What Is Parvalbumin for?
Abstract
1. Introduction
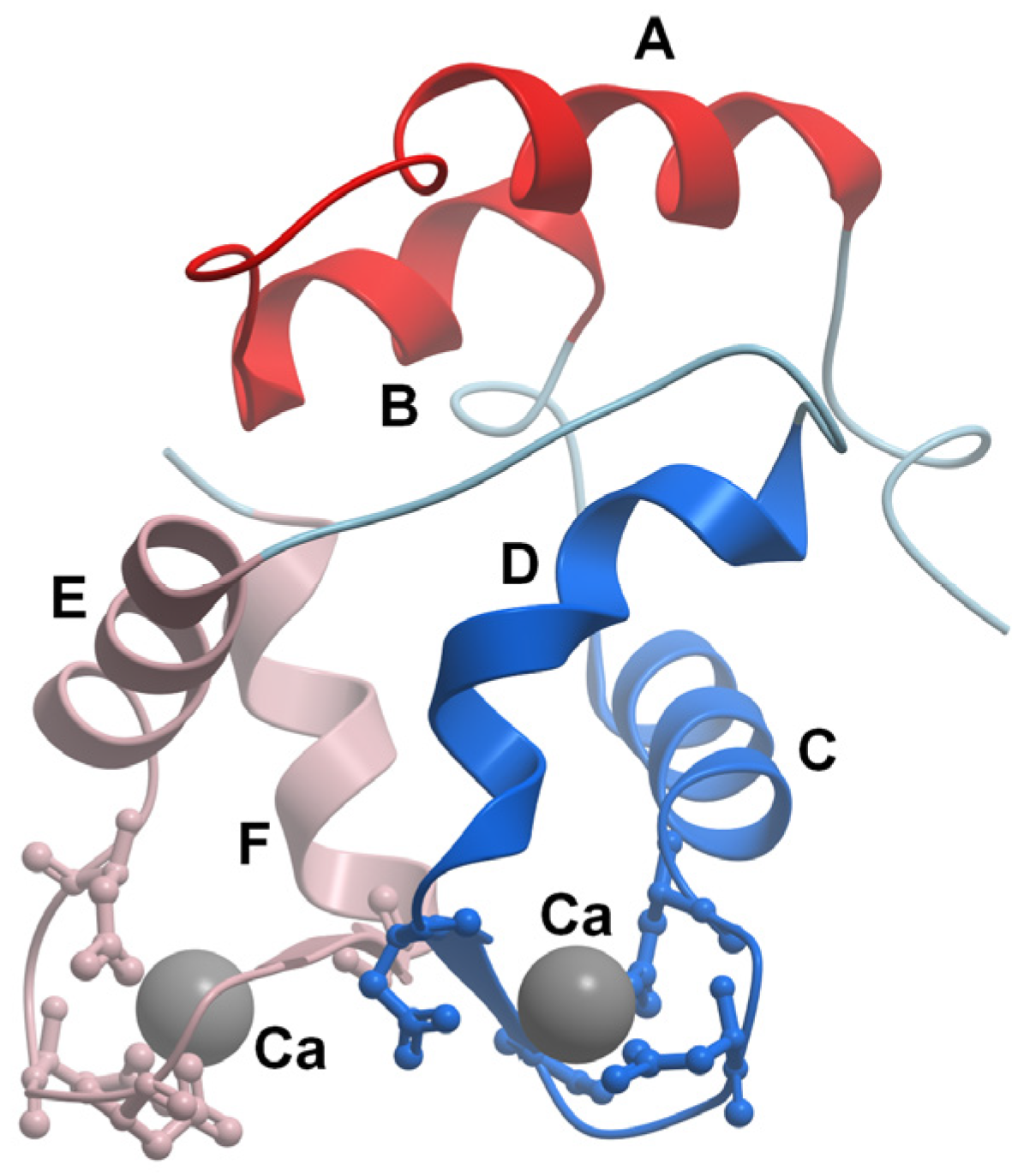
2. Parvalbumin Structure
3. Binding of Metal Ions to Parvalbumin
4. Interactions with Peptides and Membranes
5. Possible Functions of Parvalbumin
5.1. Parvalbumin in Muscle Cells
5.2. Parvalbumin in Neurons
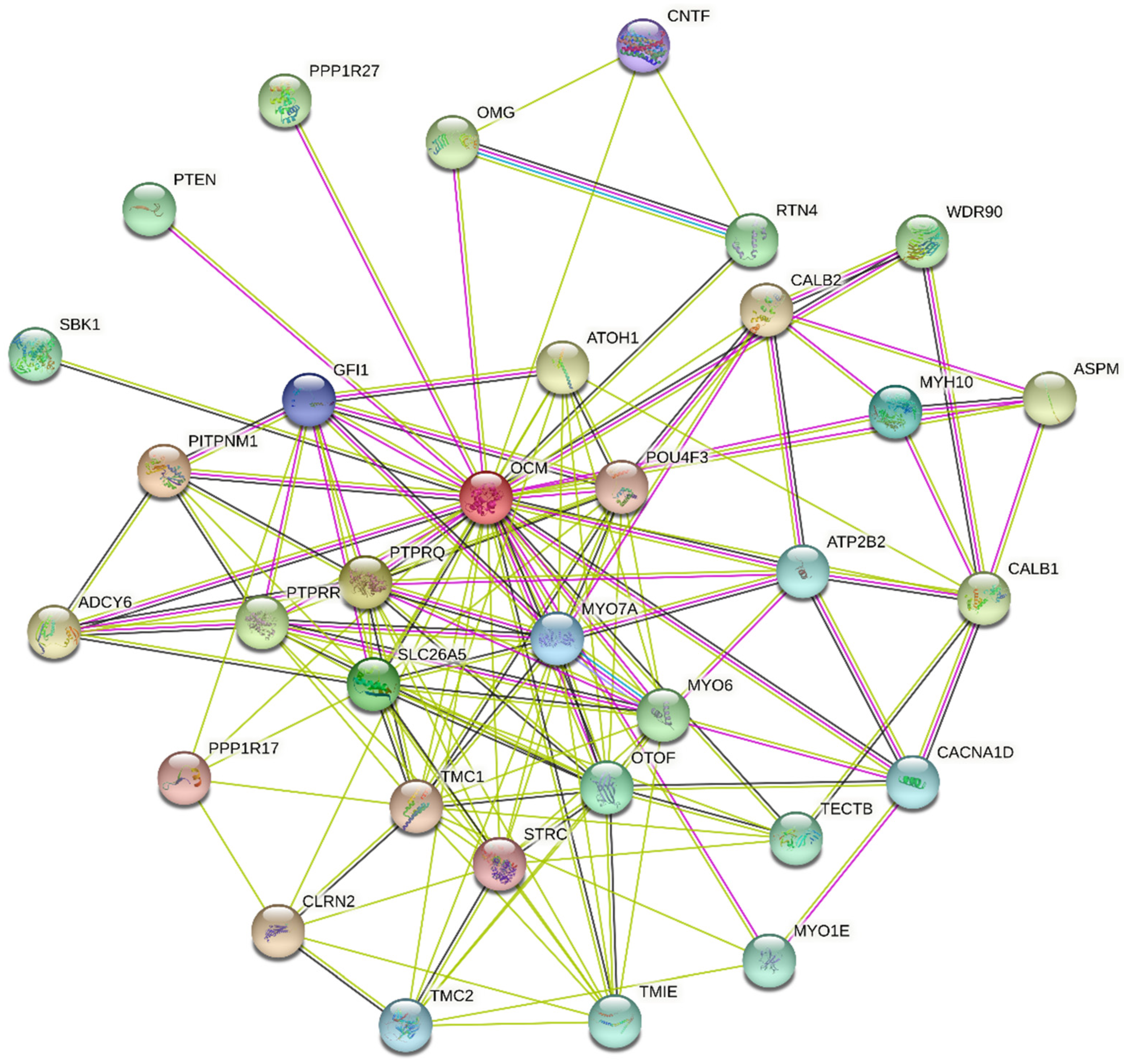
5.3. Oncomodulin
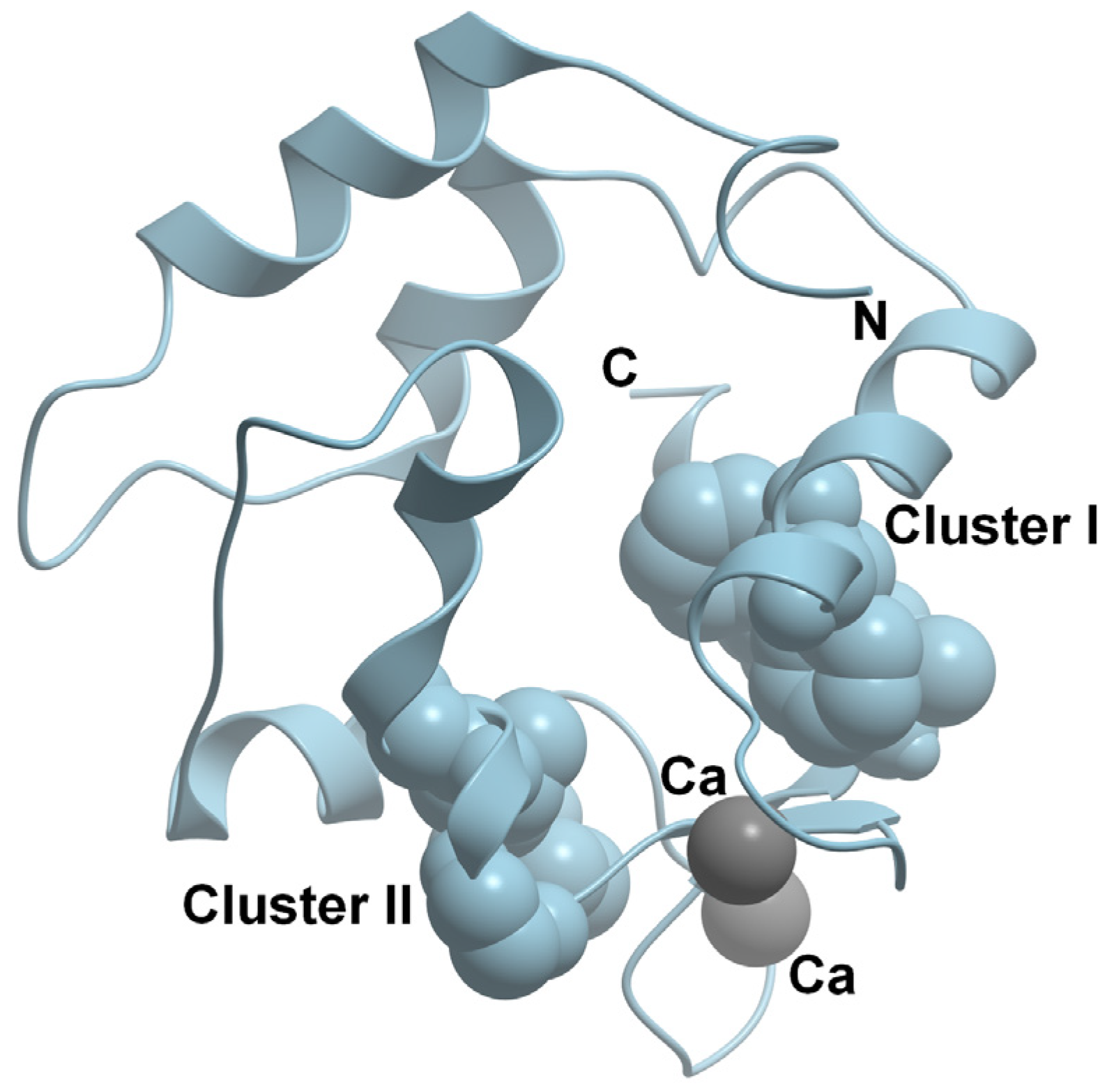
6. Intrinsic Disorder, Structural Flexibility, and Multifunctionality of Parvalbumin
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Permyakov, E.A. Parvalbumin; Nova Science Publishers, Inc.: New York, NY, USA, 2006. [Google Scholar]
- Permyakov, E.A.; Uversky, V.N.; Permyakov, S.E. Parvalbumin as a pleomorphic protein. Curr. Protein Pept. Sci. 2016, 18, 780–794. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, E.A.; Kretsinger, R.H. Calcium Binding Proteins; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010. [Google Scholar]
- Arif, S.H. A Ca2+-binding protein with numerous roles and uses: Parvalbumin in molecular biology and physiology. Bioessays 2009, 31, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Smargiassi, M.; Daghfous, G.; Leroy, B.; Legreneur, P.; Toubeau, G.; Bels, V.; Wattiez, R. Chemical basis of prey recognition in thamnophiine snakes: The unexpected new roles of parvalbumins. PLoS ONE 2012, 7, e39560. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yin, Y.; Henzl, M.T.; Lorber, B.; Nakazawa, T.; Thomas, T.T.; Jiang, F.; Langer, R.; Benowitz, L.I. Oncomodulin is a macrophage-derived signal for axon regeneration in retinal ganglion cells. Nat. Neurosci. 2006, 9, 843–852. [Google Scholar] [CrossRef]
- Novak Kujundzic, R.; Steffens, W.L.; Brewer, J.M.; Henzl, M.T.; Ragland, W.L. Characterization of avian thymic hormone and chicken parvalbumin 3 target cells. Int. Immunopharmacol. 2013, 15, 282–288. [Google Scholar] [CrossRef]
- Goodman, M.; Pechere, J.F. The evolution of muscular parvalbumins investigated by the maximum parsimony method. J. Mol. Evol. 1977, 9, 131–158. [Google Scholar] [CrossRef]
- Nakayama, S.; Moncrief, N.D.; Kretsinger, R.H. Evolution of EF hand calcium-modulated proteins. II. Domains of several subfamilies have diverse evolutionary histories. J. Mol. Evol. 1992, 34, 416–448. [Google Scholar] [CrossRef]
- Moncrief, N.D.; Kretsinger, R.H.; Goodman, M. Evolution of EF hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J. Mol. Evol. 1990, 30, 522–562. [Google Scholar] [CrossRef]
- Nockolds, C.E.; Kretsinger, R.H.; Coffee, C.J.; Bradshaw, R.A. Structure of a calcium-binding carp myogen. Proc. Natl. Acad. Sci. USA 1972, 69, 581–584. [Google Scholar] [CrossRef]
- Kretsinger, R.H.; Nockolds, C.E. Carp muscle calcium-binding protein. II. Structure determination and general description. J. Biol. Chem. 1973, 248, 3313–3326. [Google Scholar] [CrossRef]
- Permyakov, E.A.; Medvedkin, V.N.; Mitin, Y.V.; Kretsinger, R.H. Noncovalent complex between domain AB and domains CD*EF of parvalbumin. Biochim. Biophys. Acta 1991, 1076, 67–70. [Google Scholar] [CrossRef]
- Thepaut, M.; Strub, M.P.; Cave, A.; Baneres, J.L.; Berchtold, M.W.; Dumas, C.; Padilla, A. Structure of rat parvalbumin with deleted AB domain: Implications for the evolution of EF hand calcium-binding proteins and possible physiological relevance. Proteins 2001, 45, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Henzl, M.T.; Agah, S.; Larson, J.D. Association of the AB and CDEF domains from rat alpha- and beta-parvalbumin. Biochemistry 2004, 43, 10906–10917. [Google Scholar] [CrossRef] [PubMed]
- Denessiouk, K.; Permyakov, S.; Denesyuk, A.; Permyakov, E.; Johnson, M.S. Two structural motifs within canonical EF-hand calcium-binding domains identify five different classes of calcium buffers and sensors. PLoS ONE 2014, 9, e109287. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, S.E.; Vologzhannikova, A.A.; Khorn, P.A.; Shevelyova, M.P.; Kazakov, A.S.; Emelyanenko, V.I.; Denesyuk, A.I.; Denessiouk, K.A.; Uversky, V.N.; Permyakov, E.A. Comprehensive analysis of the roles of ‘black’ and ‘gray’ clusters in structure and function of rat β-parvalbumin. Cell Calcium 2018, 75, 64–78. [Google Scholar] [CrossRef] [PubMed]
- White, H.D. Kinetic mechanism of calcium binding to whiting parvalbumin. Biochemistry 1988, 27, 3357–3365. [Google Scholar] [CrossRef] [PubMed]
- Vologzhannikova, A.A.; Shevelyova, M.P.; Kazakov, A.S.; Sokolov, A.S.; Borisova, N.I.; Permyakov, E.A.; Kircheva, N.; Nikolova, V.; Dudev, T.; Permyakov, S.E. Strontium binding to α-parvalbumin, a canonical calcium-binding protein of the “EF-hand” family. Biomolecules 2021, 11, 1158. [Google Scholar] [CrossRef]
- Pauls, T.L.; Durussel, I.; Berchtold, M.W.; Cox, J.A. Inactivation of individual Ca2+-binding sites in the paired EF-hand sites of parvalbumin reveals asymmetrical metal-binding properties. Biochemistry 1994, 33, 10393–10400. [Google Scholar] [CrossRef]
- Permyakov, E.A.; Ostrovsky, A.V.; Kalinichenko, L.P. Stopped flow kinetic studies of Ca(II) and Mg(II) dissociation in cod parvalbumin and bovine alpha-lactalbumin. Biophys. Chem. 1987, 28, 225–233. [Google Scholar] [CrossRef]
- Eberspach, I.; Strassburger, W.; Glatter, U.; Gerday, C.; Wollmer, A. Interaction of parvalbumin of pike II with calcium and terbium ions. Biochim. Biophys. Acta 1988, 952, 67–76. [Google Scholar] [CrossRef]
- Laberge, M.; Wright, W.W.; Sudhakar, K.; Liebman, P.A.; Vanderkooi, J.M. Conformational effects of calcium release from parvalbumin: Comparison of computational simulations with spectroscopic investigations. Biochemistry 1997, 36, 5363–5371. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, S.E.; Bakunts, A.G.; Denesyuk, A.I.; Knyazeva, E.L.; Uversky, V.N.; Permyakov, E.A. Apo-parvalbumin as an intrinsically disordered protein. Proteins 2008, 72, 822–836. [Google Scholar] [CrossRef] [PubMed]
- Henzl, M.T.; Tanner, J.J. Solution structure of Ca2+-free rat alpha-parvalbumin. Protein Sci. 2008, 17, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Henzl, M.T.; Tanner, J.J. Solution structure of Ca2+-free rat beta-parvalbumin (oncomodulin). Protein Sci. 2007, 16, 1914–1926. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, E.A.; Yarmolenko, V.V.; Emelyanenko, V.I.; Burstein, E.A.; Closset, J.; Gerday, C. Fluorescence studies of the calcium binding to whiting (Gadus merlangus) parvalbumin. Eur. J. Biochem. 1980, 109, 307–315. [Google Scholar] [CrossRef]
- Henzl, M.T.; Larson, J.D.; Agah, S. Influence of monovalent cations on rat alpha- and beta-parvalbumin stabilities. Biochemistry 2000, 39, 5859–5867. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Bakunts, A.G.; Permyakova, M.E.; Denesyuk, A.I.; Uversky, V.N.; Permyakov, E.A. Metal-controlled interdomain cooperativity in parvalbumins. Cell Calcium 2009, 46, 163–175. [Google Scholar] [CrossRef][Green Version]
- Filimonov, V.V.; Pfeil, W.; Tsalkova, T.N.; Privalov, P.L. Thermodynamic investigations of proteins. IV. Calcium binding protein parvalbumin. Biophys. Chem. 1978, 8, 117–122. [Google Scholar] [CrossRef]
- Dolgikh, D.A.; Gilmanshin, R.I.; Brazhnikov, E.V.; Bychkova, V.E.; Semisotnov, G.V.; Venyaminov, S.; Ptitsyn, O.B. Alpha-lactalbumin: Compact state with fluctuating tertiary structure? FEBS Lett. 1981, 136, 311–315. [Google Scholar] [CrossRef]
- Kuwajima, K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins 1989, 6, 87–103. [Google Scholar] [CrossRef]
- Finkelstein, A.V.; Shakhnovich, E.I. Theory of cooperative transitions in protein molecules. II. Phase diagram for a protein molecule in solution. Biopolymers 1989, 28, 1681–1694. [Google Scholar] [CrossRef] [PubMed]
- Vologzhannikova, A.A.; Khorn, P.A.; Kazakov, A.S.; Ismailov, R.G.; Sokolov, A.S.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. In search for globally disordered apo-parvalbumins: Case of parvalbumin β-1 from coho salmon. Cell Calcium 2017, 67, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Declercq, J.P.; Tinant, B.; Parello, J.; Rambaud, J. Ionic interactions with parvalbumins. Crystal structure determination of pike 4.10 parvalbumin in four different ionic environments. J. Mol. Biol. 1991, 220, 1017–1039. [Google Scholar] [CrossRef]
- Blancuzzi, Y.; Padilla, A.; Parello, J.; Cave, A. Symmetrical rearrangement of the cation-binding sites of parvalbumin upon Ca2+/Mg2+ exchange. A study by 1H 2D NMR. Biochemistry 1993, 32, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Nara, M.; Tasumi, M.; Tanokura, M.; Hiraoki, T.; Yazawa, M.; Tsutsumi, A. Infrared studies of interaction between metal ions and Ca2+-binding proteins. Marker bands for identifying the types of coordination of the side-chain COO- groups to metal ions in pike parvalbumin (pI = 4.10). FEBS Lett. 1994, 349, 84–88. [Google Scholar] [CrossRef]
- Permyakov, E.A.; Medvedkin, V.N.; Korneichuk, G.A.; Kostrzhevskaia, E.G.; Murzin, A.G. Ca(II) inhibited binding of melittin to parvalbumin. A new function for parvalbumin. Mol. Biol. Mosc. 1989, 23, 693–698. [Google Scholar]
- Comte, M.; Maulet, Y.; Cox, J.A. Ca2+-dependent high-affinity complex formation between calmodulin and melittin. Biochem. J. 1983, 209, 269–272. [Google Scholar] [CrossRef]
- Maulet, Y.; Cox, J.A. Structural changes in melittin and calmodulin upon complex formation and their modulation by calcium. Biochemistry 1983, 22, 5680–5686. [Google Scholar] [CrossRef]
- Permyakov, E.A.; Kreimer, D.I.; Kalinichenko, L.P.; Orlova, A.A.; Shnyrov, V.L. Interaction of parvalbumins with model phospholipid vesicles. Cell Calcium 1989, 10, 71–79. [Google Scholar] [CrossRef]
- Fritz-Niggli, H.; Nievergelt-Egido, C.; Heizmann, C.W. Calcium binding parvalbumin in Drosophila testis in connection with in vivo irradiation. Radiat. Environ. Biophys. 1988, 27, 59–65. [Google Scholar] [CrossRef]
- Berchtold, M.W.; Brinkmeier, H.; Muntener, M. Calcium ion in skeletal muscle: Its crucial role for muscle function, plasticity, and disease. Physiol. Rev. 2000, 80, 1215–1265. [Google Scholar] [CrossRef]
- Hou, T.T.; Johnson, J.D.; Rall, J.A. Parvalbumin content and Ca2+ and Mg2+ dissociation rates correlated with changes in relaxation rate of frog muscle fibres. J. Physiol. Lond. 1991, 441, 285–304. [Google Scholar] [CrossRef] [PubMed]
- Leberer, E.; Pette, D. Immunochemical quantification of sarcoplasmic reticulum Ca-ATPase, of calsequestrin and of parvalbumin in rabbit skeletal muscles of defined fiber composition. Eur. J. Biochem. 1986, 156, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Haiech, J.; Derancourt, J.; Pechere, J.F.; Demaille, J.G. Magnesium and calcium binding to parvalbumins: Evidence for differences between parvalbumins and an explanation of their relaxing function. Biochemistry 1979, 18, 2752–2758. [Google Scholar] [CrossRef] [PubMed]
- Müntener, M.; Berchtold, M.W.; Heizmann, C.W. Parvalbumin in cross-reinnervated and denervated muscles. Muscle Nerve 1985, 8, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Müntener, M.; Rowlerson, A.M.; Berchtold, M.W.; Heizmann, C.W. Changes in the concentration of the calcium-binding parvalbumin in cross-reinnervated rat muscles. Comparison of biochemical with physiological and histochemical parameters. J. Biol. Chem. 1987, 262, 465–469. [Google Scholar] [CrossRef]
- Heizmann, C.W.; Berchtold, M.W.; Rowlerson, A.M. Correlation of parvalbumin concentration with relaxation speed in mammalian muscles. Proc. Natl. Acad. Sci. USA 1982, 79, 7243–7247. [Google Scholar] [CrossRef] [PubMed]
- Muntener, M.; Kaser, L.; Weber, J.; Berchtold, M.W. Increase of skeletal muscle relaxation speed by direct injection of parvalbumin cDNA. Proc. Natl. Acad. Sci. USA 1995, 92, 6504–6508. [Google Scholar] [CrossRef]
- Schwaller, B.; Dick, J.; Dhoot, G.; Carroll, S.; Vrbova, G.; Nicotera, P.; Pette, D.; Wyss, A.; Bluethmann, H.; Hunziker, W.; et al. Prolonged contraction–relaxation cycle of fast-twitch muscles in parvalbumin knockout mice. Am. J. Physiol. 1999, 276, C395–C403. [Google Scholar] [CrossRef]
- Raymackers, J.M.; Gailly, P.; Schoor, M.C.; Pette, D.; Schwaller, B.; Hunziker, W.; Celio, M.R.; Gillis, J.M. Tetanus relaxation of fast skeletal muscles of the mouse made parvalbumin deficient by gene inactivation. J. Physiol. 2000, 527, 355–364. [Google Scholar] [CrossRef]
- Rome, L.C.; Syme, D.A.; Hollingworth, S.; Lindstedt, S.L.; Baylor, S.M. The whistle and the rattle: The design of sound producing muscles. Proc. Natl. Acad. Sci. USA 1996, 93, 8095–8100. [Google Scholar] [CrossRef] [PubMed]
- Appelt, D.; Shen, V.; Franzini-Armstrong, C. Quantitation of CaATPase, feet and mitochondria in superfast muscle fibres from the toadfish, Opsanus tau. J. Muscle Res. Cell Motil. 1991, 12, 543–552. [Google Scholar] [CrossRef]
- Tikunov, B.A.; Rome, L.C. Is high concentration of parvalbumin a requirement for superfast relaxation? J. Muscle Res. Cell Motil. 2009, 30, 57–65. [Google Scholar] [CrossRef]
- Vongvatcharanon, S.; Vongvatcharanon, U.; Boonyoung, P. Immunohistochemical localization of parvalbumin calcium-binding protein in the heart tissues of various species. Acta Histochem. 2008, 110, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Vongvatcharanon, U.; Vongvatcharanon, S. Localization of parvalbumin calcium binding protein in the rat heart. Sci. Asia 2003, 29, 319–325. [Google Scholar] [CrossRef]
- Wahr, P.A.; Michele, D.E.; Metzger, J.M. Parvalbumin gene transfer corrects diastolic dysfunction in diseased cardiac myocytes. Proc. Natl. Acad. Sci. USA 1999, 96, 11982–11985. [Google Scholar] [CrossRef] [PubMed]
- Szatkowski, M.L.; Westfall, M.V.; Gomez, C.A.; Wahr, P.A.; Michele, D.E.; DelloRusso, C.; Turner, I.I.; Hong, K.E.; Albayya, F.P.; Metzger, J.M. In vivo acceleration of heart relaxation performance by parvalbumin gene delivery. J. Clin. Investig. 2001, 107, 191–198. [Google Scholar] [CrossRef][Green Version]
- Coutu, P.; Hirsch, J.C.; Szatkowski, M.L.; Metzger, J.M. Targeting diastolic dysfunction by genetic engineering of calcium handling proteins. Trends Cardiovasc. Med. 2003, 13, 63–67. [Google Scholar] [CrossRef]
- Michele, D.E.; Szatkowski, M.L.; Albayya, F.P.; Metzger, J.M. Parvalbumin gene delivery improves diastolic function in the aged myocardium in vivo. Mol. Ther. 2004, 10, 399–403. [Google Scholar] [CrossRef]
- Schmidt, U.; Zhu, X.; Lebeche, D.; Huq, F.; Guerrero, J.L.; Hajjar, R.J. In vivo gene transfer of parvalbumin improves diastolic function in aged rat hearts. Cardiovasc. Res. 2005, 66, 318–323. [Google Scholar] [CrossRef]
- Vongvatcharanon, U.; Udomuksorn, W.; Vongvatcharanon, S.; Sobhon, P. Age-related changes in parvalbumin in the heart of female rats. Acta Histochem. 2010, 112, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Asp, M.L.; Sjaastad, F.V.; Siddiqui, J.K.; Davis, J.P.; Metzger, J.M. Effects of modified parvalbumin EF-hand motifs on cardiac myocyte contractile function. Biophys. J. 2016, 110, 2094–2105. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Crocetti, J.K. Oxygen content, delivery and uptake. In Critical Care Study Guide; Criner, G.D.A., Ed.; Springer: New York, NY, USA, 2002; pp. 355–368. [Google Scholar]
- Giorgio, M.; Trinei, M.; Migliaccio, E.; Pelicci, P.G. Hydrogen peroxide: A metabolicby-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol. 2007, 8, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, S.E.; Kazakov, A.S.; Avkhacheva, N.V.; Permyakov, E.A. Parvalbumin as a metal-dependent antioxidant. Cell Calcium 2014, 55, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Hof, P.R.; Glezer, I.I.; Conde, F.; Flagg, R.A.; Rubin, M.B.; Nimchinsky, E.A.; Vogt Weisenhorn, D.M. Cellular distribution of the calcium-binding proteins parvalbumin, calbindin, and calretinin in the neocortex of mammals: Phylogenetic and developmental patterns. J. Chem. Neuroanat. 1999, 16, 77–116. [Google Scholar] [CrossRef]
- Schwaller, B. Cytosolic Ca2+ Buffers Are Inherently Ca2+ Signal Modulators. Cold Spring Harb. Perspect. Biol. 2020, 12, a035543. [Google Scholar] [CrossRef]
- Heizmann, C.W. Calcium signaling in the brain. Acta Neurobiol. Exp. (Wars) 1993, 53, 15–23. [Google Scholar]
- Celio, M.R. Parvalbumin in most gamma-aminobutyric acid containing neurons of the rat cerebral cortex. Science 1986, 231, 995–997. [Google Scholar] [CrossRef]
- Plogmann, D.; Celio, M.R. Intracellular concentration of parvalbumin in nerve cells. Brain Res. 1993, 600, 273–279. [Google Scholar] [CrossRef]
- Zuschratter, W.; Scheich, H.; Heizmann, C.W. Ultrastructural localization of the calcium-binding protein parvalbumin in neurons of the song system of the zebra finch, Poephila guttata. Cell Tissue Res. 1985, 241, 77–83. [Google Scholar] [CrossRef]
- Stichel, C.C.; Singer, W.; Heizmann, C.W.; Norman, A.W. Immunohistochemical localization of calcium-binding proteins, parvalbumin and calbindin-D 28k, in the adult and developing visual cortex of cats: A light and electron microscopic study. J. Comparat. Neurol. 1987, 262, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Caillard, O.; Moreno, H.; Schwaller, B.; Llano, I.; Celio, M.R.; Marty, A. Role of the calcium-binding protein parvalbumin in short-term synaptic plasticity. Proc. Natl. Acad. Sci. USA 2000, 97, 13372–13377. [Google Scholar] [CrossRef] [PubMed]
- Collin, T.; Chat, M.; Lucas, M.G.; Moreno, H.; Racay, P.; Schwaller, B.; Marty, A.; Llano, I. Developmental changes in parvalbumin regulate presynaptic Ca2+ signaling. J. Neurosci. 2005, 25, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Vreugdenhil, M.; Jefferys, J.G.; Celio, M.R.; Schwaller, B. Parvalbumin-deficiency facilitates repetitive IPSCs and gamma oscillations in the hippocampus. J. Neurophysiol. 2003, 89, 1414–1422. [Google Scholar] [CrossRef]
- Eggermann, E.; Jonas, P. How the ‘slow’ Ca2+ buffer parvalbumin affects transmitter release in nanodomain-coupling regimes. Nat. Neurosci. 2011, 15, 20–22. [Google Scholar] [CrossRef]
- Gonzalez-Burgos, G.; Lewis, D.A. NMDA receptor hypofunction, parvalbumin-positive neurons, and cortical γ oscillations in schizophrenia. Schizophr. Bull. 2012, 38, 950–957. [Google Scholar] [CrossRef]
- Berridge, M.J. Calcium signalling and psychiatric disease: Bipolar disorder and schizophrenia. Cell Tissue Res. 2014, 357, 477–492. [Google Scholar] [CrossRef]
- Steullet, P.; Cabungcal, J.H.; Coyle, J.; Didriksen, M.; Gill, K.; Grace, A.A.; Hensch, T.K.; LaMantia, A.S.; Lindemann, L.; Maynard, T.M.; et al. Oxidative stress-driven parvalbumin interneuron impairment as a common mechanism in models of schizophrenia. Mol. Psychiatry 2017, 22, 936–943. [Google Scholar] [CrossRef]
- Chen, G.; Carroll, S.; Racay, P.; Dick, J.; Pette, D.; Traub, I.; Vrbova, G.; Eggli, P.; Celio, M.; Schwaller, B. Deficiency in parvalbumin increases fatigue resistance in fast-twitch muscle and upregulates mitochondria. Am. J. Physiol. Cell Physiol. 2001, 281, C114–C122. [Google Scholar] [CrossRef]
- Chen, G.; Racay, P.; Bichet, S.; Celio, M.R.; Eggli, P.; Schwaller, B. Deficiency in parvalbumin, but not in calbindin D-28k upregulates mitochondrial volume and decreases smooth endoplasmic reticulum surface selectively in a peripheral, subplasmalemmal region in the soma of Purkinje cells. Neuroscience 2006, 142, 97–105. [Google Scholar] [CrossRef]
- Ducreux, S.; Gregory, P.; Schwaller, B. Inverse regulation of the cytosolic Ca2+ buffer parvalbumin and mitochondrial volume in muscle cells via SIRT1/PGC-1α axis. PLoS ONE 2012, 7, e44837. [Google Scholar] [CrossRef] [PubMed]
- Henzi, T.; Schwaller, B. Antagonistic regulation of parvalbumin expression and mitochondrial calcium handling capacity in renal epithelial cells. PLoS ONE 2015, 10, e0142005. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.R.; Gao, W.J. PV interneurons: Critical regulators of E/I balance for prefrontal cortex-dependent behavior and psychiatric disorders. Front. Neural Circuits 2018, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Wöhr, M.; Orduz, D.; Gregory, P.; Moreno, H.; Khan, U.; Vorckel, K.J.; Wolfer, D.P.; Welzl, H.; Gall, D.; Schiffmann, S.N.; et al. Lack of parvalbumin in mice leads to behavioral deficits relevant to all human autism core symptoms and related neural morphofunctional abnormalities. Transl. Psychiatry 2015, 5, e525. [Google Scholar] [CrossRef]
- Do, K.Q.; Cabungcal, J.H.; Frank, A.; Steullet, P.; Cuenod, M. Redox dysregulation, neurodevelopment, and schizophrenia. Curr. Opin. Neurobiol. 2009, 19, 220–230. [Google Scholar] [CrossRef]
- Sullivan, E.M.; O’Donnell, P. Inhibitory interneurons, oxidative stress, and schizophrenia. Schizophr. Bull. 2012, 38, 373–376. [Google Scholar] [CrossRef]
- Jiang, Z.; Cowell, R.M.; Nakazawa, K. Convergence of genetic and environmental factors on parvalbumin-positive interneurons in schizophrenia. Front. Behav. Neurosci. 2013, 7, 116. [Google Scholar] [CrossRef]
- Hidalgo, C.; Donoso, P. Crosstalk between calcium and redox signaling: From molecular mechanisms to health implications. Antioxid. Redox Signal. 2008, 10, 1275–1312. [Google Scholar] [CrossRef]
- Ermak, G.; Davies, K.J. Calcium and oxidative stress: From cell signaling to cell death. Mol. Immunol. 2002, 38, 713–721. [Google Scholar] [CrossRef]
- Berchtold, M.W. Structure and expression of genes encoding the three-domain Ca2+-binding proteins parvalbumin and oncomodulin. Biochim. Biophys. Acta 1989, 1009, 201–215. [Google Scholar] [CrossRef]
- Climer, L.K.; Cox, A.M.; Reynolds, T.J.; Simmons, D.D. Oncomodulin: The enigmatic parvalbumin protein. Front. Mol. Neurosci. 2019, 12, 235. [Google Scholar] [CrossRef] [PubMed]
- MacManus, J.P.; Whitfield, J.F.; Boynton, A.L.; Durkin, J.P.; Swierenga, S.H. Oncomodulin—A widely distributed, tumour-specific, calcium-binding protein. Oncodev. Biol. Med. 1982, 3, 79–90. [Google Scholar] [PubMed]
- Senarita, M.; Thalmann, I.; Shibasaki, O.; Thalmann, R. Calcium binding proteins in organ of Corti and basilar papilla: CBP-15, an unidentified calcium-binding protein of the inner ear. Hear. Res. 1995, 90, 169–175. [Google Scholar] [CrossRef]
- Thalmann, R.; Henzl, M.T.; Thalmann, I. Specific proteins of the organ of Corti. Acta Otolaryngol. 1997, 117, 265–268. [Google Scholar] [CrossRef]
- Henzl, M.T.; Shibasaki, O.; Comegys, T.H.; Thalmann, I.; Thalmann, R. Oncomodulin is abundant in the organ of Corti. Hear. Res. 1997, 106, 105–111. [Google Scholar] [CrossRef]
- Sakaguchi, N.; Henzl, M.T.; Thalmann, I.; Thalmann, R.; Schulte, B.A. Oncomodulin is expressed exclusively by outer hair cells in the organ of Corti. J. Histochem. Cytochem. 1998, 46, 29–40. [Google Scholar] [CrossRef]
- Tong, B.; Hornak, A.J.; Maison, S.F.; Ohlemiller, K.K.; Liberman, M.C.; Simmons, D.D. Oncomodulin, an EF-hand Ca2+ buffer, is critical for maintaining cochlear function in mice. J. Neurosci. 2016, 36, 1631–1635. [Google Scholar] [CrossRef]
- Hoffman, L.F.; Choy, K.R.; Sultemeier, D.R.; Simmons, D.D. Oncomodulin expression reveals new insights into the cellular organization of the murine utricle striola. J. Assoc. Res. Otolaryngol. 2018, 19, 33–51. [Google Scholar] [CrossRef]
- Yin, Y.; Cui, Q.; Gilbert, H.Y.; Yang, Y.; Yang, Z.; Berlinicke, C.; Li, Z.; Zaverucha-do-Valle, C.; He, H.; Petkova, V.; et al. Oncomodulin links inflammation to optic nerve regeneration. Proc. Natl. Acad. Sci. USA 2009, 106, 19587–19592. [Google Scholar] [CrossRef]
- Kurimoto, T.; Yin, Y.; Omura, K.; Gilbert, H.Y.; Kim, D.; Cen, L.P.; Moko, L.; Kügler, S.; Benowitz, L.I. Long-distance axon regeneration in the mature optic nerve: Contributions of oncomodulin, cAMP, and pten gene deletion. J. Neurosci. 2010, 30, 15654–15663. [Google Scholar] [CrossRef]
- Siawaya, J.F.D.; Capito, C.; Scharfmann, R. Oncomodulin and macrophages derived factors in pancreas injury and development paradigms. Mod. Res. Inflamm. 2013, 2, 1–8. [Google Scholar] [CrossRef]
- Hackney, C.M.; Mahendrasingam, S.; Penn, A.; Fettiplace, R. The concentrations of calcium buffering proteins in mammalian cochlear hair cells. J. Neurosci. 2005, 25, 7867–7875. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.A.; Milos, M.; MacManus, J.P. Calcium- and magnesium binding properties of oncomodulin. J. Biol. Chem. 1990, 265, 6633–6637. [Google Scholar] [CrossRef]
- Pangrsic, T.; Gabrielaitis, M.; Michanski, S.; Schwaller, B.; Wolf, F.; Strenzke, N.; Moser, T. EF-hand protein Ca2+ buffers regulate Ca2+ influx and exocytosis in sensory hair cells. Proc. Natl. Acad. Sci. USA 2015, 112, E1028–E1037. [Google Scholar] [CrossRef]
- Benowitz, L.I.; Popovich, P.G. Inflammation and axon regeneration. Curr. Opin. Neurol. 2011, 24, 577–583. [Google Scholar] [CrossRef]
- de Lima, S.; Koriyama, Y.; Kurimoto, T.; Oliveira, J.T.; Yin, Y.; Li, Y.; Gilbert, H.Y.; Fagiolini, M.; Martinez, A.M.; Benowitz, L. Full-length axon regeneration in the adult mouse optic nerve and partial recovery of simple visual behaviors. Proc. Natl. Acad. Sci. USA 2012, 109, 9149–9154. [Google Scholar] [CrossRef]
- Marin, M.A.; de Lima, S.; Gilbert, H.Y.; Giger, R.J.; Benowitz, L.; Rasband, M.N. Reassembly of excitable domains after CNS axon regeneration. J. Neurosci. 2016, 36, 9148–9160. [Google Scholar] [CrossRef]
- Huber, S.; Leuthold, M.; Sommer, E.W.; Heizmann, C.W. Human tumor cell lines express low levels of oncomodulin. Biochem. Biophys. Res. Commun. 1990, 169, 905–909. [Google Scholar] [CrossRef]
- Mes-Masson, A.M.; Masson, S.; Banville, D.; Chalifour, L. Expression of oncomodulin does not lead to the transformation or immortalization of mammalian cells in vitro. J. Cell Sci. 1989, 94, 517–525. [Google Scholar] [CrossRef]
- MacManus, J.P.; Watson, D.C.; Yaguchi, M. The complete amino acid sequence of oncomodulin—a parvalbumin-like calcium-binding protein from Morris hepatoma 5123tc. Eur. J. Biochem. 1983, 136, 9–17. [Google Scholar] [CrossRef]
- MacManus, J.P.; Watson, D.C.; Yaguchi, M. A new member of the troponin C superfamily: Comparison of the primary structures of rat oncomodulin and rat parvalbumin. Biosci. Rep. 1983, 3, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Brewer, L.M.; MacManus, J.P. Detection of oncomodulin, an oncodevelopmental protein in human placenta and choriocarcinoma cell lines. Placenta 1987, 8, 351–363. [Google Scholar] [CrossRef]
- Sommer, E.W.; Heizmann, C.W. Expression of the tumor-specific and calcium-binding protein oncomodulin during chemical transformation of rat fibroblasts. Cancer Res. 1989, 49, 899–905. [Google Scholar] [PubMed]
- Palmer, E.J.; MacManus, J.P.; Mutus, B. Inhibition of glutathione reductase by oncomodulin. Arch. Biochem. Biophys. 1990, 277, 149–154. [Google Scholar] [CrossRef]
- Klee, C.B.; Heppel, L.A. The effect of oncomodulin on cAMP phosphodiesterase activity. Biochem. Biophys. Res. Commun. 1984, 125, 420–424. [Google Scholar] [CrossRef]
- Clayshulte, T.M.; Taylor, D.F.; Henzl, M.T. Reactivity of cysteine 18 in oncomodulin. J. Biol. Chem. 1990, 265, 1800–1805. [Google Scholar] [CrossRef]
- Leroy, B.; Toubeau, G.; Falmagne, P.; Wattiez, R. Identification and characterization of new protein chemoattractants in the frog skin secretome. Mol. Cell Proteom. 2006, 5, 2114–2123. [Google Scholar] [CrossRef]
- Kägi, U.; Chafouleas, J.G.; Norman, A.W.; Heizmann, C.W. Developmental appearance of the Ca2R-binding proteins parvalbumin, calbindin D-28K, S-100 proteins and calmodulin during testicular development in the rat. Cell. Tissue Res. 1988, 252, 359–365. [Google Scholar] [CrossRef]
- Berchtold, M.W.; Celio, M.R.; Heizmann, C.W. Parvalbumin in non-muscle tissues of the rat. J. Biol. Chem. 1984, 259, 5189–5196. [Google Scholar] [CrossRef]
- Endo, T.; Takazawa, K.; Onaya, T. Parvalbumin exists in rat endocrine glands. Endocrinology 1985, 117, 527–531. [Google Scholar] [CrossRef]
- Pauls, T.L.; Portis, F.; Macrı, E.; Belser, B.; Heitz, P.; Doglioni, C.; Celio, M.R. Parvalbumin is expressed in normal and pathological human parathyroid glands. J. Histochem. Cytochem. 2000, 48, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Belge, H.; Gailly, P.; Schwaller, B.; Loffing, J.; Debaix, H.; Riveira-Munoz, E.; Beauwens, R.; Devogelaer, J.P.; Hoenderop, J.G.; Bindels, R.J.; et al. Renal expression of parvalbumin is critical for NaCl handling and response to diuretics. Proc. Natl. Acad. Sci. USA 2007, 104, 14849–14854. [Google Scholar] [CrossRef] [PubMed]
- Zacchia, M.; Capasso, G. Parvalbumin: A key protein in early tubule NaCl reabsorption. Nephrol. Dial. Transplant. 2008, 23, 1109–1111. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005, 3, 35–60. [Google Scholar] [CrossRef]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef]
- Dosztányi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005, 347, 827–839. [Google Scholar] [CrossRef]
- Necci, M.; Piovesan, D.; CAID Predictors; DisProt Curators; Tosatto, S.C.E. Critical assessment of protein intrinsic disorder prediction. Nat. Methods 2021, 18, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef] [PubMed]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D(2)P(2): Database of disordered protein predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. ESpritz: Accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, A.; Howorth, D.; Brenner, S.E.; Hubbard, T.J.; Chothia, C.; Murzin, A.G. SCOP database in 2004: Refinements integrate structure and sequence family data. Nucleic Acids Res. 2004, 32, D226–D229. [Google Scholar] [CrossRef]
- Murzin, A.G.; Brenner, S.E.; Hubbard, T.; Chothia, C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995, 247, 536–540. [Google Scholar] [CrossRef]
- de Lima Morais, D.A.; Fang, H.; Rackham, O.J.; Wilson, D.; Pethica, R.; Chothia, C.; Gough, J. SUPERFAMILY 1.75 including a domain-centric gene ontology method. Nucleic Acids Res. 2011, 39, D427–D434. [Google Scholar] [CrossRef]
- Meszaros, B.; Simon, I.; Dosztanyi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009, 5, e1000376. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Kornhauser, J.M.; Tkachev, S.; Zhang, B.; Skrzypek, E.; Murray, B.; Latham, V.; Sullivan, M. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012, 40, D261–D270. [Google Scholar] [CrossRef]
- Uversky, V.N. Protein intrinsic disorder and structure-function continuum. Prog. Mol. Biol. Transl. Sci. 2019, 166, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fonin, A.V.; Darling, A.L.; Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. Multi-functionality of proteins involved in GPCR and G protein signaling: Making sense of structure-function continuum with intrinsic disorder-based proteoforms. Cell Mol. Life Sci. 2019, 76, 4461–4492. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. p53 Proteoforms and Intrinsic Disorder: An Illustration of the Protein Structure-Function Continuum Concept. Int. J. Mol. Sci. 2016, 17, 1874. [Google Scholar] [CrossRef]
- Uversky, V.N. Looking at the recent advances in understanding α-synuclein and its aggregation through the proteoform prism. F1000Research 2017, 6, 525. [Google Scholar] [CrossRef] [PubMed]
- Malaney, P.; Uversky, V.N.; Davé, V. PTEN proteoforms in biology and disease. Cell Mol. Life Sci. 2017, 74, 2783–2794. [Google Scholar] [CrossRef]
- Marzullo, L.; Turco, M.C.; Uversky, V.N. What’s in the BAGs? Intrinsic disorder angle of the multifunctionality of the members of a family of chaperone regulators. J. Cell Biochem. 2022, 123, 22–42. [Google Scholar] [CrossRef]
- Smith, L.M.; Kelleher, N.L.; Consortium for Top Down Proteomics. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef] [PubMed]




| Parvalbumin (UniProt ID) | Number of Amino Acids | pI | Molecular Mass (Da) |
|---|---|---|---|
| Pike α-PA (P02628) | 108 | 5.0 | 11,707 |
| Pike β-PA (P02619) | 107 | 4.2 | 11,390 |
| Cod β-PA (P02622) | 113 | 4.4 | 12,108 |
| Rat α-PA (P02625) | 110 | 5.5 | 11,926 |
| Rat β-PA oncomodulin (P02631) | 109 | 4.4 | 12,188 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Permyakov, E.A.; Uversky, V.N. What Is Parvalbumin for? Biomolecules 2022, 12, 656. https://doi.org/10.3390/biom12050656
Permyakov EA, Uversky VN. What Is Parvalbumin for? Biomolecules. 2022; 12(5):656. https://doi.org/10.3390/biom12050656
Chicago/Turabian StylePermyakov, Eugene A., and Vladimir N. Uversky. 2022. "What Is Parvalbumin for?" Biomolecules 12, no. 5: 656. https://doi.org/10.3390/biom12050656
APA StylePermyakov, E. A., & Uversky, V. N. (2022). What Is Parvalbumin for? Biomolecules, 12(5), 656. https://doi.org/10.3390/biom12050656







