degradation. J. Biol. Chem. 2011, 286, 10367–10377. [Google Scholar] [CrossRef] [PubMed]
- Pancrazi, L.; Di Benedetto, G.; Colombaioni, L.; Della Sala, G.; Testa, G.; Olimpico, F.; Reyes, A.; Zeviani, M.; Pozzan, T.; Costa, M. Foxg1 localizes to mitochondria and coordinates cell differentiation and bioenergetics. Proc. Natl. Acad. Sci. USA 2015, 112, 13910–13915. [Google Scholar] [CrossRef] [PubMed]
- He, Z.-H.; Li, M.; Fang, Q.-J.; Liao, F.-L.; Zou, S.-Y.; Wu, X.; Sun, H.-Y.; Zhao, X.-Y.; Hu, Y.-J.; Xu, X.-X.; et al. FOXG1 promotes aging inner ear hair cell survival through activation of the autophagy pathway. Autophagy 2021, 17, 4341–4362. [Google Scholar] [CrossRef]
- Romaniello, R.; Saettini, F.; Panzeri, E.; Arrigoni, F.; Bassi, M.T.; Borgatti, R. A de-novo STXBP1 gene mutation in a patient showing the Rett syndrome phenotype. Neuroreport 2015, 26, 254–257. [Google Scholar] [CrossRef]
- Helbig, I.; von Deimling, M.; Marsh, E.D. Epileptic Encephalopathies as Neurodegenerative Disorders. Adv. Neurobiol. 2017, 15, 295–315. [Google Scholar] [CrossRef]
- Van Esch, H. MECP2 Duplication Syndrome; GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Froyen, G.; Corbett, M.; Vandewalle, J.; Jarvela, I.; Lawrence, O.; Meldrum, C.; Bauters, M.; Govaerts, K.; Vandeleur, L.; van Esch, H.; et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am. J. Hum. Genet. 2008, 82, 432–443. [Google Scholar] [CrossRef]
- Bosshard, M.; Aprigliano, R.; Gattiker, C.; Palibrk, V.; Markkanen, E.; Backe, P.H.; Pellegrino, S.; Raymond, F.L.; Froyen, G.; Altmeyer, M.; et al. Impaired oxidative stress response characterizes HUWE1-promoted X-linked intellectual disability. Sci. Rep. 2017, 7, 15050. [Google Scholar] [CrossRef]
- Mandemaker, I.K.; van Cuijk, L.; Janssens, R.C.; Lans, H.; Bezstarosti, K.; Hoeijmakers, J.H.; Demmers, J.A.; Vermeulen, W.; Marteijn, J.A. DNA damage-induced histone H1 ubiquitylation is mediated by HUWE1 and stimulates the RNF8-RNF168 pathway. Sci. Rep. 2017, 7, 15353. [Google Scholar] [CrossRef]
- Kim, J.; Lee, S.; Kim, H.; Lee, H.; Seong, K.M.; Youn, H.; Youn, B. Autophagic Organelles in DNA Damage Response. Front. Cell Dev. Biol. 2021, 9, 668735. [Google Scholar] [CrossRef]
- Michán, S.; Li, Y.; Chou, M.M.-H.; Parrella, E.; Ge, H.; Long, J.M.; Allard, J.S.; Lewis, K.; Miller, M.; Xu, W.; et al. SIRT1 is essential for normal cognitive function and synaptic plasticity. J. Neurosci. 2010, 30, 9695–9707. [Google Scholar] [CrossRef] [PubMed]
- Koolen, D.A.; Pfundt, R.; Linda, K.; Beunders, G.; Veenstra-Knol, H.E.; Conta, J.H.; Fortuna, A.M.; Gillessen-Kaesbach, G.; Dugan, S.; Halbach, S.; et al. The Koolen-de Vries syndrome: A phenotypic comparison of patients with a 17q21.31 microdeletion versus a KANSL1 sequence variant. Eur. J. Hum. Genet. 2016, 24, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Füllgrabe, J.; Lynch-Day, M.A.; Heldring, N.; Li, W.; Struijk, R.B.; Ma, Q.; Hermanson, O.; Rosenfeld, M.G.; Klionsky, D.J.; Joseph, B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 2013, 500, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Arbogast, T.; Iacono, G.; Chevalier, C.; Afinowi, N.O.; Houbaert, X.; van Eede, M.C.; Laliberte, C.; Birling, M.-C.; Linda, K.; Meziane, H.; et al. Mouse models of 17q21.31 microdeletion and microduplication syndromes highlight the importance of Kansl1 for cognition. PLoS Genet. 2017, 13, e1006886. [Google Scholar] [CrossRef]
- Li, T.; Lu, D.; Yao, C.; Li, T.; Dong, H.; Li, Z.; Xu, G.; Chen, J.; Zhang, H.; Yi, X.; et al. Kansl1 haploinsufficiency impairs autophagosome-lysosome fusion and links autophagic dysfunction with Koolen-de Vries syndrome in mice. Nat. Commun. 2022, 13, 931. [Google Scholar] [CrossRef]
- Di Malta, C.; Cinque, L.; Settembre, C. Transcriptional Regulation of Autophagy: Mechanisms and Diseases. Front. Cell Dev. Biol. 2019, 7, 114. [Google Scholar] [CrossRef]
- Cantó, C.; Menzies, K.J.; Auwerx, J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef]
- Wan, M.; Zhao, K.; Lee, S.S.; Francke, U. MECP2 truncating mutations cause histone H4 hyperacetylation in Rett syndrome. Hum. Mol. Genet. 2001, 10, 1085–1092. [Google Scholar] [CrossRef]
- Sbardella, D.; Tundo, G.R.; Campagnolo, L.; Valacchi, G.; Orlandi, A.; Curatolo, P.; Borsellino, G.; D’Esposito, M.; Ciaccio, C.; Di Cesare, S.; et al. Retention of Mitochondria in Mature Human Red Blood Cells as the Result of Autophagy Impairment in Rett Syndrome. Sci. Rep. 2017, 7, 12297. [Google Scholar] [CrossRef]
- Crivellari, I.; Pecorelli, A.; Cordone, V.; Marchi, S.; Pinton, P.; Hayek, J.; Cervellati, C.; Valacchi, G. Impaired mitochondrial quality control in Rett Syndrome. Arch. Biochem. Biophys. 2021, 700, 108790. [Google Scholar] [CrossRef]
- Vranas, M.; Lu, Y.; Rasool, S.; Croteau, N.; Krett, J.D.; Sauvé, V.; Gehring, K.; Fon, E.A.; Durcan, T.M.; Trempe, J.-F. Selective localization of Mfn2 near PINK1 enables its preferential ubiquitination by Parkin on mitochondria. Open Biol. 2022, 12, 210255. [Google Scholar] [CrossRef] [PubMed]
- Hamacher-Brady, A.; Brady, N.R. Mitophagy programs: Mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell. Mol. Life Sci. 2016, 73, 775–795. [Google Scholar] [CrossRef] [PubMed]
- Di Rita, A.; Peschiaroli, A.; Acunzo, P.D.; Strobbe, D.; Hu, Z.; Gruber, J.; Nygaard, M.; Lambrughi, M.; Melino, G.; Papaleo, E.; et al. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKα. Nat. Commun. 2018, 9, 3755. [Google Scholar] [CrossRef] [PubMed]
- Solt, L.A.; May, M.J. The IkappaB kinase complex: Master regulator of NF-kappaB signaling. Immunol. Res. 2008, 42, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Mochida, G.H.; Mahajnah, M.; Hill, A.D.; Basel-Vanagaite, L.; Gleason, D.; Hill, R.S.; Bodell, A.; Crosier, M.; Straussberg, R.; Walsh, C.A. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am. J. Hum. Genet. 2009, 85, 897–902. [Google Scholar] [CrossRef] [PubMed]
- Manzini, M.C.; Xiong, L.; Shaheen, R.; Tambunan, D.E.; Di Costanzo, S.; Mitisalis, V.; Tischfield, D.J.; Cinquino, A.; Ghaziuddin, M.; Christian, M.; et al. CC2D1A regulates human intellectual and social function as well as NF-κB signaling homeostasis. Cell Rep. 2014, 8, 647–655. [Google Scholar] [CrossRef]
- Gabel, H.W.; Kinde, B.; Stroud, H.; Gilbert, C.S.; Harmin, D.A.; Kastan, N.R.; Hemberg, M.; Ebert, D.H.; Greenberg, M.E. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 2015, 522, 89–93. [Google Scholar] [CrossRef]
- Kishi, N.; MacDonald, J.L.; Ye, J.; Molyneaux, B.J.; Azim, E.; Macklis, J.D. Reduction of aberrant NF-κB signalling ameliorates Rett syndrome phenotypes in Mecp2-null mice. Nat. Commun. 2016, 7, 10520. [Google Scholar] [CrossRef]
- Motil, K.J.; Barrish, J.O.; Lane, J.; Geerts, S.P.; Annese, F.; McNair, L.; Percy, A.K.; Skinner, S.A.; Neul, J.L.; Glaze, D.G. Vitamin D deficiency is prevalent in girls and women with Rett syndrome. J. Pediatr. Gastroenterol. Nutr. 2011, 53, 569–574. [Google Scholar] [CrossRef]
- Ribeiro, M.C.; Moore, S.M.; Kishi, N.; Macklis, J.D.; MacDonald, J.L. Vitamin D supplementation rescues aberrant NF-κB pathway activation and partially ameliorates Rett syndrome phenotypes in Mecp2 mutant mice. Eneuro 2020, 7. [Google Scholar] [CrossRef]
- Li, H.; Jang, W.; Kim, H.J.; Jo, K.D.; Lee, M.K.; Song, S.H.; Yang, H.O. Biochemical protective effect of 1,25-dihydroxyvitamin D3 through autophagy induction in the MPTP mouse model of Parkinson’s disease. Neuroreport 2015, 26, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, R.; Nakahira, K.; Gu, Z. Mitochondrial DNA Mutation, Diseases, and Nutrient-Regulated Mitophagy. Annu. Rev. Nutr. 2019, 39, 201–226. [Google Scholar] [CrossRef] [PubMed]
- Bhutia, S.K. Vitamin D in autophagy signaling for health and diseases: Insights on potential mechanisms and future perspectives. J. Nutr. Biochem. 2022, 99, 108841. [Google Scholar] [CrossRef] [PubMed]
- Linda, K.; Lewerissa, E.I.; Verboven, A.H.A.; Gabriele, M.; Frega, M.; Klein Gunnewiek, T.M.; Devilee, L.; Ulferts, E.; Hommersom, M.; Oudakker, A.; et al. Imbalanced autophagy causes synaptic deficits in a human model for neurodevelopmental disorders. Autophagy 2022, 18, 423–442. [Google Scholar] [CrossRef] [PubMed]
- Uray, I.P.; Uray, K. Mechanotransduction at the Plasma Membrane-Cytoskeleton Interface. Int. J. Mol. Sci. 2021, 22, 11566. [Google Scholar] [CrossRef]
- Domínguez-Romero, M.E.; Slater, P.G. Unraveling Axon Guidance during Axotomy and Regeneration. Int. J. Mol. Sci. 2021, 22, 11566. [Google Scholar] [CrossRef]
- Scapin, G.; Gasparotto, M.; Peterle, D.; Tescari, S.; Porcellato, E.; Piovesan, A.; Righetto, I.; Acquasaliente, L.; De Filippis, V.; Filippini, F. A conserved Neurite Outgrowth and Guidance motif with biomimetic potential in neuronal Cell Adhesion Molecules. Comput. Struct. Biotechnol. J. 2021, 19, 5622–5636. [Google Scholar] [CrossRef]
- Soni, N.; Gupta, S.; Rawat, S.; Krishnakumar, V.; Mohanty, S.; Banerjee, A. MicroRNA-Enriched Exosomes from Different Sources of Mesenchymal Stem Cells Can Differentially Modulate Functions of Immune Cells and Neurogenesis. Biomedicines 2021, 10, 69. [Google Scholar] [CrossRef]
- Ye, X.; Qiu, Y.; Gao, Y.; Wan, D.; Zhu, H. A Subtle Network Mediating Axon Guidance: Intrinsic Dynamic Structure of Growth Cone, Attractive and Repulsive Molecular Cues, and the Intermediate Role of Signaling Pathways. Neural Plast. 2019, 2019, 1719829. [Google Scholar] [CrossRef]
- Gurkan, C.; Koulov, A.V.; Balch, W.E. An evolutionary perspective on eukaryotic membrane trafficking. Adv. Exp. Med. Biol. 2007, 607, 73–83. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Eskelinen, E.-L.; Saftig, P. Autophagy: A lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta 2009, 1793, 664–673. [Google Scholar] [CrossRef] [PubMed]
- Valente, E.M.; Abou-Sleiman, P.M.; Caputo, V.; Muqit, M.M.K.; Harvey, K.; Gispert, S.; Ali, Z.; Del Turco, D.; Bentivoglio, A.R.; Healy, D.G.; et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004, 304, 1158–1160. [Google Scholar] [CrossRef] [PubMed]
- Kitada, T.; Asakawa, S.; Hattori, N.; Matsumine, H.; Yamamura, Y.; Minoshima, S.; Yokochi, M.; Mizuno, Y.; Shimizu, N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 1998, 392, 605–608. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Padman, B.S.; Lazarou, M. Deciphering the Molecular Signals of PINK1/Parkin Mitophagy. Trends Cell Biol. 2016, 26, 733–744. [Google Scholar] [CrossRef]
- Martinez-Vicente, M.; Talloczy, Z.; Wong, E.; Tang, G.; Koga, H.; Kaushik, S.; de Vries, R.; Arias, E.; Harris, S.; Sulzer, D.; et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat. Neurosci. 2010, 13, 567–576. [Google Scholar] [CrossRef]
- Wong, Y.C.; Holzbaur, E.L.F. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci. 2014, 34, 1293–1305. [Google Scholar] [CrossRef]
- Coffey, E.E.; Beckel, J.M.; Laties, A.M.; Mitchell, C.H. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience 2014, 263, 111–124. [Google Scholar] [CrossRef]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2503–2518. [Google Scholar] [CrossRef]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Shen, Q.; Shi, Y.; Liu, J.; Su, H.; Huang, J.; Zhang, Y.; Peng, C.; Zhou, T.; Sun, Q.; Wan, W.; et al. Acetylation of STX17 (syntaxin 17) controls autophagosome maturation. Autophagy 2021, 17, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Gao, P.; Arzberger, T.; Höllerhage, M.; Herms, J.; Höglinger, G.; Koeglsperger, T. Alpha-Synuclein defects autophagy by impairing SNAP29-mediated autophagosome-lysosome fusion. Cell Death Dis. 2021, 12, 854. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, K.; Itakura, M.; Fukutomi, T.; Nishiwaki, C.; Nakamichi, Y.; Torii, S.; Makiyama, T.; Harada, A.; Ohara-Imaizumi, M. VAMP7 Regulates Autophagosome Formation by Supporting Atg9a Functions in Pancreatic β-Cells From Male Mice. Endocrinology 2018, 159, 3674–3688. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Teng, J.; Chen, J. New insights regarding SNARE proteins in autophagosome-lysosome fusion. Autophagy 2021, 17, 2680–2688. [Google Scholar] [CrossRef] [PubMed]
- Van der Beek, J.; Jonker, C.; van der Welle, R.; Liv, N.; Klumperman, J. CORVET, CHEVI and HOPS-multisubunit tethers of the endo-lysosomal system in health and disease. J. Cell Sci. 2019, 132, jcs189134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ge, S.; Ni, B.; He, K.; Zhu, P.; Wu, X.; Shao, Y. Augmenting ATG14 alleviates atherosclerosis and inhibits inflammation via promotion of autophagosome-lysosome fusion in macrophages. Autophagy 2021, 17, 4218–4230. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Miao, G.; Xue, X.; Guo, X.; Yuan, C.; Wang, Z.; Zhang, G.; Chen, Y.; Feng, D.; Hu, J.; et al. The Vici Syndrome Protein EPG5 Is a Rab7 Effector that Determines the Fusion Specificity of Autophagosomes with Late Endosomes/Lysosomes. Mol. Cell 2016, 63, 781–795. [Google Scholar] [CrossRef]
- Fasshauer, D.; Sutton, R.B.; Brunger, A.T.; Jahn, R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA 1998, 95, 15781–15786. [Google Scholar] [CrossRef]
- Schäfer, I.B.; Hesketh, G.G.; Bright, N.A.; Gray, S.R.; Pryor, P.R.; Evans, P.R.; Luzio, J.P.; Owen, D.J. The binding of Varp to VAMP7 traps VAMP7 in a closed, fusogenically inactive conformation. Nat. Struct. Mol. Biol. 2012, 19, 1300–1309. [Google Scholar] [CrossRef]
- Li, F.; Pincet, F.; Perez, E.; Eng, W.S.; Melia, T.J.; Rothman, J.E.; Tareste, D. Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat. Struct. Mol. Biol. 2007, 14, 890–896. [Google Scholar] [CrossRef]
- Vivona, S.; Liu, C.W.; Strop, P.; Rossi, V.; Filippini, F.; Brunger, A.T. The longin SNARE VAMP7/TI-VAMP adopts a closed conformation. J. Biol. Chem. 2010, 285, 17965–17973. [Google Scholar] [CrossRef] [PubMed]
- De Franceschi, N.; Wild, K.; Schlacht, A.; Dacks, J.B.; Sinning, I.; Filippini, F. Longin and GAF domains: Structural evolution and adaptation to the subcellular trafficking machinery. Traffic 2014, 15, 104–121. [Google Scholar] [CrossRef] [PubMed]
- Nair, U.; Jotwani, A.; Geng, J.; Gammoh, N.; Richerson, D.; Yen, W.-L.; Griffith, J.; Nag, S.; Wang, K.; Moss, T.; et al. SNARE proteins are required for macroautophagy. Cell 2011, 146, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Moreau, K.; Rubinsztein, D.C. The plasma membrane as a control center for autophagy. Autophagy 2012, 8, 861–863. [Google Scholar] [CrossRef] [PubMed]
- Vats, S.; Galli, T. Introducing secretory reticulophagy/ER-phagy (SERP), a VAMP7-dependent pathway involved in neurite growth. Autophagy 2021, 17, 1037–1039. [Google Scholar] [CrossRef]
- Wojnacki, J.; Nola, S.; Bun, P.; Cholley, B.; Filippini, F.; Pressé, M.T.; Lipecka, J.; Man Lam, S.; N’guyen, J.; Simon, A.; et al. Role of VAMP7-Dependent Secretion of Reticulon 3 in Neurite Growth. Cell Rep. 2020, 33, 108536. [Google Scholar] [CrossRef]
- Herrera-Cruz, M.S.; Yap, M.C.; Tahbaz, N.; Phillips, K.; Thomas, L.; Thomas, G.; Simmen, T. Rab32 uses its effector reticulon 3L to trigger autophagic degradation of mitochondria-associated membrane (MAM) proteins. Biol. Direct 2021, 16, 22. [Google Scholar] [CrossRef]
- McGrath, K.; Agarwal, S.; Tonelli, M.; Dergai, M.; Gaeta, A.L.; Shum, A.K.; Lacoste, J.; Zhang, Y.; Wen, W.; Chung, D.; et al. A conformational switch driven by phosphorylation regulates the activity of the evolutionarily conserved SNARE Ykt6. Proc. Natl. Acad. Sci. USA 2021, 118, e2016730118. [Google Scholar] [CrossRef]
- Cuddy, L.K.; Wani, W.Y.; Morella, M.L.; Pitcairn, C.; Tsutsumi, K.; Fredriksen, K.; Justman, C.J.; Grammatopoulos, T.N.; Belur, N.R.; Zunke, F.; et al. Stress-Induced Cellular Clearance Is Mediated by the SNARE Protein ykt6 and Disrupted by α-Synuclein. Neuron 2019, 104, 869–884.e11. [Google Scholar] [CrossRef]
- Sap, K.A.; Guler, A.T.; Bury, A.; Dekkers, D.; Demmers, J.A.A.; Reits, E.A. Identification of Full-Length Wild-Type and Mutant Huntingtin Interacting Proteins by Crosslinking Immunoprecipitation in Mice Brain Cortex. J. Huntingtons. Dis. 2021, 10, 335–347. [Google Scholar] [CrossRef]
- Arasaki, K.; Shimizu, H.; Mogari, H.; Nishida, N.; Hirota, N.; Furuno, A.; Kudo, Y.; Baba, M.; Baba, N.; Cheng, J.; et al. A Role for the Ancient SNARE Syntaxin 17 in Regulating Mitochondrial Division. Dev. Cell 2015, 32, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Zheng, P.; Zhou, C.; Wang, X.; Ma, H.; Ma, W.; Zhou, X.; Teng, J.; Chen, J. DIPK2A promotes STX17- and VAMP7-mediated autophagosome-lysosome fusion by binding to VAMP7B. Autophagy 2020, 16, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Itakura, E.; Kishi-Itakura, C.; Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012, 151, 1256–1269. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Jiang, P.; Nakano, S.; Sakamaki, Y.; Yamamoto, H.; Mizushima, N. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J. Cell Biol. 2018, 217, 2633–2645. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Matsui, T.; Yamamoto, H. YKT6 as a second SNARE protein of mammalian autophagosomes. Autophagy 2019, 15, 176–177. [Google Scholar] [CrossRef]
- Kumar, S.; Jain, A.; Farzam, F.; Jia, J.; Gu, Y.; Choi, S.W.; Mudd, M.H.; Claude-Taupin, A.; Wester, M.J.; Lidke, K.A.; et al. Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J. Cell Biol. 2018, 217, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Xian, H.; Yang, Q.; Xiao, L.; Shen, H.-M.; Liou, Y.-C. STX17 dynamically regulated by Fis1 induces mitophagy via hierarchical macroautophagic mechanism. Nat. Commun. 2019, 10, 2059. [Google Scholar] [CrossRef]
- Vacca, M.; Albania, L.; Ragione, F.D.; Carpi, A.; Rossi, V.; Strazzullo, M.; De Franceschi, N.; Rossetto, O.; Filippini, F.; D’Esposito, M. Alternative splicing of the human gene SYBL1 modulates protein domain architecture of longin VAMP7/TI-VAMP, showing both non-SNARE and synaptobrevin-like isoforms. BMC Mol. Biol. 2011, 12, 26. [Google Scholar] [CrossRef]
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; et al. Autophagosomes form at ER-mitochondria contact sites. Nature 2013, 495, 389–393. [Google Scholar] [CrossRef]
- Lu, W.; Karuppagounder, S.S.; Springer, D.A.; Allen, M.D.; Zheng, L.; Chao, B.; Zhang, Y.; Dawson, V.L.; Dawson, T.M.; Lenardo, M. Genetic deficiency of the mitochondrial protein PGAM5 causes a Parkinson’s-like movement disorder. Nat. Commun. 2014, 5, 4930. [Google Scholar] [CrossRef]
- Lu, W.; Sun, J.; Yoon, J.S.; Zhang, Y.; Zheng, L.; Murphy, E.; Mattson, M.P.; Lenardo, M.J. Mitochondrial Protein PGAM5 Regulates Mitophagic Protection against Cell Necroptosis. PLoS ONE 2016, 11, e0147792. [Google Scholar] [CrossRef] [PubMed]
- Sekine, S.; Yao, A.; Hattori, K.; Sugawara, S.; Naguro, I.; Koike, M.; Uchiyama, Y.; Takeda, K.; Ichijo, H. The Ablation of Mitochondrial Protein Phosphatase Pgam5 Confers Resistance Against Metabolic Stress. EBioMedicine 2016, 5, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Han, Z.; Feng, D.; Chen, Y.; Chen, L.; Wu, H.; Huang, L.; Zhou, C.; Cai, X.; Fu, C.; et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol. Cell 2014, 54, 362–377. [Google Scholar] [CrossRef] [PubMed]
- Sugo, M.; Kimura, H.; Arasaki, K.; Amemiya, T.; Hirota, N.; Dohmae, N.; Imai, Y.; Inoshita, T.; Shiba-Fukushima, K.; Hattori, N.; et al. Syntaxin 17 regulates the localization and function of PGAM5 in mitochondrial division and mitophagy. EMBO J. 2018, 37, e98899. [Google Scholar] [CrossRef] [PubMed]
- Morales, I.; Sanchez, A.; Puertas-Avendaño, R.; Rodriguez-Sabate, C.; Perez-Barreto, A.; Rodriguez, M. Neuroglial transmitophagy and Parkinson’s disease. Glia 2020, 68, 2277–2299. [Google Scholar] [CrossRef] [PubMed]
- McLelland, G.-L.; Lee, S.A.; McBride, H.M.; Fon, E.A. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J. Cell Biol. 2016, 214, 275–291. [Google Scholar] [CrossRef]
- Poojary, S.; Shah, K.S.; Bhalala, K.B.; Hegde, A.U. CEDNIK syndrome in an Indian patient with a novel mutation of the SNAP29 gene. Pediatr. Dermatol. 2019, 36, 372–376. [Google Scholar] [CrossRef]
- Mastrodonato, V.; Beznoussenko, G.; Mironov, A.; Ferrari, L.; Deflorian, G.; Vaccari, T. A genetic model of CEDNIK syndrome in zebrafish highlights the role of the SNARE protein Snap29 in neuromotor and epidermal development. Sci. Rep. 2019, 9, 1211. [Google Scholar] [CrossRef]
- Monfrini, E.; Zech, M.; Steel, D.; Kurian, M.A.; Winkelmann, J.; Di Fonzo, A. HOPS-associated neurological disorders (HOPSANDs): Linking endolysosomal dysfunction to the pathogenesis of dystonia. Brain 2021, 144, 2610–2615. [Google Scholar] [CrossRef]
- Peter-Ross, E.M. Molecular hypotheses to explain the shared pathways and underlying pathobiological causes in catatonia and in catatonic presentations in neuropsychiatric disorders. Med. Hypotheses 2018, 113, 54–64. [Google Scholar] [CrossRef]
- Monfrini, E.; Cogiamanian, F.; Salani, S.; Straniero, L.; Fagiolari, G.; Garbellini, M.; Carsana, E.; Borellini, L.; Biella, F.; Moggio, M.; et al. A Novel Homozygous VPS11 Variant May Cause Generalized Dystonia. Ann. Neurol. 2021, 89, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Kamińska, J.; Kolakowski, D. Proteins from Vps13 family: From molecular function to pathogenesis of neurodegenerative disorders. Postepy Biochem. 2018, 64, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Wang, J.; Xiong, J.; Fang, N.; Ji, W.-K. VPS13D interacts with VCP/p97 and negatively regulates endoplasmic reticulum-mitochondria interactions. Mol. Biol. Cell 2021, 32, 1474–1486. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.-L.; Liu, W.; Hu, J.-X.; Erion, J.R.; Ye, J.; Mei, L.; Xiong, W.-C. VPS35 Deficiency or Mutation Causes Dopaminergic Neuronal Loss by Impairing Mitochondrial Fusion and Function. Cell Rep. 2015, 12, 1631–1643. [Google Scholar] [CrossRef]
- Wu, Y.-R.; Lin, C.-H.; Chao, C.-Y.; Chang, C.-W.; Chen, C.-M.; Lee-Chen, G.-J. Rare VPS35 A320V Variant in Taiwanese Parkinson’s Disease Indicates Disrupted CI-MPR Sorting and Impaired Mitochondrial Morphology. Brain Sci. 2020, 10, 783. [Google Scholar] [CrossRef]
- Li, G.; Marlin, M.C. Rab family of GTPases. Methods Mol. Biol. 2015, 1298, 1–15. [Google Scholar] [CrossRef]
- Pfeffer, S.R. Rab GTPases: Master regulators that establish the secretory and endocytic pathways. Mol. Biol. Cell 2017, 28, 712–715. [Google Scholar] [CrossRef]
- Zhen, Y.; Stenmark, H. Cellular functions of Rab GTPases at a glance. J. Cell Sci. 2015, 128, 3171–3176. [Google Scholar] [CrossRef]
- Sorkin, A.; von Zastrow, M. Endocytosis and signalling: Intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 2009, 10, 609–622. [Google Scholar] [CrossRef]
- Novick, P. Regulation of membrane traffic by Rab GEF and GAP cascades. Small GTPases 2016, 7, 252–256. [Google Scholar] [CrossRef]
- Hsu, F.; Spannl, S.; Ferguson, C.; Hyman, A.A.; Parton, R.G.; Zerial, M. Rab5 and Alsin regulate stress-activated cytoprotective signaling on mitochondria. Elife 2018, 7, e32282. [Google Scholar] [CrossRef] [PubMed]
- Otomo, A.; Hadano, S.; Okada, T.; Mizumura, H.; Kunita, R.; Nishijima, H.; Showguchi-Miyata, J.; Yanagisawa, Y.; Kohiki, E.; Suga, E.; et al. ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum. Mol. Genet. 2003, 12, 1671–1687. [Google Scholar] [CrossRef] [PubMed]
- Topp, J.D.; Gray, N.W.; Gerard, R.D.; Horazdovsky, B.F. Alsin is a Rab5 and Rac1 guanine nucleotide exchange factor. J. Biol. Chem. 2004, 279, 24612–24623. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Wong, Y.C.; Gao, F.; Krainc, D. Dysregulation of mitochondria-lysosome contacts by GBA1 dysfunction in dopaminergic neuronal models of Parkinson’s disease. Nat. Commun. 2021, 12, 1807. [Google Scholar] [CrossRef]
- Do, J.; McKinney, C.; Sharma, P.; Sidransky, E. Glucocerebrosidase and its relevance to Parkinson disease. Mol. Neurodegener. 2019, 14, 36. [Google Scholar] [CrossRef]
- Hegedűs, K.; Takáts, S.; Boda, A.; Jipa, A.; Nagy, P.; Varga, K.; Kovács, A.L.; Juhász, G. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol. Biol. Cell 2016, 27, 3132–3142. [Google Scholar] [CrossRef]
- Heo, J.-M.; Ordureau, A.; Swarup, S.; Paulo, J.A.; Shen, K.; Sabatini, D.M.; Harper, J.W. RAB7A phosphorylation by TBK1 promotes mitophagy via the PINK-PARKIN pathway. Sci. Adv. 2018, 4, eaav0443. [Google Scholar] [CrossRef]
- Ahmad, L.; Zhang, S.-Y.; Casanova, J.-L.; Sancho-Shimizu, V. Human TBK1: A Gatekeeper of Neuroinflammation. Trends Mol. Med. 2016, 22, 511–527. [Google Scholar] [CrossRef]
- Kim, C.Y.; Alcalay, R.N. Genetic Forms of Parkinson’s Disease. Semin. Neurol. 2017, 37, 135–146. [Google Scholar] [CrossRef]
- Vieweg, S.; Mulholland, K.; Bräuning, B.; Kachariya, N.; Lai, Y.-C.; Toth, R.; Singh, P.K.; Volpi, I.; Sattler, M.; Groll, M.; et al. PINK1-dependent phosphorylation of Serine111 within the SF3 motif of Rab GTPases impairs effector interactions and LRRK2-mediated phosphorylation at Threonine72. Biochem. J. 2020, 477, 1651–1668. [Google Scholar] [CrossRef]
- Shibata, S.; Teshima, Y.; Niimi, K.; Inagaki, S. Involvement of ARHGEF10, GEF for RhoA, in Rab6/Rab8-mediating membrane traffic. Small GTPases 2019, 10, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Peränen, J. Rab8 GTPase as a regulator of cell shape. Cytoskeleton 2011, 68, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Ryan, T.A.; Tumbarello, D.A. Optineurin: A Coordinator of Membrane-Associated Cargo Trafficking and Autophagy. Front. Immunol. 2018, 9, 1024. [Google Scholar] [CrossRef] [PubMed]
- Blümer, J.; Rey, J.; Dehmelt, L.; Mazel, T.; Wu, Y.-W.; Bastiaens, P.; Goody, R.S.; Itzen, A. RabGEFs are a major determinant for specific Rab membrane targeting. J. Cell Biol. 2013, 200, 287–300. [Google Scholar] [CrossRef]
- Lai, Y.-C.; Kondapalli, C.; Lehneck, R.; Procter, J.B.; Dill, B.D.; Woodroof, H.I.; Gourlay, R.; Peggie, M.; Macartney, T.J.; Corti, O.; et al. Phosphoproteomic screening identifies Rab GTPases as novel downstream targets of PINK1. EMBO J. 2015, 34, 2840–2861. [Google Scholar] [CrossRef]
- Li, X.; Standley, C.; Sapp, E.; Valencia, A.; Qin, Z.-H.; Kegel, K.B.; Yoder, J.; Comer-Tierney, L.A.; Esteves, M.; Chase, K.; et al. Mutant huntingtin impairs vesicle formation from recycling endosomes by interfering with Rab11 activity. Mol. Cell. Biol. 2009, 29, 6106–6116. [Google Scholar] [CrossRef]
- Li, X.; Sapp, E.; Chase, K.; Comer-Tierney, L.A.; Masso, N.; Alexander, J.; Reeves, P.; Kegel, K.B.; Valencia, A.; Esteves, M.; et al. Disruption of Rab11 activity in a knock-in mouse model of Huntington’s disease. Neurobiol. Dis. 2009, 36, 374–383. [Google Scholar] [CrossRef]
- Aoyama, K.; Suh, S.W.; Hamby, A.M.; Liu, J.; Chan, W.Y.; Chen, Y.; Swanson, R.A. Neuronal glutathione deficiency and age-dependent neurodegeneration in the EAAC1 deficient mouse. Nat. Neurosci. 2006, 9, 119–126. [Google Scholar] [CrossRef]
- Li, X.; Valencia, A.; Sapp, E.; Masso, N.; Alexander, J.; Reeves, P.; Kegel, K.B.; Aronin, N.; Difiglia, M. Aberrant Rab11-dependent trafficking of the neuronal glutamate transporter EAAC1 causes oxidative stress and cell death in Huntington’s disease. J. Neurosci. 2010, 30, 4552–4561. [Google Scholar] [CrossRef]
- Ortiz-Sandoval, C.G.; Hughes, S.C.; Dacks, J.B.; Simmen, T. Interaction with the effector dynamin-related protein 1 (Drp1) is an ancient function of Rab32 subfamily proteins. Cell. Logist. 2014, 4, e986399. [Google Scholar] [CrossRef]
- Haile, Y.; Deng, X.; Ortiz-Sandoval, C.; Tahbaz, N.; Janowicz, A.; Lu, J.-Q.; Kerr, B.J.; Gutowski, N.J.; Holley, J.E.; Eggleton, P.; et al. Rab32 connects ER stress to mitochondrial defects in multiple sclerosis. J. Neuroinflamm. 2017, 14, 19. [Google Scholar] [CrossRef] [PubMed]
- Rybnicek, J.; Samtleben, S.; Herrera-Cruz, M.S.; Simmen, T. Expression of a T39N mutant Rab32 protein arrests mitochondria movement within neurites of differentiated SH-SY5Y cells. Small GTPases 2020, 11, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Errea, O.; Moreno, B.; Gonzalez-Franquesa, A.; Garcia-Roves, P.M.; Villoslada, P. The disruption of mitochondrial axonal transport is an early event in neuroinflammation. J. Neuroinflamm. 2015, 12, 152. [Google Scholar] [CrossRef] [PubMed]
- D’Aquila, P.; Montesanto, A.; De Rango, F.; Guarasci, F.; Passarino, G.; Bellizzi, D. Epigenetic signature: Implications for mitochondrial quality control in human aging. Aging 2019, 11, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Waschbüsch, D.; Michels, H.; Strassheim, S.; Ossendorf, E.; Kessler, D.; Gloeckner, C.J.; Barnekow, A. LRRK2 transport is regulated by its novel interacting partner Rab32. PLoS ONE 2014, 9, e111632. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wilson, G.R.; Stephenson, S.E.M.; Bozaoglu, K.; Farrer, M.J.; Lockhart, P.J. The emerging role of Rab GTPases in the pathogenesis of Parkinson’s disease. Mov. Disord. 2018, 33, 196–207. [Google Scholar] [CrossRef]
- Li, Q.; Lei, F.; Tang, Y.; Pan, J.S.-C.; Tong, Q.; Sun, Y.; Sheikh-Hamad, D. Megalin mediates plasma membrane to mitochondria cross-talk and regulates mitochondrial metabolism. Cell. Mol. Life Sci. 2018, 75, 4021–4040. [Google Scholar] [CrossRef]
- Sheikh-Hamad, D.; Holliday, M.; Li, Q. Megalin-Mediated Trafficking of Mitochondrial Intracrines: Relevance to Signaling and Metabolism. J. Cell. Immunol. 2021, 3, 364–369. [Google Scholar]
- Basso, J.M.V.; Yurchenko, I.; Wiens, M.R.; Staii, C. Neuron dynamics on directional surfaces. Soft Matter 2019, 15, 9931–9941. [Google Scholar] [CrossRef]
- Fang, D.; Yan, S.; Yu, Q.; Chen, D.; Yan, S.S. Mfn2 is Required for Mitochondrial Development and Synapse Formation in Human Induced Pluripotent Stem Cells/hiPSC Derived Cortical Neurons. Sci. Rep. 2016, 6, 31462. [Google Scholar] [CrossRef]
- Wu, Z.; Zhu, Y.; Cao, X.; Sun, S.; Zhao, B. Mitochondrial toxic effects of Aβ through mitofusins in the early pathogenesis of Alzheimer’s disease. Mol. Neurobiol. 2014, 50, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.-H.; Lee, H.S.; Cha, D.M.; Kang, S.-Y. Hereditary Motor and Sensory Neuropathy Type VI with Bilateral Middle Cerebellar Peduncle Involvement. Exp. Neurobiol. 2014, 23, 266–269. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pareyson, D.; Saveri, P.; Sagnelli, A.; Piscosquito, G. Mitochondrial dynamics and inherited peripheral nerve diseases. Neurosci. Lett. 2015, 596, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Di Meglio, C.; Bonello-Palot, N.; Boulay, C.; Milh, M.; Ovaert, C.; Levy, N.; Chabrol, B. Clinical and allelic heterogeneity in a pediatric cohort of 11 patients carrying MFN2 mutation. Brain Dev. 2016, 38, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Oyarzabal, A.; Bravo-Alonso, I.; Sánchez-Aragó, M.; Rejas, M.T.; Merinero, B.; García-Cazorla, A.; Artuch, R.; Ugarte, M.; Rodríguez-Pombo, P. Mitochondrial response to the BCKDK-deficiency: Some clues to understand the positive dietary response in this form of autism. Biochim. Biophys. Acta 2016, 1862, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Filadi, R.; Greotti, E.; Turacchio, G.; Luini, A.; Pozzan, T.; Pizzo, P. Presenilin 2 Modulates Endoplasmic Reticulum-Mitochondria Coupling by Tuning the Antagonistic Effect of Mitofusin 2. Cell Rep. 2016, 15, 2226–2238. [Google Scholar] [CrossRef]
- Klacanova, K.; Kovalska, M.; Chomova, M.; Pilchova, I.; Tatarkova, Z.; Kaplan, P.; Racay, P. Global brain ischemia in rats is associated with mitochondrial release and downregulation of Mfn2 in the cerebral cortex, but not the hippocampus. Int. J. Mol. Med. 2019, 43, 2420–2428. [Google Scholar] [CrossRef]
- Han, S.; Nandy, P.; Austria, Q.; Siedlak, S.L.; Torres, S.; Fujioka, H.; Wang, W.; Zhu, X. Mfn2 Ablation in the Adult Mouse Hippocampus and Cortex Causes Neuronal Death. Cells 2020, 9, 116. [Google Scholar] [CrossRef]
- Maiti, P.; Lavdovskaia, E.; Barrientos, A.; Richter-Dennerlein, R. Role of GTPases in Driving Mitoribosome Assembly. Trends Cell Biol. 2021, 31, 284–297. [Google Scholar] [CrossRef]
- Hillen, H.S.; Lavdovskaia, E.; Nadler, F.; Hanitsch, E.; Linden, A.; Bohnsack, K.E.; Urlaub, H.; Richter-Dennerlein, R. Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling. Nat. Commun. 2021, 12, 3672. [Google Scholar] [CrossRef]
- Wang, X.-L.; Liu, Q.; Chen, G.-J.; Li, M.-L.; Ding, Y.-H. Overexpression of MTERF4 promotes the amyloidogenic processing of APP by inhibiting ADAM10. Biochem. Biophys. Res. Commun. 2017, 482, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Vawter, M.P.; Harvey, P.D.; DeLisi, L.E. Dysregulation of X-linked gene expression in Klinefelter’s syndrome and association with verbal cognition. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2007, 144, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Simonetti, L.; Ferreira, L.G.A.; Vidi, A.C.; de Souza, J.S.; Kunii, I.S.; Melaragno, M.I.; de Mello, C.B.; Carvalheira, G.; Dias da Silva, M.R. Intelligence Quotient Variability in Klinefelter Syndrome Is Associated With GTPBP6 Expression Under Regulation of X-Chromosome Inactivation Pattern. Front. Genet. 2021, 12, 724625. [Google Scholar] [CrossRef]
- Alberts, P.; Rudge, R.; Hinners, I.; Muzerelle, A.; Martinez-Arca, S.; Irinopoulou, T.; Marthiens, V.; Tooze, S.; Rathjen, F.; Gaspar, P.; et al. Cross talk between tetanus neurotoxin-insensitive vesicle-associated membrane protein-mediated transport and L1-mediated adhesion. Mol. Biol. Cell 2003, 14, 4207–4220. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, M.; Thompson, P.; Lemmon, V. CRASH Syndrome: Mutations in L1CAM Correlate with Severity of the Disease. Neuropediatrics 1997, 28, 175–178. [Google Scholar] [CrossRef]
- Fransen, E.; van Camp, G.; Vits, L.; Willems, P.J. L1-associated diseases: Clinical geneticists divide, molecular geneticists unite. Hum. Mol. Genet. 1997, 6, 1625–1632. [Google Scholar] [CrossRef]
- Gasparotto, M.; Hernandez Gomez, Y.S.; Peterle, D.; Grinzato, A.; Zen, F.; Pontarollo, G.; Acquasaliente, L.; Scapin, G.; Bergantino, E.; De Filippis, V.; et al. NOG-Derived Peptides Can Restore Neuritogenesis on a CRASH Syndrome Cell Model. Biomedicines 2022, 10, 102. [Google Scholar] [CrossRef]
- Kraus, K.; Kleene, R.; Braren, I.; Loers, G.; Lutz, D.; Schachner, M. A fragment of adhesion molecule L1 is imported into mitochondria, and regulates mitochondrial metabolism and trafficking. J. Cell Sci. 2018, 131, jcs210500. [Google Scholar] [CrossRef]
- Hu, J.; Lin, S.L.; Schachner, M. A fragment of cell adhesion molecule L1 reduces amyloid-β plaques in a mouse model of Alzheimer’s disease. Cell Death Dis. 2022, 13, 48. [Google Scholar] [CrossRef]
- Kamemura, K.; Chihara, T. Multiple functions of the ER-resident VAP and its extracellular role in neural development and disease. J. Biochem. 2019, 165, 391–400. [Google Scholar] [CrossRef]
- Borgese, N.; Iacomino, N.; Colombo, S.F.; Navone, F. The Link between VAPB Loss of Function and Amyotrophic Lateral Sclerosis. Cells 2021, 10, 1865. [Google Scholar] [CrossRef] [PubMed]
- Kamemura, K.; Chen, C.-A.; Okumura, M.; Miura, M.; Chihara, T. Amyotrophic lateral sclerosis-associated Vap33 is required for maintaining neuronal dendrite morphology and organelle distribution in Drosophila. Genes Cells 2021, 26, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, S.; Sillence, D.J. Niemann-Pick type C disease: Cellular pathology and pharmacotherapy. J. Neurochem. 2020, 153, 674–692. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, K.; Hutt, D.M.; Scott, S.M.; Gupta, V.; Mao, S.; Balch, W.E. Correction of Niemann-Pick type C1 trafficking and activity with the histone deacetylase inhibitor valproic acid. J. Biol. Chem. 2020, 295, 8017–8035. [Google Scholar] [CrossRef]
- LoPresti, P. HDAC6 in Diseases of Cognition and of Neurons. Cells 2020, 10, 12. [Google Scholar] [CrossRef]
- English, K.; Barton, M.C. HDAC6: A Key Link Between Mitochondria and Development of Peripheral Neuropathy. Front. Mol. Neurosci. 2021, 14, 684714. [Google Scholar] [CrossRef]
- Gold, W.A.; Lacina, T.A.; Cantrill, L.C.; Christodoulou, J. MeCP2 deficiency is associated with reduced levels of tubulin acetylation and can be restored using HDAC6 inhibitors. J. Mol. Med. 2015, 93, 63–72. [Google Scholar] [CrossRef]
- Adalbert, R.; Kaieda, A.; Antoniou, C.; Loreto, A.; Yang, X.; Gilley, J.; Hoshino, T.; Uga, K.; Makhija, M.T.; Coleman, M.P. Novel HDAC6 Inhibitors Increase Tubulin Acetylation and Rescue Axonal Transport of Mitochondria in a Model of Charcot-Marie-Tooth Type 2F. ACS Chem. Neurosci. 2020, 11, 258–267. [Google Scholar] [CrossRef]
- Fazal, R.; Boeynaems, S.; Swijsen, A.; De Decker, M.; Fumagalli, L.; Moisse, M.; Vanneste, J.; Guo, W.; Boon, R.; Vercruysse, T.; et al. HDAC6 inhibition restores TDP-43 pathology and axonal transport defects in human motor neurons with TARDBP mutations. EMBO J. 2021, 40, e106177. [Google Scholar] [CrossRef]
- Henley, J.M.; Carmichael, R.E.; Wilkinson, K.A. Extranuclear SUMOylation in Neurons. Trends Neurosci. 2018, 41, 198–210. [Google Scholar] [CrossRef]
- Guerra de Souza, A.C.; Prediger, R.D.; Cimarosti, H. SUMO-regulated mitochondrial function in Parkinson’s disease. J. Neurochem. 2016, 137, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Juncker, M.; Kim, C.; Reed, R.; Haas, A.; Schwartzenburg, J.; Desai, S. ISG15 attenuates post-translational modifications of mitofusins and congression of damaged mitochondria in Ataxia Telangiectasia cells. Biochim. Biophys. Acta. Mol. Basis Dis. 2021, 1867, 166102. [Google Scholar] [CrossRef] [PubMed]
- Iorio, R.; Celenza, G.; Petricca, S. Mitophagy: Molecular Mechanisms, New Concepts on Parkin Activation and the Emerging Role of AMPK/ULK1 Axis. Cells 2021, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Barazzuol, L.; Giamogante, F.; Brini, M.; Calì, T. PINK1/Parkin Mediated Mitophagy, Ca(2+) Signalling, and ER-Mitochondria Contacts in Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 1772. [Google Scholar] [CrossRef]
- Singh, P.K.; Muqit, M.M.K. Parkinson’s: A Disease of Aberrant Vesicle Trafficking. Annu. Rev. Cell Dev. Biol. 2020, 36, 237–264. [Google Scholar] [CrossRef]
- O’Hara, D.M.; Pawar, G.; Kalia, S.K.; Kalia, L.V. LRRK2 and α-Synuclein: Distinct or Synergistic Players in Parkinson’s Disease? Front. Neurosci. 2020, 14, 577. [Google Scholar] [CrossRef]
- Calió, M.L.; Henriques, E.; Siena, A.; Bertoncini, C.R.A.; Gil-Mohapel, J.; Rosenstock, T.R. Mitochondrial Dysfunction, Neurogenesis, and Epigenetics: Putative Implications for Amyotrophic Lateral Sclerosis Neurodegeneration and Treatment. Front. Neurosci. 2020, 14, 679. [Google Scholar] [CrossRef]
- Julian, L.M.; Stanford, W.L. Organelle Cooperation in Stem Cell Fate: Lysosomes as Emerging Regulators of Cell Identity. Front. Cell Dev. Biol. 2020, 8, 591. [Google Scholar] [CrossRef]
- Hanashima, C.; Li, S.C.; Shen, L.; Lai, E.; Fishell, G. Foxg1 suppresses early cortical cell fate. Science 2004, 303, 56–59. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Zoghbi, H.Y. Rett Syndrome and the Ongoing Legacy of Close Clinical Observation. Cell 2016, 167, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gasparotto, M.; Lee, Y.-S.; Palazzi, A.; Vacca, M.; Filippini, F. Nuclear and Cytoplasmatic Players in Mitochondria-Related CNS Disorders: Chromatin Modifications and Subcellular Trafficking. Biomolecules 2022, 12, 625. https://doi.org/10.3390/biom12050625
Gasparotto M, Lee Y-S, Palazzi A, Vacca M, Filippini F. Nuclear and Cytoplasmatic Players in Mitochondria-Related CNS Disorders: Chromatin Modifications and Subcellular Trafficking. Biomolecules. 2022; 12(5):625. https://doi.org/10.3390/biom12050625
Chicago/Turabian StyleGasparotto, Matteo, Yi-Shin Lee, Alessandra Palazzi, Marcella Vacca, and Francesco Filippini. 2022. "Nuclear and Cytoplasmatic Players in Mitochondria-Related CNS Disorders: Chromatin Modifications and Subcellular Trafficking" Biomolecules 12, no. 5: 625. https://doi.org/10.3390/biom12050625
APA StyleGasparotto, M., Lee, Y.-S., Palazzi, A., Vacca, M., & Filippini, F. (2022). Nuclear and Cytoplasmatic Players in Mitochondria-Related CNS Disorders: Chromatin Modifications and Subcellular Trafficking. Biomolecules, 12(5), 625. https://doi.org/10.3390/biom12050625






