Abstract
Phosphorylation facilitates the regulation of all fundamental biological processes, which has triggered extensive research of protein kinases and their roles in human health and disease. In addition to their phosphotransferase activity, certain kinases have evolved to adopt additional catalytic functions, while others have completely lost all catalytic activity. We searched the Universal Protein Resource Knowledgebase (UniProtKB) database for bifunctional protein kinases and focused on kinases that are critical for bacterial and human cellular homeostasis. These kinases engage in diverse functional roles, ranging from environmental sensing and metabolic regulation to immune-host defense and cell cycle control. Herein, we describe their dual catalytic activities and how they contribute to disease pathogenesis.
1. Introduction
Protein phosphorylation regulates prokaryotic and eukaryotic cellular processes and signal transduction [1]. In humans, protein phosphorylation is mediated by 538 protein kinases, which comprise 3% of the human genome [2,3]. Protein kinases control metabolism, transcription, translation, cell cycle progression, cytoskeletal rearrangement, apoptosis, differentiation, intracellular communication, and homeostasis; thus, their dysregulation drives many human diseases. Development of small molecule kinase inhibitors has yielded 71 FDA-approved compounds used in the treatment of various cancers and inflammatory diseases [4,5].
Most FDA-approved kinase inhibitors bind to the highly conserved adenosine triphosphate (ATP)-binding pocket and prevent binding of substrate ATP. Surprisingly, some kinase targets have been found to retain or even increase their downstream signaling despite binding to ATP-competitive inhibitors. For example, certain BRAF inhibitors and kinase-dead BRAF mutants paradoxically activate downstream MEK phosphorylation by inducing dimerization and activation of uninhibited BRAF and CRAF [6,7,8]. The RAF family kinases exemplify the many noncatalytic functions of kinases, which extend to scaffolding, allosteric regulation, protein-DNA interactions, subcellular targeting, and beyond [9,10].
Protein kinases can exhibit catalytic activities other than phosphotransfer. For example, Ire1α is an evolutionarily conserved member of the unfolded protein response pathway (UPR) and contains both a protein kinase domain and an endoribonuclease domain. The UPR is a stress response pathway that responds to the accumulation of unfolded proteins in the endoplasmic reticulum (ER) by producing molecular chaperones and halting translation. Misfolded proteins induce Ire1α dimerization, which subsequently activates the ribonuclease domain (RNase) domain to splice transcription factor XBP1 and upregulate UPR response genes. Feldman and coworkers explored the allosteric relationship between Ire1α’s kinase and RNase domains by screening commercially available ATP-competitive kinase inhibitors and imidazopyrazine-based kinase-inhibiting RNase attenuators (KIRAs) [11]. Inhibitors of Ire1α kinase were found to bind and stabilize specific kinase domain conformations. Some kinase inhibitors promoted dimerization and activated the enzyme’s RNase activity, whereas the KIRAs stabilized monomeric Ire1α and inhibited both kinase and RNase activity. This work provides a proof-of-concept that ATP-competitive inhibitors can influence the non-phosphotransferase functions of their target kinases.
Bifunctional kinases such as Ire1α have the potential to reveal unique modes of allosteric regulation between different protein domains, as well as provide mechanistic insights into the functional benefits of having two signaling activities within the same enzyme. This knowledge can be further employed to guide drug discovery or drug repurposing campaigns toward finding unique strategies for specific and effective kinase inhibition. There may also be functional benefits to having two signaling activities on one enzyme, as is seen in other multi-domain enzyme complexes like fatty acid synthase.
In this review, we identify protein kinases documented with multiple catalytic activities in the UniProtKB database. We present a synopsis of our findings and highlight the bifunctionalities of bacterial and human kinases that are critical for cellular homeostasis.
2. Materials and Methods
We mined the UniProtKB (release version 2022_01), SwissProt, and TrEMBL databases [12] with a search strategy designed to identify Ser/Thr, Tyr, and dual-specificity kinases based on Enzyme Commission (EC) classification numbers. Although histidine kinases comprise much of the bacterial proteome, we restricted ourselves to only Ser/Thr and Tyr protein kinases, the critical mediators of signal transduction in humans.
The UniProtKB search algorithm employed in this study was as follows:
((ec:2.7.10.* OR ec:2.7.11.* OR ec:2.7.12.*) (ec:1.*.*.* OR ec:3.*.*.* OR ec:4.*.*.* OR ec:5.*.*.* OR ec:6.*.*.* OR ec:2.1.*.* OR ec:2.2.*.* OR ec:2.3.*.* OR ec:2.4.*.* OR ec:2.5.*.* OR ec:2.6.*.* OR ec:2.8.*.* OR ec:2.9.*.* OR ec:2.10.*.* OR ec:2.7.1.* OR ec:2.7.2.* OR ec:2.7.3.* OR ec:2.7.4.* OR ec:2.7.5.* OR ec:2.7.6.* OR ec:2.7.7.* OR ec:2.7.8.* OR ec:2.7.9.* OR ec:2.7.13.* OR ec:2.7.14.* OR ec:2.7.99.*)) OR (ec:2.7.10.* (ec:2.7.11.* OR ec:2.7.12.*)) OR (ec:2.7.11.* ec:2.7.12.*)
Our search returned 36,160 protein kinases with documented bifunctionality (Table S1). Of these kinases, 879 were Swiss-Prot Reviewed, with records extracted from the literature and curator-evaluated computational analysis. The remaining 35,281 kinases were TrEMBL Unreviewed, indicating that they have been computationally analyzed but still require full manual annotation by UniProtKB curators. We focused our efforts on the reviewed kinases to ensure that our selection contained verified bifunctional protein kinases (Table S2). We manually inspected all 879 entries to identify unique groups of homologous kinases.
3. Results
Most of the kinases are dual-specificity Ser/Thr and Tyr protein kinases or protein kinases that predominantly exhibit other nucleotide triphosphatase activities. However, our search also returned examples of protein kinase activity paired with phosphatase, phosphorylase, endoribonuclease, acetyltransferase, carboxylyase, and sugar kinase catalytic functions. Given our interest in disease-relevant prokaryotic and eukaryotic kinases, we removed plant bifunctional kinases from the species Arabidopsis thaliana (22 kinases), Oryza sativa (2 kinases), and Petunia integrifolia (1 kinase), winnowing our initial 879 kinases down to 854 kinases. In addition, we removed two kinases found only in Dictyostelium discoideum, the bifunctional serine/threonine-protein kinase/NEDD4-like E3 ubiquitin-protein ligase and hybrid signal transduction histidine kinase G (dhkG). These kinases do not have homologs in humans, and their functions are inferred only from homology. Replicates across different bacterial strains made up most of the remaining 852 kinases, which we divided into 22 unique kinases (Table S3). General overviews of these kinases and their domain architectures are given in Table 1 and Figure 1, respectively.

Table 1.
Unique bifunctional kinases in the UniProtKB database.
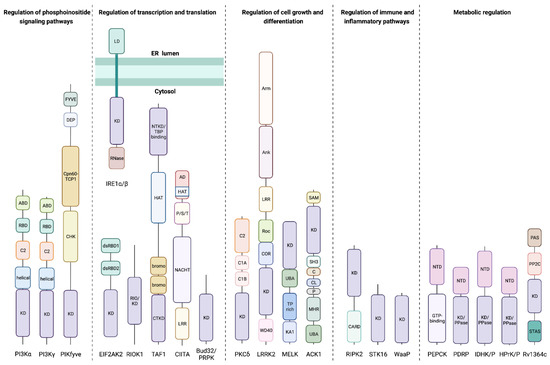
Figure 1.
Domain architecture of bifunctional kinases. Representative structural domain schematic of 22 unique bifunctional kinases annotated in the UniProtKB database. Abbreviations denote the following: kinase domain (KD), Ras-binding domain (RBD), adaptor-binding domain (ABD), Cys-His-Lys homology domain (CHK), chaperonin-60–T-complex protein 1 subunit alpha domain (Cpn60-TCP1), disheveled–EGL10–pleckstrin domain (DEP), Fab 1–YOTB–Vac 1–EEA1 zinc finger domain (FYVE), endoribonuclease (RNase), luminal domain (LD), double-stranded RNA binding domain (dsRBD), C-terminal kinase domain (CTKD), N-terminal kinase domain (NTKD), TATA-box binding protein (TBP), histone acetyltransferase domain (HAT), bromodomain (bromo), leucine-rich repeat (LRR), GTP-binding domain (GTP), Pro-Ser-Thr rich domain (P/S/T), acidic domain (AD), β-transducin repeat (WD40), C-terminal of Roc domain (COR), Ras of complex G-domain (Roc), ankyrin domain (Ank), Armadillo domain (Arm), kinase-associated 1 domain (KA1), TP dipeptide-rich domain (TP rich), ubiquitin-associated region (UBA), Mig6 homology region (MHR), PPXY motif (P), clathrin-interacting region (CL), Cdc42/GTPase binding domain (C), Src homology 3 domain (SH3), sterile alpha motif (SAM), caspase activation/recruitment domain (CARD), N-terminal domain (NTD), kinase and phosphatase/phosphorylase/pyrophosphorylase domain (KD/PPase), Per-Arnt-Sim domain (PAS), PP2C-like phosphatase domain (PP2C).
4. Regulation of Phosphoinositide Signaling Pathways
4.1. Phosphatidylinositol 3-Kinases (PI3K)
PI3Ks are ubiquitously expressed intracellular lipid kinases that phosphorylate the 3′ hydroxyl groups of phosphatidylinositol and also harbor Ser/Thr protein kinase activity. These proteins regulate several critical cellular functions including cell survival, proliferation, motility, and vesicle trafficking [13,14]. PI3Ks are divided into three classes (class I, II, and III) based on their primary structure and substrate specificity. Class-I PI3Ks are further separated into subclasses IA (PI3Kα, PI3Kβ, PI3Kδ) and IB (PI3Kγ) based on their regulatory proteins and signaling pathway involvement.
PI3Ks function as heterodimers, consisting of a regulatory subunit and a catalytic subunit that converts the phosphatidylinositol second messenger PI(3,4)P2 (PIP2) to PI(3,4,5)P3 (PIP3) in response to extracellular stimuli on the activation of upstream receptor tyrosine kinases (RTKs) and G-protein coupled receptors (GPCRs) [15,16]. Conversion of PIP2 to PIP3 promotes the activation of downstream proteins AKT and mTOR [17]. PI3K activity is antagonized by the PTEN phosphatase, which hydrolyzes PIP3 to PIP2 [18]. As PIP3 plays a critical role in cell growth and replication, aberrant lipid kinase activity can promote oncogenic signaling. Class-I proteins have been recognized as promising drug targets due to their involvement in cancer and immune disease pathogenesis via the PI3K/mTOR pathway. The structural conservation of the ATP-binding pocket has complicated the development of isoform-specific inhibitors. Pan-PI3K inhibitors are active against all class-I isoforms and result in off-target toxicity and side effects. To date, there are five FDA-approved PI3K inhibitors (alpelisib, copanlisib, duvelisib, idelalisib, and umbralisib). While PI3K kinase inhibitors have been successfully applied to certain malignancies, their use and development has been hindered by poor drug tolerance and toxicity [19].
4.2. PI3Kα (PIK3CA)
PIK3CA encodes the phosphatidylinositol 3-kinase catalytic subunit alpha (PI3Kα), also known as p110α. PI3Kα is the predominant catalytic isoform for regulation of glucose homeostasis and is commonly mutated and amplified in a variety of cancers [20]. The catalytic p110α subunit is composed of five domains, including an adaptor binding domain (ABD) that binds to the Class-IA regulatory domains, a Ras binding domain (RBD), a C2 domain that binds to cell membranes, a helical domain with unknown function, and a catalytic kinase domain. PI3Kα activation is mediated by receptor tyrosine kinases (RTKs), Ras proteins, and additional small molecules including calmodulin [21,22].
The catalytic p110α subunit can couple to regulatory subunit p85α, the SH2 domain of which contains high-affinity binding sites to the phosphorylated tyrosine motif (pYXXM) found in the C-terminus of RTKs [23]. The RTK pYXXM motif disrupts interactions between the catalytic p110α subunit and regulatory p85α subunit and activates PI3Kα by releasing the inhibitory p85α SH2 domains from the catalytic p110α subunit. This release triggers a conformational change in PI3Kα that exposes the kinase domain and permits membrane binding [24]. It has been shown that p110α phosphorylates Ser608 of the p85α regulatory subunit and decreases catalytic activity in vitro [25]. Mutation of Ser608 to Ala or Glu reduced lipid kinase activity, as well as the interactions between the p110α catalytic subunit and p85α regulatory subunit. Thus, phosphorylation of Ser608 may reveal a mechanism for regulating activity by stabilizing the autoinhibited state.
Mutations in PI3Kα are frequently found in brain, breast, head and neck, endometrial, cervical, and gastric cancers [17,26,27]. Most mutations cause gain-of-function (GOF) and are found within the helical or kinase domains. It has been reported that GOF mutations in the helical domain require interactions with Ras-GTP, whereas kinase domain mutations require interactions with the p85 regulatory subunit [28]. Co-existing mutations in both domains have been found to synergistically increase the catalytic function and tumorigenic activity [29]. In addition, deletions in the C2 domain have been found to activate PI3K signaling while also increasing the sensitivity to PI3Kα inhibitors, suggesting that residues within the C2 domain are critical for for PI3Kα function [30]. There are several hotspot mutations in p110α that confer a gain of function, including helical domain mutations E545K and E542K and the catalytic domain mutation H1047R. Helical domain mutations E545K and E542K have been shown to suppress inhibition of p110α by the p85 regulatory subunit, which results in mutation-driven signaling that promotes glucose metabolism and cervical cancer cell proliferation [31]. Catalytic domain mutation H1047R has been shown to enhance interactions between p110α and the lipid membrane, thereby enhancing its lipid kinase activity and downstream signaling.
The importance of PI3Kα in disease has been well-established; in May 2019, alpelisib (Piqray™, Novartis, Morris County, NJ, USA) was the first PI3Kα inhibitor approved by the FDA for the treatment of breast cancer [32]. Alpelisib has specific activity for PI3Kα over other isoforms, despite their nearly identical active sites, and potently inhibits common mutations such as E545K and H1047R.
4.3. PI3Kγ (PIK3CG)
PIK3CG encodes for the phosphatidylinositol 3-kinase catalytic subunit gamma (PI3Kγ), also known as p110γ. PI3Kγ belongs to class-IB PI3Ks and has both lipid and Ser/Thr protein kinase activity. It regulates immune stimulation and suppression in inflammation and cancer [33]. The PI3Kγ catalytic subunit has five domains, consisting of a putative uncharacterized adaptor binding domain (ABD), a Ras-binding domain (RBD), a C2 domain for binding cell membranes, an α-helical domain, and a catalytic kinase domain [34,35]. PI3Kγ activation is primarily regulated through interactions with GPCRs, which occur through association of the PI3Kγ regulatory subunit with the G-protein βγ subunits [36,37]. It can also be activated through interaction between the PI3Kγ RBD domain and Ras GTPases [38]. In the absence of lipid membrane binding, PI3Kγ maintains an inactive conformation [39].
In contrast to PI3Kα, PI3Kγ is commonly upregulated or overexpressed in cancer rather than mutated; however, mutations have been identified in cancer patients [40,41]. Loss-of-function mutations in PI3Kγ cause severe immunodeficiency, highlighting PI3Kγ’s critical role in promoting appropriate adaptive immune responses [42]. PI3Kγ is also a driver of inflammatory and metabolic disorders including rheumatoid arthritis, atherosclerosis, lupus, obesity, and pulmonary fibrosis [43].
4.4. PIKfyve (PIKFYVE)
PIKfyve is a bifunctional lipid kinase with Ser/Thr protein kinase activity. PIKfyve and its enzymatic products regulate cellular processes including membrane trafficking, ion channel activity, cytoskeletal dynamics, nuclear transport, stress- and hormone-induced signaling, transcription, and cell cycle progression [44,45].
As a lipid kinase, it synthesizes phosphoinositides (PIs) PtdIns(3,5)P2 from PtdIns3P and PtdIns5P from PtdIns. Synthesis of PtdIns(3,5)P2 by PIKfyve is negatively regulated by the formation of a multi-protein complex with scaffolding regulator ArPIKfyve and phosphatase Sac3 [46]. The cryo-EM structure revealed that formation of this complex sterically hinders PIKfyve from accessing membrane-associated PIs [45]. Serine residue autophosphorylation of PIKfyve represses its own lipid kinase activity and simultaneously activates Sac3 lipid phosphatase activity to downregulate lipid product synthesis [45]. Sac3 is also a serine phosphatase that acts on PIKfyve to increase the lipid kinase activity [45]. PIKfyve kinase activity is required for Sac3 lipid phosphatase activity [45], meaning that its dual function is critical for maintaining the delicate balance of lipid homeostasis. However, given the presence of multiple phospho-sites on PIKfyve and its role as a target in multiple signaling pathways, how the delicate balance between the dual activities of PIKfyve and Sac3 is maintained remains unknown.
PIKfyve has four structured domains including a FYVE finger domain, which targets the protein to PtdIns3P-enriched endosome membranes, and a DEP domain with unknown function. The middle of the protein interacts with several binding partners and contains two domains, a Cpn60-TCP1 domain, which has sequence similarity to chaperonins, and a CHK homology region containing the conserved Cys, His, and Lys residues found in all PIKfyve orthologs. The PIKfyve catalytic domain controls all three of its catalytic functions (PtdIns(3,5)P2/PtdIns5P synthesis and protein phosphorylation) and contains a single ATP-binding site. Mutation of the catalytic Lys-1831 abrogates all three kinase activities [47]. Mutations in PIKfyve cause functional defects in endosomal sorting, leading to Francois-Neetens fleck corneal dystrophy [48].
5. Regulation of Transcription and Translation
5.1. Ire1α (ERN1)
Ire1α is a ubiquitously expressed transmembrane ER stress sensor that functions as a Ser/Thr protein kinase and endoribonuclease. It is the best-studied branch of the unfolded protein response (UPR), an integrated intracellular signal transduction pathway that is activated in response to the accumulation of unfolded proteins in the ER [49]. The UPR initially responds to ER stress by blocking translation and upregulating molecular chaperones and folding enzymes. Prolonged cell stress causes a switch to pro-apoptotic and pro-inflammatory signaling cascades. The UPR has become an attractive pathway for drug discovery efforts due to its involvement in cancer, inflammation, neurological disorders, diabetes, and ischemia-reperfusion injury [50].
Ire1α contains an N-terminal ER luminal domain bound to cytoplasmic Ser/Thr kinase and endoribonuclease domains through a transmembrane linker [51]. The accumulation of misfolded protein causes Ire1α to dimerize in a “back-to-back” orientation [52], resulting in the formation of high-order oligomers and release of the ER Hsp70 chaperone BiP from the inactive Ire1α monomer. Thus, Ire1α activation is facilitated by luminal domain dimerization and trans-autophosphorylation of the cytosolic kinase domains [53,54]. Phosphorylation of the Ire1α activation segment stabilizes the RNase domain, which excises a 26-nucleotide intron from XBP1 [52,55]. The two spliced exons are then ligated by RNA ligase RtcB to form XBP1s [56], which encode an essential transcription factor for downstream activation of UPR response genes [57,58].
5.2. Ire1β (ERN2)
Ire1β is a close paralog of Ire1α. It has dual Ser/Thr protein kinase and endoribonuclease activities and induces the unfolded protein response (UPR) through transcription factor XBP1 [55,58], albeit less effectively than Ire1α [59]. Like Ire1α, it contains separate kinase and endoribonuclease domains. Ire1β serves as a direct and dominant-negative suppressor of Ire1α, dampening the UPR and ER stress response in epithelial cells of the intestine and other mucosal surfaces [53,60].
5.3. Protein Kinase R (EIF2AK2)
The eukaryotic translation initiation factor 2α kinase 2 (EIF2AK2), also known as protein kinase R (PKR) and interferon-induced double-stranded RNA-activated protein kinase, is a dual-specificity Ser/Thr and Tyr protein kinase. It contains two dsRNA binding motifs (dsRBD) and a kinase domain. PKR has been extensively studied as a regulator of the integrated stress response to RNA and DNA viruses, working as both a translational repressor through phosphorylation of Ser-51 on EIF2α and as a transcriptional upregulator of innate immune pathway proteins including Iκβ and NF-κβ [61,62,63]. On DNA damage, PKR halts the G2-M cell cycle transition by phosphorylating Tyr-4 of CDK1, causing its ubiquitination and proteasomal degradation [64]. PKR is a critical signaling mediator of cell proliferation and apoptosis through its interactions with p38 MAP kinase, NF-κβ, and the insulin signaling pathways [63,65,66,67]. It is ubiquitously expressed in vertebrates [68] and is implicated in cancer, neurodegeneration, inflammation, aging, and metabolic disorders [69,70].
The most widely used PKR inhibitor is C16, an imidazo-oxindole inhibitor with an in vitro IC50 of 210 nM; however, less potent inhibitors including 2-aminopurine have also been developed [70]. As an ATP-competitive inhibitor, C16 inhibits both the Ser/Thr and Tyr kinase functions of PKR and has been investigated for use in memory enhancement [71], neurodegeneration [72], inflammation [73], and metabolic disorders [74].
5.4. RIOK1 (RIOK1)
RIOK1 is a Ser/Thr protein kinase and ATPase in the Rio (right open reading frame) family of atypical protein kinases. In humans, there are three Rio subfamilies: Rio1 and Rio2 are conserved across all kingdoms of life, while Rio3 is only found in multicellular eukaryotes [75]. Specifically, RIOK1 functions in processes essential for cell proliferation, cell cycle progression, and chromosome maintenance. Yeast and human RIOK1 kinases are essential for the biogenesis of small ribosomal subunits, and depletion of this enzyme causes cell cycle arrest in yeast. Although RIOK1 is classified as a Ser/Thr kinase [76], it predominantly acts as an ATPase and regulates pre-40S ribosomal subunit association [77].
The RIO kinase domain adopts the conserved bilobal kinase domain structure but lacks an activation loop and substrate recognition sites. It instead contains a C-terminal RIO-kinase specific αR helix and a flexible loop between β3 and helix αC, which together allow it to accommodate a phospho-Asp substrate and serve as an ATPase [78]. At this time, there are no specific RIOK1 inhibitors. However, RNAi-mediated knockdown of RIOK1 reduced cell proliferation in RAS-driven cancer cell lines [79], making it an alluring target for drug discovery efforts.
5.5. TATA-Box Binding Protein Associated Factor 1/Transcription Initiation Factor TFIID Subunit 1 (TAF1)
TATA-box binding protein associated factor 1 (TAF1) is a Ser/Thr protein kinase and histone acetyltransferase with ubiquitin-activation and -conjugation activities [80]. TAF1 is the largest subunit of TFIID and binds core promoter sequences and transcriptional regulators. TFIID, which initiates RNA-polymerase II-dependent transcription, is the core scaffold for the TFIID basal transcription factor complex [81,82]. TFIID is comprised of the evolutionarily conserved TATA binding protein (TBP) and multiple TBP-associated factors (TAFs).
TAF1 is a bipartite kinase composed of N- and C-terminal kinase domains (NTK and CTK) and a centrally located histone acetyltransferase domain (HAT) [83]. Between the HAT and CTK domains are two bromodomains that selectively bind to multiply acetylated histone H4 peptides [84]. TAF1 acetylates histones H3 and H4 [85], and TAF1-mediated acetylation of H3 is required for Sp1 activation of cyclin D1 transcription and G1 to S-phase cell cycle progression [86]. Both TAF1 kinase domains are necessary for Ser phosphorylation of the TFIIF subunit transcription factor RAP74 and initiation of the RNA polymerase II transcription complex assembly [83,87]. TAF1 also phosphorylates tumor suppressor p53 on Thr-55, resulting in Mdm2-mediated p53 degradation and progression through G1 of the cell cycle [88]. Interaction between retinoblastoma protein Rb and TAF1 inhibits its kinase activity, which suggests that TAF1 may play a role in tumor suppression [89]. Mutations to the TAF1 gene cause X-linked dystonia parkinsonism and X-linked syndromic intellectual development disorder-33 [90,91]; however, the specific mechanisms by which TAF1 mutations drive these diseases are unknown.
5.6. MHC Class II Transactivator (CIITA)
MHC class-II transactivator (CIITA) is a Ser/Thr protein kinase and histone acetyltransferase that functions as a transcriptional coactivator and as the master regulator of major histone compatibility complex (MHC) class-II gene expression [92]. CIITA is a functional homolog of TAF1 and uses multiple mechanisms to activate transcription initiation and elongation [93,94]. CIITA phosphorylates the C-terminal of TAF7, a component of the TFIID promoter complex, and Ser-36 of histone H2B [95]. Its acetyltransferase domain is required for de novo transcription of MHC class-II genes and enhanced transcription of MHC class-I genes [96]. Autophosphorylation of CIITA markedly increases its acetyltransferase activity, which suggests that the acetyltransferase domain is regulated by CIITA’s intrinsic kinase activity [95].
CIITA is the only documented transcription factor in the nucleotide-binding oligomerization (NOD) family and contains a conserved tripartite structure consisting of a variable N-terminal effector-binding domain (EBD), a central NOD domain, and a C-terminal region with a variable number of leucine-rich repeats [93]. Its N-terminal acidic domain for transcriptional transactivation contains its histone acetyltransferase domain (HAT), followed by a proline-, serine-, and threonine-rich domain (P/S/T domain) with multiple phosphorylation sites [97]. The central nucleotide-binding domain (NACHT domain) acts as a kinase domain, and in conjunction with the C-terminal leucine-rich repeats, it affects self-association and nuclear import [98,99]. Mutations in the CIITA gene cause severe immunodeficiency syndrome, also known as bare lymphocyte syndrome [100].
5.7. p53-Related Protein Kinase (TP53RK)
The p53-related protein kinase (PRPK) and its homologs Bud32/Kae1 are Ser/Thr protein kinases and ATPases present in eukaryotes including yeast, humans, and pathogenic fungi including Candida albicans, Coccidioides immitis, and Cryptococcus neoformans. PRPK is one component of the highly conserved EKC/KEOPS complex (endopeptidase-like and kinase associated to transcribed chromatin/kinase, endopeptidase, and other proteins of small size). The EKC/KEOPS complex is required for the universal threonyl carbamoyl adenosine (t6a) tRNA modification, which is found in all tRNAs that pair with ANN codons where N is any of the four bases [101]. The EKC/KEOPS complex and t6a modification are critical for life, with remarkable sequence conservation from archaea to mammals [102,103].
PRPK maintains the conserved human kinase domain architecture including the regulatory and catalytic spines, a DFG motif, and a salt bridge between the invariant lysine in β3 and the invariant glutamate in helix αC [104]. There is high functional conservation between PRPK and its yeast homolog Bud32 [105]. Bud32′s regulatory partner Kae1 (OSGEP in humans) switches its intrinsic kinase activity to ATPase activity, which is required for EKC/KEOPS complex function [106]. Kae1 binds to the C-terminal lobe of Bud32 while Cgi121 (TPRKB in humans) binds to the N-terminal lobe, with the catalytic residues Lys-52, Asp-161, and Asp-182 interacting between the two lobes [107]. Based on the similarity between Bud32 and the Rio2 atypical kinase, it is hypothesized that its ATPase activity is required for the dissociation of tRNA from the complex.
Mutations in PRPK and the EKC/KEOPS complex cause Galloway-Mowat syndrome, an autosomal recessive disease characterized by a combination of early-onset nephrotic syndrome and microcephaly with brain anomalies [108]. PRPK is phosphorylated at Ser-250 by AKT [109] and is a promising therapeutic target in colon adenocarcinomas, cutaneous squamous carcinomas [108], and multiple myeloma [110].
6. Regulation of Cell Growth and Differentiation
6.1. Protein Kinase C δ (PRKCD)
Protein kinase C delta type (PKCδ) is a dual-specificity Ser/Thr and Tyr protein kinase. It is a member of the protein kinase C (PKC) superfamily and regulates neutrophil and endothelial cell proinflammatory signaling through Ser/Thr/Tyr phosphorylation of a myriad of substrates [111,112]. Thus, it serves as a critical regulator of the inflammatory response in cancer, diabetes, heart disease, sepsis, and autoimmune disease [112]. PKCδ contains an N-terminal Ca2+-sensing domain, a kinase domain, and an AGC-kinase family C-terminal regulatory tail that stabilizes the kinase domain and serves as a docking surface for regulatory proteins [113].
Unlike other PKC isoforms, PKCδ is uniquely regulated by phosphorylation of tyrosine residues within the regulatory domain (Tyr-52, -64, -155, -187), hinge region (Tyr-311, -332), and catalytic domain (Tyr-505, -512, -523). Tyr phosphorylation within the catalytic domain increases PKCδ activity, whereas Tyr phosphorylation within the regulatory domain primarily influences downstream signaling [112]. Phosphorylation of regulatory domain Tyr-155 and hinge region Tyr-311 are both important for PKCδ-mediated proinflammatory signaling and the initiation of cytotoxic and apoptotic pathways [112,114]. Tyr-155 phosphorylation regulates gene expression and apoptosis, whereas Tyr-311 phosphorylation causes a conformational change that exposes the PKCδ caspase cleavage site [115]. Mutations of Tyr-155 increase cell proliferation in response to its substrate phorbol 12-myristate 13-acetate (PMA). In addition to regulating kinase activity, tyrosine phosphorylation also amplifies nuclear localization of PKCδ during the genotoxic stress response and induces apoptosis [115].
6.2. Leucine-Rich Repeat Ser/Thr Protein Kinase 2 (LRRK2)
Leucine-rich repeat serine/threonine-protein kinase 2 (LRRK2) is a Ser/Thr protein kinase and GTPase involved in neuronal plasticity [116], autophagy [117], and ciliogenesis [118]. LRRK2 is a multi-domain protein that belongs to the Roco family of G-proteins. This protein family contains a central Ras of complex proteins (Roc) G-domain with an adjacent C-terminal of Roc (COR) dimerization domain. LRRK2 also contains N-terminal armadillo, ankyrin, and leucine-rich repeat (LRR) scaffold domains, a kinase domain, and a C-terminal WD40 scaffold [119]. The Roc domain is a functional GTPase [120] responsible for activation and regulation of the kinase domain on binding to guanine nucleotides [121]. In the cytosol, GTP-bound LRRK2 is primarily monomeric and exhibits low kinase activity, whereas at the membrane it is dimeric and active [122]. The cytosolic localization of LRKK2 is stabilized by phosphorylation-dependent interactions between its N-terminal segment and the ubiquitous regulatory protein 14-3-3 [123]. LRRK2 primarily acts as a Ser/Thr kinase through phosphorylation of Rab GTPases, which are master regulators of intracellular vesicle trafficking and membrane transport [118]. GTP-bound Rab proteins bind to the N-terminus of LRRK2 and recruit it to the membrane, where it undergoes a multi-step hydrolysis cycle and results in dimeric GDP-bound LRRK2 [122]. LRRK2 dimerization at the membrane is mediated by its Roc/COR domains and is critical for GTP hydrolysis and achieving maximal kinase activity. On LRRK2 activation, it phosphorylates GTP-bound substrates. The interplay between GTPase activity and kinase activity reveals that these activities are clearly linked.
Mutations in LRRK2 are the most common genetic cause of Parkinson’s disease (PD) [124,125]. LRRK2 phosphorylates amyloid precursor protein at Thr-668 and cause loss of dopaminergic neurons [126,127] in PD. It also impairs proteasomal degradation of Tau and results in the accumulation of high-molecular-weight Tau neurofibrillary tangles, linking LRRK2 to dementia in PD and Alzheimer’s disease [128]. Interestingly, pathogenic mutations are found in both the kinase domain (G2019S, I2020T) and the Roc/COR GTPase domains (R1441C/G/H, Y1699C), suggesting that both enzymatic activities are crucial for PD pathogenesis [129,130,131]. The roles of LRKK2 in disease pathogenesis have stimulated the development of potent and selective inhibitors, such as third-generation ATP-competitive inhibitors MLi-2 and PF-06685360 [132,133]. Although inhibitors of LRRK2 have been identified, the lack of safety and side effects of these ATP-competitive compounds remain a challenge. Since the GTPase and kinase functions of LRRK2 depend on one another, its GTPase activity may be critical for pathogenicity. Thus, recent progress has been made toward developing allosteric LRRK2 inhibitors that mainly act by interfering with its GTPase activity rather than inhibiting kinase activity directly.
6.3. Maternal Embryonic Leucine Zipper Kinase (MELK)
The maternal embryonic leucine zipper kinase (MELK) is a dual-specificity Ser/Thr and Tyr protein kinase [134] from the adenosine monophosphate-activated protein kinase (AMPK)-related family. This protein family is highly upregulated in breast [135], brain [136], colorectal, gastric, and ovarian cancers [137]. MELK is predominantly a Ser/Thr kinase with broad substrate specificity and only phosphorylates tyrosine autocatalytically [134]. It is composed of a kinase domain followed by a ubiquitin-associated region, an autoinhibitory region, and a kinase-associated-1 domain. MELK mediates cell cycle progression through phosphorylation of CDC25B, promoting its localization to the centrosome and spindle poles during mitosis [138]. High MELK expression is associated with aggressive undifferentiated tumors; however, there is conflicting evidence regarding its role as an oncogenic driver [139].
6.4. ACK1 (TNK2)
Activated Cdc42 kinase 1 (ACK1) is a dual-specificity non-receptor Tyr and Ser/Thr protein kinase that functions as a specific effector kinase for the GTP-bound form of Cdc42. ACK1 is involved in cell survival, migration, spreading, growth, proliferation, and epigenetic regulation. It has eight distinct domains: a sterile alpha motif (SAM), a catalytic kinase domain, an SH3 domain, a Cdc42/GTPase binding domain, a clathrin interacting region, a PPXY motif, a Mig6 homology region (MHR) or EGFR binding domain, and a ubiquitin-association domain. The ACK1 multi-domain architecture facilitates its localization to different cellular compartments and association with binding partners [140].
ACK1 acts as a dual-specificity kinase toward Wiskott-Aldrich syndrome protein (WASP), an effector protein of Cdc42 that plays an integral role in actin filament formation. ACK1 phosphorylates WASP on Ser-242 and Tyr-256; Ser-242 phosphorylation was found to enhance WASP-mediated actin polymerization in cell lysates [141]. In addition, ACK1 acts as a protein tyrosine kinase toward AKT, a critical mediator of cell survival through inhibition of pro-apoptotic proteins. ACK1 directly phosphorylates AKT on the evolutionarily conserved kinase domain residue Tyr-176 [142]. Phosphorylation of AKT Tyr-176 localizes it to the plasma membrane and promotes Ser/Thr phosphorylation that leads to its activation [142].
ACK1 promotes tumorigenesis and metastasis in prostate cancer and tamoxifen-resistant breast cancer [143,144,145,146], and it is also implicated in systemic lupus erythematosus [147]. To identify potent and selective inhibitors of Ack1 kinase activity, Lawrence et al. used a fragment-based approach and identified (R)-9b [148], which has proven effective in suppressing the growth of both prostate cancer and triple-negative breast cancer cell lines [149]. (R)-9b potently inhibits Ack1-mediated phosphorylation of Tyr-176 on AKT, though its effects on WASP Ser phosphorylation and other substrates have not yet been reported.
7. Regulation of Immune and Inflammatory Pathways
7.1. RIPK2 (RIPK2)
Receptor-interacting protein kinase 2 (RIPK2) is a dual-specificity Ser/Thr and Tyr protein kinase that plays an essential role in inflammation and innate immunity through the NOD/RIPK2/NF-κB signaling pathway [150,151]. RIPK2 acts downstream of the NOD-like receptors NOD1 and NOD2 (nucleotide-binding oligomerization domains 1 and 2), which together regulate cytokine production in response to intracellular bacterial infection [152,153]. NOD1 and NOD2 bind distinct components of bacterial peptidoglycan and ATP and form a signaling complex with RIPK2. Aberrant NOD/RIPK2 signaling is implicated in the pathogenesis of numerous inflammatory diseases. NOD1 polymorphisms have been linked to susceptibility to early-onset inflammatory bowel disease [154], sarcoidosis [155], and asthma [156]. Specific NOD2 polymorphisms within the leucine-rich repeat (LRR) domain have been linked to Crohn’s disease [157]. The expression levels of both NOD2 and activated/phosphorylated RIPK2 are upregulated in pediatric patients with active Crohn’s disease or ulcerative colitis [158,159]. RIPK2′s involvement in inflammatory diseases makes it a promising drug target.
RIPK2 contains two domains, an N-terminal catalytic kinase domain with homology to the other RIPK family members and a unique caspase activation/recruitment domain (CARD), which facilitates interactions with NOD1 and NOD2 [160,161]. RIPK2 binding to NOD1 or NOD2 induces its autophosphorylation on residues Tyr-474 and Ser-176 [162,163], leading to Lys-63-linked polyubiquitination at Lys-209 [164]. Subsequent activation of the NF-κB pathway and mitogen-activated protein kinases (MAPKs) thereby induce autophagy [165] and transcription of proinflammatory cytokines and chemokines [166]. Several inhibitors have been found to effectively inhibit RIPK2 kinase activity, with WEHI-345 [167], OD36, and OD38 [168] exhibiting improved specificity. WEHI-345 was found to delay activation of NF-κB on NOD stimulation and prevented cytokine productive both in vitro and in vivo. Interestingly, the crystal structure of FDA-approved ATP-competitive inhibitor ponatinib revealed an allosteric site that may provide an avenue for designing more specific RIPK2 inhibitors [169].
7.2. Ser/Thr Protein Kinase 16 (STK16)
Serine-threonine protein kinase 16 (STK16), also known as myristoylated/palmitoylated serine/threonine-protein kinase, is a dual-specificity Ser/Thr and Tyr protein kinase with a C-terminal protein kinase domain and N-terminal myristoylation and palmitoylation site at Cys-6 and -8 [170]. It is part of the Numb-associated kinases (NAK) family, with only 25% sequence identity to its closest structural homolog Aurora kinase A [171]. STK16 is constitutively active, which is partially explained by its unique activation segment, which contains a β sheet at the tip of the activation loop and a large α-helical insert [171]. Strikingly, it replaces the conserved DFG motif with a DLG motif, a sequence variation found in less than 6% of human kinases [171].
STK16 directly regulates actin polymerization and depolymerization, thereby controlling the Golgi complex integrity [172]. It enhances the pro-angiogenic capacity of tumors as a transcriptional co-activator of VEGF secretion [173]. It phosphorylates developmentally-regulated GTP-binding protein 1 (DRG1) and 4EBP1 and is involved in the TGF-β signaling pathway, linking it to the regulation of cell growth and homeostasis [171,172]. Discovery of the highly selective STK16 inhibitor STK16-IN-1, which has potent activity both in vitro and in live cells, has provided a useful tool for further investigation of this underexplored kinase and its disease involvement [174].
7.3. Lipopolysaccharide Kinase WaaP (rfaP)
WaaP is a bifunctional sugar kinase and Tyr protein kinase in Pseudomonas aeruginosa, E. coli, and Salmonella species that is responsible for phosphorylation of the inner core heptose (HepI) of lipopolysaccharide (LPS) [175]. LPS is a major virulence factor for P. aeruginosa, an opportunistic Gram-negative pathogen commonly associated with nosocomial infections, pneumonia in cystic fibrosis patients, soft-tissue infections in burn victims, osteomyelitis, sepsis, and ecthyma gangrenosum in immunocompromised patients. P. aeruginosa LPS is composed of lipid A endotoxin, a core oligosaccharide divided into an inner and outer core, and O-antigen. The inner oligosaccharide core contains heptose and octulosonic acid and is the most highly phosphorylated among Gram-negative bacteria [176]. LPS contributes to the low outer-membrane permeability of P. aeruginosa, which in combination with multiple efflux pumps and production of antibiotic-inactivating genes, confers a high intrinsic antibiotic resistance [177].
WaaP is essential to P. aeruginosa viability; knockout is lethal and can only be tolerated if another copy of WaaP is added in trans [175]. The Virulence Factors Database lists WaaP as an essential virulence factor for P. aeruginosa pathogenicity [178]. Mutation of WaaP in Salmonella enterica serovar Typhimurium causes loss of virulence in vivo and increased antibiotic susceptibility [179]. Inducible WaaP depletion in P. aeruginosa blocks core phosphorylation, causing truncation of the LPS core, marked invaginations of the LPS inner membrane, and increased susceptibility to polymyxin B and novobiocin [180].
Although WaaP does not demonstrate true in vivo protein kinase activity, it shares sequence homology with conserved functional residues of Ser/Thr protein kinases protein kinase Cα and SNF1 and Tyr protein kinases Src and EGFR [181]. It autophosphorylates on Tyr residues and phosphorylates exogenous poly(GluTyr) peptide. Its crystal structure revealed structural homology with eukaryotic kinases including an activation loop DFG motif, a catalytic HRD motif, a glycine-rich P-loop, and a catalytically invariant Lys at position 51 [182]. Mutation of structurally predicted essential kinase residues increased the P. aeruginosa susceptibility to EDTA or completed the abrogated cellular viability [182], indicating a potential avenue for therapeutic development. WaaP’s structural and functional similarity to eukaryotic protein kinases and its integral role in LPS synthesis and P. aeruginosa growth make it a ripe target for existing small molecule protein kinase inhibitors, which may orthogonally inhibit its sugar kinase activity. Disruption of the outer membrane’s stability and the resulting increase in permeability could potentially re-sensitize P. aeruginosa to treatment with first-line antibiotics.
8. Regulation of Metabolism
8.1. PEPCK (PCK1)
Phosphoenolpyruvate carboxykinase (PEPCK) is a carboxylyase and Ser/Thr protein kinase that has canonically been described as a gluconeogenic enzyme. PEPCK is the rate-limiting enzyme of gluconeogenesis, where it catalyzes reversible decarboxylation and phosphorylation of oxaloacetate [183]. Its role as a protein kinase in lipogenesis was only recently uncovered. PEPCK phosphorylation at Ser-90 by AKT results in its translocation from the cytosol to the ER, where it transfers a phosphate group from GTP to ER-anchor proteins INSIG1 and -2 on Ser-207 and Ser-151, respectively. INSIG phosphorylation disrupts the binding of cholesterol-derived oxysterols, resulting in activation of sterol regulatory element-binding proteins (SREBPs), transcription of lipogenesis-related genes, and tumorigenesis in mice. Notably, staining of patient-derived hepatocellular carcinoma (HCC) tumors showed a correlation between high levels of phosphorylation at PEPCK1 Ser-90, INSIG1 Ser-207, and INSIG2 Ser-151 with decreased overall durations of survival, linking PEPCK’s alternative catalytic function to the clinical aggressiveness of HCC [184]. Metabolic reprogramming of PEPCK has also been found in pancreatic [185] and esophageal carcinomas [186], warranting further investigation of its role as a protein kinase. GTP-competitive PEPCK inhibitors have previously been developed as anti-diabetic agents [187,188]. Given that GTP serves as the main phosphate donor for PEPCK, these pre-existing inhibitors may also abrogate its protein kinase activity.
8.2. Pyruvate Orthophosphate Dikinase Regulatory Protein (PDRP)/Phosphoenolpyruvate Synthetase Regulatory Protein (ppsR)
Pyruvate orthophosphate dikinase regulatory protein (PDRP) is a bifunctional Ser/Thr protein kinase and pyrophosphorylase involved in the regulation of gluconeogenesis protein pyruvate orthophosphate dikinase (PPDK). PPDK regenerates phosphoenolpyruvate from pyruvate and ATP in C4 plant photosynthesis [189,190]. PDRP phosphorylates Thr-527 on PPDK during inactivation and dephosphorylates it during activation in response to changes in lighting [191]. Although it is primarily found in plants, PDRP also shares sequence homology with the domain of unknown function (DUF)299 genes present in some bacteria, which led to the identification of PDRP homologs in bacteria as well as an analogous protein phosphoenolpyruvate synthetase regulatory protein (PSRP) [192]. PSRP is a bifunctional Ser/Thr kinase and pyrophosphorylase involved in the regulation of phosphoenolpyruvate synthetase (PEPS), a critical metabolic switch for bacterial survival on small carbon substrates that catalyzes the phosphorylation of pyruvate to PEP. Based on our UniProtKB search results, PDRP is present in various bacterial species including pathogens Bacillus anthracis, Bacillus cereus, Bartonella henselae, Clostridium botulinum, Rickettsia prowazekii, Listeria monocytogenes, and Staphylococcus aureus, whereas PSRP is found primarily in Escherichia coli, Salmonella species, and Shigella species. The bacterial kinases have been insufficiently researched, but the mechanism has been well-characterized in Arabidopsis thaliana and Zea mays.
PDRP phosphorylates and dephosphorylates PPDK active site Thr-527 in Zea mays using ADP as its phosphoryl donor [190,193]. The resulting phosphothreonine residue interferes with the activity of the catalytic His-529 residue. Zea mays PDRP forms a homodimer with an N-terminal domain and C-terminal domain that share the same unique protein fold and contain phosphate-binding P-loops [194]. Structural superimposition with M. pneumoniae HPrK/P and enzymatic activity assays of mutant PDRP revealed that the C-terminal domain contains a single active site used for both kinase and phosphorylase functions, while the N-terminal domain is proposed to have allosteric regulatory effects. Further characterization of these proteins is necessary given their high prevalence in our search results.
8.3. Isocitrate Dehydrogenase Kinase/Phosphatase (aceK)
Isocitrate dehydrogenase kinase/phosphatase (IDHK/P) is a bifunctional Ser/Thr protein kinase and phosphatase with specific ATPase activity. It phosphorylates and dephosphorylates isocitrate dehydrogenase, a key enzyme in the Krebs cycle responsible for the conversion of isocitrate to α-ketoglutarate, based on nutrient availability [195,196,197]. It was the first example of prokaryotic protein phosphorylation [198,199] and is only seen in Gram-negative bacteria including Escherichia coli, Klebsiella, Pseudomonas, Yersinia, Shigella, and Salmonella species. It serves as the metabolic switch between the Krebs cycle and the glyoxylate bypass, an anaplerotic pathway that allows net conversion of acetyl-CoA to carbohydrates via isocitrate lyase and malate synthase. This allows bacteria to survive on acetate and fatty acids without the loss of carbon as CO2. IDHK/P phosphorylates isocitrate dehydrogenase on Ser-113, completely inactivating it through occlusion of the isocitrate binding site [200].
IDHK/P contains two distinct domains, a catalytic C-terminal domain that contains the kinase, phosphatase, and ATPase functions, and an N-terminal regulatory domain, which contains small binding pockets for modulating the enzymatic activity [199]. The kinase domain resembles eukaryotic Ser/Thr and Tyr protein kinases including the IL-1 receptor and FGFR, while the regulatory domain contains a unique protein fold [201]. The ATP-binding site contains an invariant Lys-336, which is conserved among protein kinases [202]. Loop β3αC is homologous with the eukaryotic activation loop and controls accessibility to this ATP-binding site. Residues 484–510 form a substrate recognition loop (SRL) that is critical for the highly specific binding to isocitrate dehydrogenase homodimers [203]. A kinetic analysis of WT and two mutant IDHK/P proteins indicated that the kinase, phosphatase, and ATPase catalytic functions of the enzyme share the same active site [204,205].
8.4. HPr Kinase/Phosphorylase (hprK)
HPr kinase/phosphorylase (HPrK/P) is a bifunctional Ser/Thr protein kinase and phosphorylase that is a central sensor for carbon catabolite repression in bacteria. It was first identified as a Ser protein kinase sensitive to cellular concentrations of metabolic intermediates. HPrK/P phosphorylates and dephosphorylates Ser-46 of HPr, a phosphocarrier protein of the phosphoenolpyruvate:carbohydrate phosphotransferase system [206,207,208]. In the presence of glucose, catabolite control protein CcpA forms a ternary complex with Ser(P)-46-HPr and the catabolite responsive element DNA sequence to repress transcription of alternative sugar pathway operons [209,210]. HPrK/P is also linked to the transcriptional regulation of cell adhesion and virulence of Gram-negative pathogens [211]. In Listeria monocytogenes, formation of Ser(P)-46-HPr inhibits PrfA, a transcriptional activator of virulence genes including the hemolysin-encoded hly [212].
The crystal structures of the HPrK/P catalytic domain from Lactobacillus casei and the full-length kinase/phosphorylase from Mycoplasma pneumoniae and Staphlycoccus xylosus revealed a structure unrelated to eukaryotic protein kinases [213,214,215]. HPrK/P is a homohexamer with a three-fold axis orthogonal to three two-fold axes, with each half of a hexamer subunit containing a central loop [216]. Each subunit contains a Walker A ATP-binding motif forming a typical phosphate-binding loop (P-loop). The C-terminal domain, which contains the P-loop, carries both kinase and phosphorylase activities, and the N-terminal domain is currently poorly understood but hypothesized to have an allosteric regulatory role [216]. Co-crystallization of the L. casei HPrK/P catalytic domain and B. subtilis HPr demonstrated substrate binding at sites of the hexamer where two subunits overlap. Superposition of the L. casei V267F HPrK/P mutant, which adopts the kinase conformation, with WT HPrK/P in the phosphorylase conformation, demonstrated that the same active site catalyzes both enzymatic functions [217]. The HPrK/P catalytic function is allosterically mediated by ATP concentration; the presence of ATP favors the kinase conformation whereas the absence of ATP favors the phosphorylase conformation. Ramstrom and coworkers identified a bacteriostatic inhibitor of B. subtilis HPrK/P, Do9, with micromolar activity against both its kinase and phosphorylase activities [216,218]. Intriguingly, competition experiments with Do9 indicate that it is not ATP-competitive, suggesting an allosteric binding mode that may yield more selective inhibition of microbial growth.
8.5. Multidomain Regulatory Protein Rv1364c (Rv1364c)
Rv1364c is a bifunctional Ser/Thr protein kinase and phosphatase only found in pathogenic Mycobacterium tuberculosis. Bacteria respond to environmental stressors including changes in temperature, oxygen, and osmotic stress by modifying their patterns of gene expression with alternative σ factors, which provide promoter recognition sites for RNA polymerase. σ factors are fine-tuned by anti-σ factors and anti-σ factor antagonists. In Mycobacterium tuberculosis, the general stress response σ factor is σF, which is regulated in part by Rv1364c, a unique multi-domain polypeptide only found in pathogenic mycobacteria [219]. In contrast to the homologous multiprotein system present in Bacillus subtilis, Rv1364c encodes the entire toolbox for σF regulation on a single polypeptide. It contains an N-terminal Per-Arnt-Sim (PAS) domain, which senses the energy levels in the cells, followed by a PP2C-like phosphatase domain, a kinase domain, and a sulfate transporter and AntiSigma factor antagonist (STAS) domain, which binds to σF on phosphorylation [220]. The STAS signaling domain is phosphorylated on Ser-600 by the kinase domain and dephosphorylated by the phosphatase domain [221,222]. Phosphorylation promotes dimerization and suggests a system where phosphorylation-dependent partner-switching of domains controls the bacterial transcriptional response [221]. Misra et al. proposed an orthogonal model in which the eukaryotic-like Ser/Thr kinase protein kinase D serves as an osmotic sensor to phosphorylate Rv1364c and release σF, linking extracellular sensing to the σF regulon [220]. Further investigation of this virulence factor is necessary given the high worldwide mortality of M. tuberculosis and the emergence of drug-resistant strains.
9. Discussion
In this review, we sought to identify serine/threonine and tyrosine kinases that harbor additional enzymatic activities. We probed the UniProtKB database for kinases with multiple EC annotations to uncover the frequency at which these dual activities are encoded by the same polypeptide chain. From our findings, we highlight seventeen human kinases and five bacterial kinases that have multiple catalytic activities and that are involved in the regulation of phosphoinositide signaling, transcription/translation, cell growth and differentiation, immunity, and metabolism. The search returned some kinases that are well-established as critical signaling mediators and drivers of human disease, making them promising drug targets. On the other hand, the remaining kinases require further functional characterization. Nine of these bifunctional kinases have separate catalytic functions within the same polypeptide chain, with their functions tightly controlled through upstream signaling effectors and allosteric regulation. The other 13 kinases are dual-specificity protein kinases and nucleoside triphosphatases with promiscuous substrate specificity that (i) may have no biological relevance, or (ii) could be critical for their signaling roles.
There are broad implications for kinases with multiple catalytic activities. For example, how do the evolutionary benefits differ between one protein with a single multifunctional domain (e.g., IDHK/P), one protein with two domains with two distinct functions (e.g., LRRK2), and two proteins with two distinct functions? Both HPrK/P and IDHK/P couple their contrasting kinase and phosphatase activities to the same multifunctional domain, and furthermore, to the same active site. Given that these proteins serve as key bacterial metabolic switches, their bifunctional active sites suggest an evolutionary benefit to harboring exquisite allosteric control. In addition, Rv1364c combines a multi-polypeptide signaling system from B. subtilis into a single polypeptide in M. tuberculosis. Consolidation of its kinase and phosphatase activities to a single protein may impart a pathogenic advantage by controlling localization to ensure the rapid temporal control of these contrasting activities.
The results in this study only represent 879 of the total 36,160 protein kinases from our search. As we continue to study existing proteins in more detail with more sensitive biochemical, structural, and computational methods, we can expect to identify additional enzymatic activities from well-characterized enzymes. For example, PEPCK, long-known as the rate-limiting enzyme of gluconeogenesis, has recently been identified as a protein kinase in an orthogonal and clinically relevant signaling pathway. Given the other metabolic enzymes identified in our search, this suggests that there are other metabolic kinases with non-canonical protein kinase activity [223].
In our study, we identified bifunctional kinases with dual phosphatase, phosphorylase, endoribonuclease, acetyltransferase, carboxylyase, and lipid/sugar kinase enzymatic activities. Given our findings, it is plausible that proteins within these functional groups could exhibit secondary kinase activities that have yet to be elucidated. Furthermore, it would be illuminating to extend this study to the human proteome and classify all bifunctional enzymes, which could potentially reveal evolutionary benefits of pairing certain enzymatic functions. We theorize that bifunctionality within the kinome and the proteome at large represents an exciting field of study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biom12050685/s1. Table S1: Full list of UniProtKB results from search algorithm. Table S2: Manually curated and reviewed UniProtKB results. Table S3: Grouped list of final 22 unique bifunctional kinases across species/strains from the UniProtKB search results.
Author Contributions
A.M.R. and V.R.M. developed the search algorithm, interpreted the results, created the figure, and wrote the manuscript with supervision from G.G. and M.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by NIH R35 GM119437 (M.A.S.), NIH F30 CA260771 (A.M.R.), NIH T32 GM008444 (A.M.R.), and NIH T32 GM127253 (V.R.M.). This content is solely our responsibility and does not necessarily reflect the official views of the NIH.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
V.R.M. is grateful for support from the Stony Brook University Renaissance School of Medicine Scholars in the Biomedical Sciences program. Figure 1 was created with BioRender.com (accessed on 4 April 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Endicott, J.A.; Noble, M.E.; Johnson, L.N. The structural basis for control of eukaryotic protein kinases. Annu. Rev. Biochem. 2012, 81, 587–613. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Fabbro, D.; Cowan-Jacob, S.W.; Moebitz, H. Ten things you should know about protein kinases: IUPHAR Review 14. Br. J. Pharmacol. 2015, 172, 2675–2700. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2022 update. Pharmacol. Res. 2022, 175, 106037. [Google Scholar] [CrossRef] [PubMed]
- Attwood, M.M.; Fabbro, D.; Sokolov, A.V.; Knapp, S.; Schioth, H.B. Trends in kinase drug discovery: Targets, indications and inhibitor design. Nat. Rev. Drug Discov. 2021, 20, 839–861. [Google Scholar] [CrossRef] [PubMed]
- Hatzivassiliou, G.; Song, K.; Yen, I.; Brandhuber, B.J.; Anderson, D.J.; Alvarado, R.; Ludlam, M.J.; Stokoe, D.; Gloor, S.L.; Vigers, G.; et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 2010, 464, 431–435. [Google Scholar] [CrossRef]
- Heidorn, S.J.; Milagre, C.; Whittaker, S.; Nourry, A.; Niculescu-Duvas, I.; Dhomen, N.; Hussain, J.; Reis-Filho, J.S.; Springer, C.J.; Pritchard, C.; et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 2010, 140, 209–221. [Google Scholar] [CrossRef]
- Poulikakos, P.I.; Zhang, C.; Bollag, G.; Shokat, K.M.; Rosen, N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 2010, 464, 427–430. [Google Scholar] [CrossRef]
- Kung, J.E.; Jura, N. Structural Basis for the Non-catalytic Functions of Protein Kinases. Structure 2016, 24, 7–24. [Google Scholar] [CrossRef]
- Rauch, J.; Volinsky, N.; Romano, D.; Kolch, W. The secret life of kinases: Functions beyond catalysis. Cell Commun. Signal. 2011, 9, 23. [Google Scholar] [CrossRef]
- Feldman, H.C.; Tong, M.; Wang, L.; Meza-Acevedo, R.; Gobillot, T.A.; Lebedev, I.; Gliedt, M.J.; Hari, S.B.; Mitra, A.K.; Backes, B.J.; et al. Structural and Functional Analysis of the Allosteric Inhibition of IRE1alpha with ATP-Competitive Ligands. ACS Chem. Biol. 2016, 11, 2195–2205. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Cantley, L.C. The phosphoinositide 3-kinase pathway. Science 2002, 296, 1655–1657. [Google Scholar] [CrossRef] [PubMed]
- Vivanco, I.; Sawyers, C.L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer 2002, 2, 489–501. [Google Scholar] [CrossRef]
- Locke, M.N.; Thorner, J. Regulation of TORC2 function and localization by Rab5 GTPases in Saccharomyces cerevisiae. Cell Cycle 2019, 18, 1084–1094. [Google Scholar] [CrossRef]
- Martinez Marshall, M.N.; Emmerstorfer-Augustin, A.; Leskoske, K.L.; Zhang, L.H.; Li, B.; Thorner, J. Analysis of the roles of phosphatidylinositol-4,5-bisphosphate and individual subunits in assembly, localization, and function of Saccharomyces cerevisiae target of rapamycin complex 2. Mol. Biol. Cell 2019, 30, 1555–1574. [Google Scholar] [CrossRef]
- Cheng, H.; Orr, S.T.M.; Bailey, S.; Brooun, A.; Chen, P.; Deal, J.G.; Deng, Y.L.; Edwards, M.P.; Gallego, G.M.; Grodsky, N.; et al. Structure-Based Drug Design and Synthesis of PI3Kalpha-Selective Inhibitor (PF-06843195). J. Med. Chem. 2021, 64, 644–661. [Google Scholar] [CrossRef]
- Maehama, T.; Dixon, J.E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998, 273, 13375–13378. [Google Scholar] [CrossRef]
- Hanker, A.B.; Kaklamani, V.; Arteaga, C.L. Challenges for the Clinical Development of PI3K Inhibitors: Strategies to Improve Their Impact in Solid Tumors. Cancer Discov. 2019, 9, 482–491. [Google Scholar] [CrossRef]
- Molinaro, A.; Becattini, B.; Mazzoli, A.; Bleve, A.; Radici, L.; Maxvall, I.; Sopasakis, V.R.; Molinaro, A.; Backhed, F.; Solinas, G. Insulin-Driven PI3K-AKT Signaling in the Hepatocyte Is Mediated by Redundant PI3Kalpha and PI3Kbeta Activities and Is Promoted by RAS. Cell Metab. 2019, 29, 1400–1409.E5. [Google Scholar] [CrossRef]
- Fruman, D.A.; Rommel, C. PI3K and cancer: Lessons, challenges and opportunities. Nat. Rev. Drug Discov. 2014, 13, 140–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jang, H.; Gaponenko, V.; Nussinov, R. Phosphorylated Calmodulin Promotes PI3K Activation by Binding to the SH2 Domains. Biophys. J. 2017, 113, 1956–1967. [Google Scholar] [CrossRef] [PubMed]
- Nolte, R.T.; Eck, M.J.; Schlessinger, J.; Shoelson, S.E.; Harrison, S.C. Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat. Struct. Biol. 1996, 3, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jang, H.; Nussinov, R. The mechanism of PI3Kalpha activation at the atomic level. Chem. Sci. 2019, 10, 3671–3680. [Google Scholar] [CrossRef]
- Foukas, L.C.; Beeton, C.A.; Jensen, J.; Phillips, W.A.; Shepherd, P.R. Regulation of phosphoinositide 3-kinase by its intrinsic serine kinase activity in vivo. Mol. Cell. Biol. 2004, 24, 966–975. [Google Scholar] [CrossRef][Green Version]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef]
- Samuels, Y.; Wang, Z.; Bardelli, A.; Silliman, N.; Ptak, J.; Szabo, S.; Yan, H.; Gazdar, A.; Powell, S.M.; Riggins, G.J.; et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004, 304, 554. [Google Scholar] [CrossRef]
- Zhao, L.; Vogt, P.K. Hot-spot mutations in p110alpha of phosphatidylinositol 3-kinase (pI3K): Differential interactions with the regulatory subunit p85 and with RAS. Cell Cycle 2010, 9, 596–600. [Google Scholar] [CrossRef]
- Zhao, L.; Vogt, P.K. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. Proc. Natl. Acad. Sci. USA 2008, 105, 2652–2657. [Google Scholar] [CrossRef]
- Croessmann, S.; Sheehan, J.H.; Lee, K.M.; Sliwoski, G.; He, J.; Nagy, R.; Riddle, D.; Mayer, I.A.; Balko, J.M.; Lanman, R.; et al. PIK3CA C2 Domain Deletions Hyperactivate Phosphoinositide 3-kinase (PI3K), Generate Oncogene Dependence, and Are Exquisitely Sensitive to PI3Kalpha Inhibitors. Clin. Cancer Res. 2018, 24, 1426–1435. [Google Scholar] [CrossRef]
- Jiang, W.; He, T.; Liu, S.; Zheng, Y.; Xiang, L.; Pei, X.; Wang, Z.; Yang, H. The PIK3CA E542K and E545K mutations promote glycolysis and proliferation via induction of the beta-catenin/SIRT3 signaling pathway in cervical cancer. J. Hematol. Oncol. 2018, 11, 139. [Google Scholar] [CrossRef] [PubMed]
- Markham, A. Alpelisib: First Global Approval. Drugs 2019, 79, 1249–1253. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, M.M.; Messer, K.S.; Ralainirina, N.; Li, H.; Leem, C.J.; Gorjestani, S.; Woo, G.; Nguyen, A.V.; Figueiredo, C.C.; Foubert, P.; et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature 2016, 539, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, K.; Yu, L.; Chen, Y.; Cai, Y.; Jin, J.; Hou, T. Targeting phosphatidylinositol 3-kinase gamma (PI3Kgamma): Discovery and development of its selective inhibitors. Med. Res. Rev. 2021, 41, 1599–1621. [Google Scholar] [CrossRef] [PubMed]
- Walker, E.H.; Perisic, O.; Ried, C.; Stephens, L.; Williams, R.L. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature 1999, 402, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Deladeriere, A.; Gambardella, L.; Pan, D.; Anderson, K.E.; Hawkins, P.T.; Stephens, L.R. The regulatory subunits of PI3Kgamma control distinct neutrophil responses. Sci. Signal. 2015, 8, ra8. [Google Scholar] [CrossRef]
- Rynkiewicz, N.K.; Anderson, K.E.; Suire, S.; Collins, D.M.; Karanasios, E.; Vadas, O.; Williams, R.; Oxley, D.; Clark, J.; Stephens, L.R.; et al. Gbetagamma is a direct regulator of endogenous p101/p110gamma and p84/p110gamma PI3Kgamma complexes in mouse neutrophils. Sci. Signal. 2020, 13, eaaz4003. [Google Scholar] [CrossRef]
- Pacold, M.E.; Suire, S.; Perisic, O.; Lara-Gonzalez, S.; Davis, C.T.; Walker, E.H.; Hawkins, P.T.; Stephens, L.; Eccleston, J.F.; Williams, R.L. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 2000, 103, 931–943. [Google Scholar] [CrossRef]
- Gangadhara, G.; Dahl, G.; Bohnacker, T.; Rae, R.; Gunnarsson, J.; Blaho, S.; Oster, L.; Lindmark, H.; Karabelas, K.; Pemberton, N.; et al. A class of highly selective inhibitors bind to an active state of PI3Kgamma. Nat. Chem. Biol. 2019, 15, 348–357. [Google Scholar] [CrossRef]
- Kang, S.; Denley, A.; Vanhaesebroeck, B.; Vogt, P.K. Oncogenic transformation induced by the p110beta, -gamma, and -delta isoforms of class I phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. USA 2006, 103, 1289–1294. [Google Scholar] [CrossRef]
- Thorpe, L.M.; Yuzugullu, H.; Zhao, J.J. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 2015, 15, 7–24. [Google Scholar] [CrossRef]
- Takeda, A.J.; Maher, T.J.; Zhang, Y.; Lanahan, S.M.; Bucklin, M.L.; Compton, S.R.; Tyler, P.M.; Comrie, W.A.; Matsuda, M.; Olivier, K.N.; et al. Human PI3Kgamma deficiency and its microbiota-dependent mouse model reveal immunodeficiency and tissue immunopathology. Nat. Commun. 2019, 10, 4364. [Google Scholar] [CrossRef] [PubMed]
- Rathinaswamy, M.K.; Dalwadi, U.; Fleming, K.D.; Adams, C.; Stariha, J.T.B.; Pardon, E.; Baek, M.; Vadas, O.; DiMaio, F.; Steyaert, J.; et al. Structure of the phosphoinositide 3-kinase (PI3K) p110gamma-p101 complex reveals molecular mechanism of GPCR activation. Sci. Adv. 2021, 7, eabj4282. [Google Scholar] [CrossRef] [PubMed]
- Shisheva, A. PIKfyve and its Lipid products in health and in sickness. Curr. Top. Microbiol. Immunol. 2012, 362, 127–162. [Google Scholar] [CrossRef] [PubMed]
- Lees, J.A.; Li, P.; Kumar, N.; Weisman, L.S.; Reinisch, K.M. Insights into Lysosomal PI(3,5)P2 Homeostasis from a Structural-Biochemical Analysis of the PIKfyve Lipid Kinase Complex. Mol. Cell 2020, 80, 736–743.E4. [Google Scholar] [CrossRef]
- Sbrissa, D.; Ikonomov, O.C.; Fu, Z.; Ijuin, T.; Gruenberg, J.; Takenawa, T.; Shisheva, A. Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J. Biol. Chem. 2007, 282, 23878–23891. [Google Scholar] [CrossRef]
- Ikonomov, O.C.; Sbrissa, D.; Shisheva, A. Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J. Biol. Chem. 2001, 276, 26141–26147. [Google Scholar] [CrossRef]
- Li, S.; Tiab, L.; Jiao, X.; Munier, F.L.; Zografos, L.; Frueh, B.E.; Sergeev, Y.; Smith, J.; Rubin, B.; Meallet, M.A.; et al. Mutations in PIP5K3 are associated with Francois-Neetens mouchetee fleck corneal dystrophy. Am. J. Hum. Genet. 2005, 77, 54–63. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef]
- Hetz, C.; Chevet, E.; Harding, H.P. Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 2013, 12, 703–719. [Google Scholar] [CrossRef]
- Tirasophon, W.; Welihinda, A.A.; Kaufman, R.J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998, 12, 1812–1824. [Google Scholar] [CrossRef] [PubMed]
- Korennykh, A.V.; Egea, P.F.; Korostelev, A.A.; Finer-Moore, J.; Zhang, C.; Shokat, K.M.; Stroud, R.M.; Walter, P. The unfolded protein response signals through high-order assembly of Ire1. Nature 2009, 457, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Bertolotti, A.; Zhang, Y.; Hendershot, L.M.; Harding, H.P.; Ron, D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000, 2, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Bagratuni, T.; Davenport, E.L.; Nowak, P.R.; Silva-Santisteban, M.C.; Hardcastle, A.; McAndrews, C.; Rowlands, M.G.; Morgan, G.J.; Aherne, W.; et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011, 30, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Calfon, M.; Zeng, H.; Urano, F.; Till, J.H.; Hubbard, S.R.; Harding, H.P.; Clark, S.G.; Ron, D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 2002, 415, 92–96. [Google Scholar] [CrossRef]
- Lu, Y.; Liang, F.X.; Wang, X. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol. Cell 2014, 55, 758–770. [Google Scholar] [CrossRef]
- Lee, A.H.; Iwakoshi, N.N.; Glimcher, L.H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 2003, 23, 7448–7459. [Google Scholar] [CrossRef]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Imagawa, Y.; Hosoda, A.; Sasaka, S.; Tsuru, A.; Kohno, K. RNase domains determine the functional difference between IRE1alpha and IRE1beta. FEBS Lett. 2008, 582, 656–660. [Google Scholar] [CrossRef]
- Grey, M.J.; Cloots, E.; Simpson, M.S.; LeDuc, N.; Serebrenik, Y.V.; De Luca, H.; De Sutter, D.; Luong, P.; Thiagarajah, J.R.; Paton, A.W.; et al. IRE1beta negatively regulates IRE1alpha signaling in response to endoplasmic reticulum stress. J. Cell Biol. 2020, 219, e201904048. [Google Scholar] [CrossRef]
- McAllister, C.S.; Taghavi, N.; Samuel, C.E. Protein kinase PKR amplification of interferon beta induction occurs through initiation factor eIF-2alpha-mediated translational control. J. Biol. Chem. 2012, 287, 36384–36392. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.A.; Gil, J.; Ventoso, I.; Guerra, S.; Domingo, E.; Rivas, C.; Esteban, M. Impact of protein kinase PKR in cell biology: From antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 2006, 70, 1032–1060. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.C.; Weil, R.; Dam, E.; Hovanessian, A.G.; Meurs, E.F. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol. Cell. Biol. 2000, 20, 4532–4542. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.H.; Miah, M.A.; Kim, K.P.; Bae, Y.S. New Cdc2 Tyr 4 phosphorylation by dsRNA-activated protein kinase triggers Cdc2 polyubiquitination and G2 arrest under genotoxic stresses. EMBO Rep. 2010, 11, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Garcia, M.A.; Gomez-Puertas, P.; Guerra, S.; Rullas, J.; Nakano, H.; Alcami, J.; Esteban, M. TRAF family proteins link PKR with NF-kappa B activation. Mol. Cell. Biol. 2004, 24, 4502–4512. [Google Scholar] [CrossRef]
- Silva, A.M.; Whitmore, M.; Xu, Z.; Jiang, Z.; Li, X.; Williams, B.R. Protein kinase R (PKR) interacts with and activates mitogen-activated protein kinase kinase 6 (MKK6) in response to double-stranded RNA stimulation. J. Biol. Chem. 2004, 279, 37670–37676. [Google Scholar] [CrossRef]
- Yang, X.; Nath, A.; Opperman, M.J.; Chan, C. The double-stranded RNA-dependent protein kinase differentially regulates insulin receptor substrates 1 and 2 in HepG2 cells. Mol. Biol. Cell 2010, 21, 3449–3458. [Google Scholar] [CrossRef]
- Taniuchi, S.; Miyake, M.; Tsugawa, K.; Oyadomari, M.; Oyadomari, S. Integrated stress response of vertebrates is regulated by four eIF2alpha kinases. Sci. Rep. 2016, 6, 32886. [Google Scholar] [CrossRef]
- Garcia-Ortega, M.B.; Lopez, G.J.; Jimenez, G.; Garcia-Garcia, J.A.; Conde, V.; Boulaiz, H.; Carrillo, E.; Peran, M.; Marchal, J.A.; Garcia, M.A. Clinical and therapeutic potential of protein kinase PKR in cancer and metabolism. Expert Rev. Mol. Med. 2017, 19, E9. [Google Scholar] [CrossRef]
- Gal-Ben-Ari, S.; Barrera, I.; Ehrlich, M.; Rosenblum, K. PKR: A Kinase to Remember. Front. Mol. Neurosci. 2018, 11, 480. [Google Scholar] [CrossRef]
- Stern, E.; Chinnakkaruppan, A.; David, O.; Sonenberg, N.; Rosenblum, K. Blocking the eIF2alpha kinase (PKR) enhances positive and negative forms of cortex-dependent taste memory. J. Neurosci. 2013, 33, 2517–2525. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Paccalin, M.; Lafay-Chebassier, C.; Chalon, S.; Ingrand, I.; Pinguet, J.; Pontcharraud, R.; Guillard, O.; Fauconneau, B.; Page, G. Pharmacological inhibition of PKR in APPswePS1dE9 mice transiently prevents inflammation at 12 months of age but increases Abeta42 levels in the late stages of the Alzheimer’s disease. Curr. Alzheimer Res. 2012, 9, 344–360. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Yin, S.J.; Rong, R.Q. PKR and HMGB1 expression and function in rheumatoid arthritis. Genet. Mol. Res. 2015, 14, 17864–17870. [Google Scholar] [CrossRef] [PubMed]
- Udumula, M.P.; Babu, M.S.; Bhat, A.; Dhar, I.; Sriram, D.; Dhar, A. High glucose impairs insulin signaling via activation of PKR pathway in L6 muscle cells. Biochem. Biophys. Res. Commun. 2017, 486, 645–651. [Google Scholar] [CrossRef]
- LaRonde, N.A. The ancient microbial RIO kinases. J. Biol. Chem. 2014, 289, 9488–9492. [Google Scholar] [CrossRef]
- Angermayr, M.; Bandlow, W. RIO1, an extraordinary novel protein kinase. FEBS Lett. 2002, 524, 31–36. [Google Scholar] [CrossRef]
- Widmann, B.; Wandrey, F.; Badertscher, L.; Wyler, E.; Pfannstiel, J.; Zemp, I.; Kutay, U. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol. Biol. Cell 2012, 23, 22–35. [Google Scholar] [CrossRef]
- Ferreira-Cerca, S.; Kiburu, I.; Thomson, E.; LaRonde, N.; Hurt, E. Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic Acids Res. 2014, 42, 8635–8647. [Google Scholar] [CrossRef]
- Weinberg, F.; Reischmann, N.; Fauth, L.; Taromi, S.; Mastroianni, J.; Kohler, M.; Halbach, S.; Becker, A.C.; Deng, N.; Schmitz, T.; et al. The Atypical Kinase RIOK1 Promotes Tumor Growth and Invasive Behavior. EBioMedicine 2017, 20, 79–97. [Google Scholar] [CrossRef]
- Kloet, S.L.; Whiting, J.L.; Gafken, P.; Ranish, J.; Wang, E.H. Phosphorylation-dependent regulation of cyclin D1 and cyclin A gene transcription by TFIID subunits TAF1 and TAF7. Mol. Cell. Biol. 2012, 32, 3358–3369. [Google Scholar] [CrossRef]
- Wang, H.; Curran, E.C.; Hinds, T.R.; Wang, E.H.; Zheng, N. Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module. Cell Res. 2014, 24, 1433–1444. [Google Scholar] [CrossRef] [PubMed]
- Louder, R.K.; He, Y.; Lopez-Blanco, J.R.; Fang, J.; Chacon, P.; Nogales, E. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 2016, 531, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Dikstein, R.; Ruppert, S.; Tjian, R. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell 1996, 84, 781–790. [Google Scholar] [CrossRef]
- Jacobson, R.H.; Ladurner, A.G.; King, D.S.; Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 2000, 288, 1422–1425. [Google Scholar] [CrossRef] [PubMed]
- Mizzen, C.A.; Yang, X.J.; Kokubo, T.; Brownell, J.E.; Bannister, A.J.; Owen-Hughes, T.; Workman, J.; Wang, L.; Berger, S.L.; Kouzarides, T.; et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 1996, 87, 1261–1270. [Google Scholar] [CrossRef]
- Hilton, T.L.; Li, Y.; Dunphy, E.L.; Wang, E.H. TAF1 histone acetyltransferase activity in Sp1 activation of the cyclin D1 promoter. Mol. Cell. Biol. 2005, 25, 4321–4332. [Google Scholar] [CrossRef]
- O’Brien, T.; Tjian, R. Functional analysis of the human TAFII250 N-terminal kinase domain. Mol. Cell 1998, 1, 905–911. [Google Scholar] [CrossRef]
- Li, H.H.; Li, A.G.; Sheppard, H.M.; Liu, X. Phosphorylation on Thr-55 by TAF1 mediates degradation of p53: A role for TAF1 in cell G1 progression. Mol. Cell 2004, 13, 867–878. [Google Scholar] [CrossRef]
- Siegert, J.L.; Robbins, P.D. Rb inhibits the intrinsic kinase activity of TATA-binding protein-associated factor TAFII250. Mol. Cell. Biol. 1999, 19, 846–854. [Google Scholar] [CrossRef]
- Makino, S.; Kaji, R.; Ando, S.; Tomizawa, M.; Yasuno, K.; Goto, S.; Matsumoto, S.; Tabuena, M.D.; Maranon, E.; Dantes, M.; et al. Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am. J. Hum. Genet. 2007, 80, 393–406. [Google Scholar] [CrossRef]
- O’Rawe, J.A.; Wu, Y.; Dorfel, M.J.; Rope, A.F.; Au, P.Y.; Parboosingh, J.S.; Moon, S.; Kousi, M.; Kosma, K.; Smith, C.S.; et al. TAF1 Variants Are Associated with Dysmorphic Features, Intellectual Disability, and Neurological Manifestations. Am. J. Hum. Genet. 2015, 97, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Singer, D.S. CIITA and Its Dual Roles in MHC Gene Transcription. Front. Immunol. 2013, 4, 476. [Google Scholar] [CrossRef] [PubMed]
- Reith, W.; LeibundGut-Landmann, S.; Waldburger, J.M. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 2005, 5, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Choi, N.M.; Majumder, P.; Boss, J.M. Regulation of major histocompatibility complex class II genes. Curr. Opin. Immunol. 2011, 23, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Soe, K.C.; Devaiah, B.N.; Singer, D.S. Transcriptional coactivator CIITA, a functional homolog of TAF1, has kinase activity. Biochim. Biophys. Acta 2013, 1829, 1184–1190. [Google Scholar] [CrossRef]
- Raval, A.; Howcroft, T.K.; Weissman, J.D.; Kirshner, S.; Zhu, X.S.; Yokoyama, K.; Ting, J.; Singer, D.S. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol. Cell 2001, 7, 105–115. [Google Scholar] [CrossRef]
- Fontes, J.D.; Kanazawa, S.; Jean, D.; Peterlin, B.M. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol. Cell. Biol. 1999, 19, 941–947. [Google Scholar] [CrossRef][Green Version]
- Kretsovali, A.; Spilianakis, C.; Dimakopoulos, A.; Makatounakis, T.; Papamatheakis, J. Self-association of class II transactivator correlates with its intracellular localization and transactivation. J. Biol. Chem. 2001, 276, 32191–32197. [Google Scholar] [CrossRef]
- Ting, J.P.; Trowsdale, J. Genetic control of MHC class II expression. Cell 2002, 109 (Suppl. 1), S21–S33. [Google Scholar] [CrossRef]
- Steimle, V.; Otten, L.A.; Zufferey, M.; Mach, B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell 1993, 75, 135–146. [Google Scholar] [CrossRef]
- Srinivasan, M.; Mehta, P.; Yu, Y.; Prugar, E.; Koonin, E.V.; Karzai, A.W.; Sternglanz, R. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 2011, 30, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Downey, M.; Houlsworth, R.; Maringele, L.; Rollie, A.; Brehme, M.; Galicia, S.; Guillard, S.; Partington, M.; Zubko, M.K.; Krogan, N.J.; et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 2006, 124, 1155–1168. [Google Scholar] [CrossRef] [PubMed]
- Kisseleva-Romanova, E.; Lopreiato, R.; Baudin-Baillieu, A.; Rousselle, J.C.; Ilan, L.; Hofmann, K.; Namane, A.; Mann, C.; Libri, D. Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J. 2006, 25, 3576–3585. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, X.; Banerjee, S.; Chen, H.; Ma, W.; Bode, A.M.; Dong, Z. Crystal structure of the human PRPK-TPRKB complex. Commun. Biol. 2021, 4, 167. [Google Scholar] [CrossRef] [PubMed]
- Facchin, S.; Lopreiato, R.; Ruzzene, M.; Marin, O.; Sartori, G.; Gotz, C.; Montenarh, M.; Carignani, G.; Pinna, L.A. Functional homology between yeast piD261/Bud32 and human PRPK: Both phosphorylate p53 and PRPK partially complements piD261/Bud32 deficiency. FEBS Lett. 2003, 549, 63–66. [Google Scholar] [CrossRef]
- Perrochia, L.; Guetta, D.; Hecker, A.; Forterre, P.; Basta, T. Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t6A tRNA modification. Nucleic Acids Res. 2013, 41, 9484–9499. [Google Scholar] [CrossRef]
- Zhang, W.; Collinet, B.; Graille, M.; Daugeron, M.C.; Lazar, N.; Libri, D.; Durand, D.; van Tilbeurgh, H. Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex. Nucleic Acids Res. 2015, 43, 3358–3372. [Google Scholar] [CrossRef]
- Braun, D.A.; Rao, J.; Mollet, G.; Schapiro, D.; Daugeron, M.C.; Tan, W.; Gribouval, O.; Boyer, O.; Revy, P.; Jobst-Schwan, T.; et al. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet. 2017, 49, 1529–1538. [Google Scholar] [CrossRef]
- Facchin, S.; Ruzzene, M.; Peggion, C.; Sartori, G.; Carignani, G.; Marin, O.; Brustolon, F.; Lopreiato, R.; Pinna, L.A. Phosphorylation and activation of the atypical kinase p53-related protein kinase (PRPK) by Akt/PKB. Cell. Mol. Life Sci. 2007, 64, 2680–2689. [Google Scholar] [CrossRef]
- Hideshima, T.; Cottini, F.; Nozawa, Y.; Seo, H.S.; Ohguchi, H.; Samur, M.K.; Cirstea, D.; Mimura, N.; Iwasawa, Y.; Richardson, P.G.; et al. p53-related protein kinase confers poor prognosis and represents a novel therapeutic target in multiple myeloma. Blood 2017, 129, 1308–1319. [Google Scholar] [CrossRef]
- Kilpatrick, L.E.; Lee, J.Y.; Haines, K.M.; Campbell, D.E.; Sullivan, K.E.; Korchak, H.M. A role for PKC-delta and PI 3-kinase in TNF-alpha-mediated antiapoptotic signaling in the human neutrophil. Am. J. Physiol. Cell Physiol. 2002, 283, C48–C57. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Langston, J.C.; Tang, Y.; Kiani, M.F.; Kilpatrick, L.E. The Role of Tyrosine Phosphorylation of Protein Kinase C Delta in Infection and Inflammation. Int. J. Mol. Sci. 2019, 20, 1498. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.C. Protein kinase C: Perfectly balanced. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 208–230. [Google Scholar] [CrossRef] [PubMed]
- Humphries, M.J.; Ohm, A.M.; Schaack, J.; Adwan, T.S.; Reyland, M.E. Tyrosine phosphorylation regulates nuclear translocation of PKCdelta. Oncogene 2008, 27, 3045–3053. [Google Scholar] [CrossRef]
- Steinberg, S.F. Distinctive activation mechanisms and functions for protein kinase Cdelta. Biochem. J. 2004, 384, 449–459. [Google Scholar] [CrossRef]
- Zach, S.; Felk, S.; Gillardon, F. Signal transduction protein array analysis links LRRK2 to Ste20 kinases and PKC zeta that modulate neuronal plasticity. PLoS ONE 2010, 5, e13191. [Google Scholar] [CrossRef]
- Gomez-Suaga, P.; Luzon-Toro, B.; Churamani, D.; Zhang, L.; Bloor-Young, D.; Patel, S.; Woodman, P.G.; Churchill, G.C.; Hilfiker, S. Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Hum. Mol. Genet. 2012, 21, 511–525. [Google Scholar] [CrossRef]
- Steger, M.; Diez, F.; Dhekne, H.S.; Lis, P.; Nirujogi, R.S.; Karayel, O.; Tonelli, F.; Martinez, T.N.; Lorentzen, E.; Pfeffer, S.R.; et al. Systematic proteomic analysis of LRRK2-mediated Rab GTPase phosphorylation establishes a connection to ciliogenesis. Elife 2017, 6, 831. [Google Scholar] [CrossRef]
- Guaitoli, G.; Raimondi, F.; Gilsbach, B.K.; Gomez-Llorente, Y.; Deyaert, E.; Renzi, F.; Li, X.; Schaffner, A.; Jagtap, P.K.; Boldt, K.; et al. Structural model of the dimeric Parkinson’s protein LRRK2 reveals a compact architecture involving distant interdomain contacts. Proc. Natl. Acad. Sci. USA 2016, 113, E4357–E4366. [Google Scholar] [CrossRef]
- Guo, L.; Gandhi, P.N.; Wang, W.; Petersen, R.B.; Wilson-Delfosse, A.L.; Chen, S.G. The Parkinson’s disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp. Cell Res. 2007, 313, 3658–3670. [Google Scholar] [CrossRef]
- Deng, J.; Lewis, P.A.; Greggio, E.; Sluch, E.; Beilina, A.; Cookson, M.R. Structure of the ROC domain from the Parkinson’s disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc. Natl. Acad. Sci. USA 2008, 105, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Berger, Z.; Smith, K.A.; Lavoie, M.J. Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry 2010, 49, 5511–5523. [Google Scholar] [CrossRef] [PubMed]
- Nichols, R.J.; Dzamko, N.; Morrice, N.A.; Campbell, D.G.; Deak, M.; Ordureau, A.; Macartney, T.; Tong, Y.; Shen, J.; Prescott, A.R.; et al. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson’s disease-associated mutations and regulates cytoplasmic localization. Biochem. J. 2010, 430, 393–404. [Google Scholar] [CrossRef]
- Simon-Sanchez, J.; Schulte, C.; Bras, J.M.; Sharma, M.; Gibbs, J.R.; Berg, D.; Paisan-Ruiz, C.; Lichtner, P.; Scholz, S.W.; Hernandez, D.G.; et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009, 41, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Satake, W.; Nakabayashi, Y.; Mizuta, I.; Hirota, Y.; Ito, C.; Kubo, M.; Kawaguchi, T.; Tsunoda, T.; Watanabe, M.; Takeda, A.; et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat. Genet. 2009, 41, 1303–1307. [Google Scholar] [CrossRef]
- Chen, Z.C.; Zhang, W.; Chua, L.L.; Chai, C.; Li, R.; Lin, L.; Cao, Z.; Angeles, D.C.; Stanton, L.W.; Peng, J.H.; et al. Phosphorylation of amyloid precursor protein by mutant LRRK2 promotes AICD activity and neurotoxicity in Parkinson’s disease. Sci. Signal. 2017, 10, aam6790. [Google Scholar] [CrossRef]
- Chen, C.Y.; Weng, Y.H.; Chien, K.Y.; Lin, K.J.; Yeh, T.H.; Cheng, Y.P.; Lu, C.S.; Wang, H.L. (G2019S) LRRK2 activates MKK4-JNK pathway and causes degeneration of SN dopaminergic neurons in a transgenic mouse model of PD. Cell Death Differ. 2012, 19, 1623–1633. [Google Scholar] [CrossRef]
- Guerreiro, P.S.; Gerhardt, E.; Lopes da Fonseca, T.; Bahr, M.; Outeiro, T.F.; Eckermann, K. LRRK2 Promotes Tau Accumulation, Aggregation and Release. Mol. Neurobiol. 2016, 53, 3124–3135. [Google Scholar] [CrossRef]
- Rudenko, I.N.; Cookson, M.R. Heterogeneity of leucine-rich repeat kinase 2 mutations: Genetics, mechanisms and therapeutic implications. Neurotherapeutics 2014, 11, 738–750. [Google Scholar] [CrossRef]
- Sheng, Z.; Zhang, S.; Bustos, D.; Kleinheinz, T.; Le Pichon, C.E.; Dominguez, S.L.; Solanoy, H.O.; Drummond, J.; Zhang, X.; Ding, X.; et al. Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci. Transl. Med. 2012, 4, 164ra161. [Google Scholar] [CrossRef]
- Steger, M.; Tonelli, F.; Ito, G.; Davies, P.; Trost, M.; Vetter, M.; Wachter, S.; Lorentzen, E.; Duddy, G.; Wilson, S.; et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. Elife 2016, 5, 879. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.D.; DeMong, D.E.; Greshock, T.J.; Basu, K.; Dai, X.; Harris, J.; Hruza, A.; Li, S.W.; Lin, S.I.; Liu, H.; et al. Discovery of a 3-(4-Pyrimidinyl) Indazole (MLi-2), an Orally Available and Selective Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitor that Reduces Brain Kinase Activity. J. Med. Chem. 2017, 60, 2983–2992. [Google Scholar] [CrossRef] [PubMed]
- Fell, M.J.; Mirescu, C.; Basu, K.; Cheewatrakoolpong, B.; DeMong, D.E.; Ellis, J.M.; Hyde, L.A.; Lin, Y.; Markgraf, C.G.; Mei, H.; et al. MLi-2, a Potent, Selective, and Centrally Active Compound for Exploring the Therapeutic Potential and Safety of LRRK2 Kinase Inhibition. J. Pharmacol. Exp. Ther. 2015, 355, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Beullens, M.; Vancauwenbergh, S.; Morrice, N.; Derua, R.; Ceulemans, H.; Waelkens, E.; Bollen, M. Substrate specificity and activity regulation of protein kinase MELK. J. Biol. Chem. 2005, 280, 40003–40011. [Google Scholar] [CrossRef]
- Lin, M.L.; Park, J.H.; Nishidate, T.; Nakamura, Y.; Katagiri, T. Involvement of maternal embryonic leucine zipper kinase (MELK) in mammary carcinogenesis through interaction with Bcl-G, a pro-apoptotic member of the Bcl-2 family. Breast Cancer Res. 2007, 9, R17. [Google Scholar] [CrossRef]
- Nakano, I.; Masterman-Smith, M.; Saigusa, K.; Paucar, A.A.; Horvath, S.; Shoemaker, L.; Watanabe, M.; Negro, A.; Bajpai, R.; Howes, A.; et al. Maternal embryonic leucine zipper kinase is a key regulator of the proliferation of malignant brain tumors, including brain tumor stem cells. J. Neurosci. Res. 2008, 86, 48–60. [Google Scholar] [CrossRef]
- Gray, D.; Jubb, A.M.; Hogue, D.; Dowd, P.; Kljavin, N.; Yi, S.; Bai, W.; Frantz, G.; Zhang, Z.; Koeppen, H.; et al. Maternal embryonic leucine zipper kinase/murine protein serine-threonine kinase 38 is a promising therapeutic target for multiple cancers. Cancer Res. 2005, 65, 9751–9761. [Google Scholar] [CrossRef]
- Mirey, G.; Chartrain, I.; Froment, C.; Quaranta, M.; Bouche, J.P.; Monsarrat, B.; Tassan, J.P.; Ducommun, B. CDC25B phosphorylated by pEg3 localizes to the centrosome and the spindle poles at mitosis. Cell Cycle 2005, 4, 806–811. [Google Scholar] [CrossRef]
- Huang, H.T.; Seo, H.S.; Zhang, T.; Wang, Y.; Jiang, B.; Li, Q.; Buckley, D.L.; Nabet, B.; Roberts, J.M.; Paulk, J.; et al. MELK is not necessary for the proliferation of basal-like breast cancer cells. Elife 2017, 6, 002. [Google Scholar] [CrossRef]
- Mahajan, K.; Mahajan, N.P. Shepherding AKT and androgen receptor by Ack1 tyrosine kinase. J. Cell. Physiol. 2010, 224, 327–333. [Google Scholar] [CrossRef]
- Yokoyama, N.; Lougheed, J.; Miller, W.T. Phosphorylation of WASP by the Cdc42-associated kinase ACK1: Dual hydroxyamino acid specificity in a tyrosine kinase. J. Biol. Chem. 2005, 280, 42219–42226. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, K.; Coppola, D.; Challa, S.; Fang, B.; Chen, Y.A.; Zhu, W.; Lopez, A.S.; Koomen, J.; Engelman, R.W.; Rivera, C.; et al. Ack1 mediated AKT/PKB tyrosine 176 phosphorylation regulates its activation. PLoS ONE 2010, 5, e9646. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, K.; Lawrence, H.R.; Lawrence, N.J.; Mahajan, N.P. ACK1 tyrosine kinase interacts with histone demethylase KDM3A to regulate the mammary tumor oncogene HOXA1. J. Biol. Chem. 2014, 289, 28179–28191. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, K.; Malla, P.; Lawrence, H.R.; Chen, Z.; Kumar-Sinha, C.; Malik, R.; Shukla, S.; Kim, J.; Coppola, D.; Lawrence, N.J.; et al. ACK1/TNK2 Regulates Histone H4 Tyr88-phosphorylation and AR Gene Expression in Castration-Resistant Prostate Cancer. Cancer Cell 2017, 31, 790–803.E8. [Google Scholar] [CrossRef]
- Mahajan, N.P.; Liu, Y.; Majumder, S.; Warren, M.R.; Parker, C.E.; Mohler, J.L.; Earp, H.S.; Whang, Y.E. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proc. Natl. Acad. Sci. USA 2007, 104, 8438–8443. [Google Scholar] [CrossRef]
- Van der Horst, E.H.; Degenhardt, Y.Y.; Strelow, A.; Slavin, A.; Chinn, L.; Orf, J.; Rong, M.; Li, S.; See, L.H.; Nguyen, K.Q.; et al. Metastatic properties and genomic amplification of the tyrosine kinase gene ACK1. Proc. Natl. Acad. Sci. USA 2005, 102, 15901–15906. [Google Scholar] [CrossRef]
- Lanata, C.M.; Chung, S.A.; Criswell, L.A. DNA methylation 101: What is important to know about DNA methylation and its role in SLE risk and disease heterogeneity. Lupus Sci. Med. 2018, 5, e000285. [Google Scholar] [CrossRef]
- Lawrence, H.R.; Mahajan, K.; Luo, Y.; Zhang, D.; Tindall, N.; Huseyin, M.; Gevariya, H.; Kazi, S.; Ozcan, S.; Mahajan, N.P.; et al. Development of novel ACK1/TNK2 inhibitors using a fragment-based approach. J. Med. Chem. 2015, 58, 2746–2763. [Google Scholar] [CrossRef]
- Wu, X.; Zahari, M.S.; Renuse, S.; Kelkar, D.S.; Barbhuiya, M.A.; Rojas, P.L.; Stearns, V.; Gabrielson, E.; Malla, P.; Sukumar, S.; et al. The non-receptor tyrosine kinase TNK2/ACK1 is a novel therapeutic target in triple negative breast cancer. Oncotarget 2017, 8, 2971–2983. [Google Scholar] [CrossRef]
- Boyle, J.P.; Parkhouse, R.; Monie, T.P. Insights into the molecular basis of the NOD2 signalling pathway. Open Biol. 2014, 4, 25520185. [Google Scholar] [CrossRef]
- He, S.; Wang, X. RIP kinases as modulators of inflammation and immunity. Nat. Immunol. 2018, 19, 912–922. [Google Scholar] [CrossRef] [PubMed]
- Abbott, D.W.; Wilkins, A.; Asara, J.M.; Cantley, L.C. The Crohn’s disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr. Biol. 2004, 14, 2217–2227. [Google Scholar] [CrossRef] [PubMed]
- Strober, W.; Watanabe, T. NOD2, an intracellular innate immune sensor involved in host defense and Crohn’s disease. Mucosal. Immunol. 2011, 4, 484–495. [Google Scholar] [CrossRef] [PubMed]
- McGovern, D.P.; Hysi, P.; Ahmad, T.; van Heel, D.A.; Moffatt, M.F.; Carey, A.; Cookson, W.O.; Jewell, D.P. Association between a complex insertion/deletion polymorphism in NOD1 (CARD4) and susceptibility to inflammatory bowel disease. Hum. Mol. Genet. 2005, 14, 1245–1250. [Google Scholar] [CrossRef]
- Tanabe, T.; Ishige, I.; Suzuki, Y.; Aita, Y.; Furukawa, A.; Ishige, Y.; Uchida, K.; Suzuki, T.; Takemura, T.; Ikushima, S.; et al. Sarcoidosis and NOD1 variation with impaired recognition of intracellular Propionibacterium acnes. Biochim. Biophys. Acta 2006, 1762, 794–801. [Google Scholar] [CrossRef]
- Hysi, P.; Kabesch, M.; Moffatt, M.F.; Schedel, M.; Carr, D.; Zhang, Y.; Boardman, B.; von Mutius, E.; Weiland, S.K.; Leupold, W.; et al. NOD1 variation, immunoglobulin E and asthma. Hum. Mol. Genet. 2005, 14, 935–941. [Google Scholar] [CrossRef]
- Hugot, J.P.; Chamaillard, M.; Zouali, H.; Lesage, S.; Cezard, J.P.; Belaiche, J.; Almer, S.; Tysk, C.; O’Morain, C.A.; Gassull, M.; et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001, 411, 599–603. [Google Scholar] [CrossRef]
- Stronati, L.; Negroni, A.; Merola, P.; Pannone, V.; Borrelli, O.; Cirulli, M.; Annese, V.; Cucchiara, S. Mucosal NOD2 expression and NF-kappaB activation in pediatric Crohn’s disease. Inflamm. Bowel Dis. 2008, 14, 295–302. [Google Scholar] [CrossRef]
- Stronati, L.; Negroni, A.; Pierdomenico, M.; D’Ottavio, C.; Tirindelli, D.; Di Nardo, G.; Oliva, S.; Viola, F.; Cucchiara, S. Altered expression of innate immunity genes in different intestinal sites of children with ulcerative colitis. Dig. Liver Dis. 2010, 42, 848–853. [Google Scholar] [CrossRef]
- Thome, M.; Hofmann, K.; Burns, K.; Martinon, F.; Bodmer, J.L.; Mattmann, C.; Tschopp, J. Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr. Biol. 1998, 8, 885–888. [Google Scholar] [CrossRef]
- McCarthy, J.V.; Ni, J.; Dixit, V.M. RIP2 is a novel NF-kappaB-activating and cell death-inducing kinase. J. Biol. Chem. 1998, 273, 16968–16975. [Google Scholar] [CrossRef] [PubMed]
- Dorsch, M.; Wang, A.; Cheng, H.; Lu, C.; Bielecki, A.; Charron, K.; Clauser, K.; Ren, H.; Polakiewicz, R.D.; Parsons, T.; et al. Identification of a regulatory autophosphorylation site in the serine-threonine kinase RIP2. Cell Signal. 2006, 18, 2223–2229. [Google Scholar] [CrossRef] [PubMed]
- Tigno-Aranjuez, J.T.; Asara, J.M.; Abbott, D.W. Inhibition of RIP2’s tyrosine kinase activity limits NOD2-driven cytokine responses. Genes. Dev. 2010, 24, 2666–2677. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Fujimoto, Y.; Lucas, P.C.; Nakano, H.; Fukase, K.; Nunez, G.; Inohara, N. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 2008, 27, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Travassos, L.H.; Carneiro, L.A.; Ramjeet, M.; Hussey, S.; Kim, Y.G.; Magalhaes, J.G.; Yuan, L.; Soares, F.; Chea, E.; Le Bourhis, L.; et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010, 11, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Windheim, M.; Lang, C.; Peggie, M.; Plater, L.A.; Cohen, P. Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem. J. 2007, 404, 179–190. [Google Scholar] [CrossRef]
- Nachbur, U.; Stafford, C.A.; Bankovacki, A.; Zhan, Y.; Lindqvist, L.M.; Fiil, B.K.; Khakham, Y.; Ko, H.J.; Sandow, J.J.; Falk, H.; et al. A RIPK2 inhibitor delays NOD signalling events yet prevents inflammatory cytokine production. Nat. Commun. 2015, 6, 6442. [Google Scholar] [CrossRef]
- Tigno-Aranjuez, J.T.; Benderitter, P.; Rombouts, F.; Deroose, F.; Bai, X.; Mattioli, B.; Cominelli, F.; Pizarro, T.T.; Hoflack, J.; Abbott, D.W. In vivo inhibition of RIPK2 kinase alleviates inflammatory disease. J. Biol. Chem. 2014, 289, 29651–29664. [Google Scholar] [CrossRef]
- Canning, P.; Ruan, Q.; Schwerd, T.; Hrdinka, M.; Maki, J.L.; Saleh, D.; Suebsuwong, C.; Ray, S.; Brennan, P.E.; Cuny, G.D.; et al. Inflammatory Signaling by NOD-RIPK2 Is Inhibited by Clinically Relevant Type II Kinase Inhibitors. Chem. Biol. 2015, 22, 1174–1184. [Google Scholar] [CrossRef]
- Berson, A.E.; Young, C.; Morrison, S.L.; Fujii, G.H.; Sheung, J.; Wu, B.; Bolen, J.B.; Burkhardt, A.L. Identification and characterization of a myristylated and palmitylated serine/threonine protein kinase. Biochem. Biophys. Res. Commun. 1999, 259, 533–538. [Google Scholar] [CrossRef]
- Eswaran, J.; Bernad, A.; Ligos, J.M.; Guinea, B.; Debreczeni, J.E.; Sobott, F.; Parker, S.A.; Najmanovich, R.; Turk, B.E.; Knapp, S. Structure of the human protein kinase MPSK1 reveals an atypical activation loop architecture. Structure 2008, 16, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, X.; Li, B.; Wang, J.; Wang, W.; Liu, J.; Liu, Q.; Zhang, X. STK16 regulates actin dynamics to control Golgi organization and cell cycle. Sci. Rep. 2017, 7, 44607. [Google Scholar] [CrossRef] [PubMed]
- Guinea, B.; Ligos, J.M.; Lain de Lera, T.; Martin-Caballero, J.; Flores, J.; Gonzalez de la Pena, M.; Garcia-Castro, J.; Bernad, A. Nucleocytoplasmic shuttling of STK16 (PKL12), a Golgi-resident serine/threonine kinase involved in VEGF expression regulation. Exp. Cell Res. 2006, 312, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, J.; Yang, X.; Li, B.; Wu, H.; Qi, S.; Chen, C.; Liu, X.; Yu, K.; Wang, W.; et al. Discovery of a Highly Selective STK16 Kinase Inhibitor. ACS Chem. Biol. 2016, 11, 1537–1543. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.G.; Matewish, M.J.; Burrows, L.L.; Monteiro, M.A.; Perry, M.B.; Lam, J.S. Lipopolysaccharide core phosphates are required for viability and intrinsic drug resistance in Pseudomonas aeruginosa. Mol. Microbiol. 2000, 35, 718–727. [Google Scholar] [CrossRef]
- Sadovskaya, I.; Brisson, J.R.; Thibault, P.; Richards, J.C.; Lam, J.S.; Altman, E. Structural characterization of the outer core and the O-chain linkage region of lipopolysaccharide from Pseudomonas aeruginosa serotype O5. Eur. J. Biochem. 2000, 267, 1640–1650. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Yethon, J.A.; Gunn, J.S.; Ernst, R.K.; Miller, S.I.; Laroche, L.; Malo, D.; Whitfield, C. Salmonella enterica serovar typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence In vivo. Infect. Immun. 2000, 68, 4485–4491. [Google Scholar] [CrossRef]
- Delucia, A.M.; Six, D.A.; Caughlan, R.E.; Gee, P.; Hunt, I.; Lam, J.S.; Dean, C.R. Lipopolysaccharide (LPS) inner-core phosphates are required for complete LPS synthesis and transport to the outer membrane in Pseudomonas aeruginosa PAO1. mBio 2011, 2, e00142-11. [Google Scholar] [CrossRef]
- Zhao, X.; Lam, J.S. WaaP of Pseudomonas aeruginosa is a novel eukaryotic type protein-tyrosine kinase as well as a sugar kinase essential for the biosynthesis of core lipopolysaccharide. J. Biol. Chem. 2002, 277, 4722–4730. [Google Scholar] [CrossRef] [PubMed]
- Kreamer, N.N.K.; Chopra, R.; Caughlan, R.E.; Fabbro, D.; Fang, E.; Gee, P.; Hunt, I.; Li, M.; Leon, B.C.; Muller, L.; et al. Acylated-acyl carrier protein stabilizes the Pseudomonas aeruginosa WaaP lipopolysaccharide heptose kinase. Sci. Rep. 2018, 8, 14124. [Google Scholar] [CrossRef] [PubMed]
- Latorre-Muro, P.; Baeza, J.; Armstrong, E.A.; Hurtado-Guerrero, R.; Corzana, F.; Wu, L.E.; Sinclair, D.A.; Lopez-Buesa, P.; Carrodeguas, J.A.; Denu, J.M. Dynamic Acetylation of Phosphoenolpyruvate Carboxykinase Toggles Enzyme Activity between Gluconeogenic and Anaplerotic Reactions. Mol. Cell 2018, 71, 718–732.E9. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Wang, Z.; Xia, Y.; Shao, F.; Xia, W.; Wei, Y.; Li, X.; Qian, X.; Lee, J.H.; Du, L.; et al. The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature 2020, 580, 530–535. [Google Scholar] [CrossRef]
- Zhu, X.R.; Peng, S.Q.; Wang, L.; Chen, X.Y.; Feng, C.X.; Liu, Y.Y.; Chen, M.B. Identification of phosphoenolpyruvate carboxykinase 1 as a potential therapeutic target for pancreatic cancer. Cell Death Dis. 2021, 12, 918. [Google Scholar] [CrossRef]
- Shao, F.; Bian, X.; Jiang, H.; Zhao, G.; Zhu, L.; Xu, D.; Wang, S.; Guo, W.; Xing, D.; Xue, Q.; et al. Association of phosphoenolpyruvate carboxykinase 1 protein kinase activity-dependent sterol regulatory element-binding protein 1 activation with prognosis of oesophageal carcinoma. Eur. J. Cancer 2021, 142, 123–131. [Google Scholar] [CrossRef]
- Foley, L.H.; Wang, P.; Dunten, P.; Ramsey, G.; Gubler, M.L.; Wertheimer, S.J. X-ray structures of two xanthine inhibitors bound to PEPCK and N-3 modifications of substituted 1,8-dibenzylxanthines. Bioorg. Med. Chem. Lett. 2003, 13, 3871–3874. [Google Scholar] [CrossRef]
- Foley, L.H.; Wang, P.; Dunten, P.; Ramsey, G.; Gubler, M.L.; Wertheimer, S.J. Modified 3-alkyl-1,8-dibenzylxanthines as GTP-competitive inhibitors of phosphoenolpyruvate carboxykinase. Bioorg. Med. Chem. Lett. 2003, 13, 3607–3610. [Google Scholar] [CrossRef]
- Burnell, J.N.; Hatch, M.D. Light-dark modulation of leaf pyruvate, Pi dikinase. Trends Biochem. Sci. 1985, 10, 288–291. [Google Scholar] [CrossRef]
- Chastain, C.J.; Botschner, M.; Harrington, G.E.; Thompson, B.J.; Mills, S.E.; Sarath, G.; Chollet, R. Further Analysis of Maize C4 Pyruvate, Orthophosphate Dikinase Phosphorylation by Its Bifunctional Regulatory Protein Using Selective Substitutions of the Regulatory Thr-456 and Catalytic His-458 Residues. Arch. Biochem. Biophys. 2000, 375, 165–170. [Google Scholar] [CrossRef]
- Chen, Y.B.; Lu, T.C.; Wang, H.X.; Shen, J.; Bu, T.T.; Chao, Q.; Gao, Z.F.; Zhu, X.G.; Wang, Y.F.; Wang, B.C. Posttranslational Modification of Maize Chloroplast Pyruvate Orthophosphate Dikinase Reveals the Precise Regulatory Mechanism of Its Enzymatic Activity. Plant Physiol. 2014, 165, 534–549. [Google Scholar] [CrossRef] [PubMed]
- Burnell, J.N. Cloning and characterization of Escherichia coli DUF299: A bifunctional ADP-dependent kinase--Pi-dependent pyrophosphorylase from bacteria. BMC Biochem. 2010, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Roeske, C.A.; Kutny, R.M.; Budde, R.J.; Chollet, R. Sequence of the phosphothreonyl regulatory site peptide from inactive maize leaf pyruvate, orthophosphate dikinase. J. Biol. Chem. 1988, 263, 6683–6687. [Google Scholar] [CrossRef]
- Jiang, L.; Chen, Y.B.; Zheng, J.; Chen, Z.; Liu, Y.; Tao, Y.; Wu, W.; Chen, Z.; Wang, B.C. Structural Basis of Reversible Phosphorylation by Maize Pyruvate Orthophosphate Dikinase Regulatory Protein. Plant Physiol. 2016, 170, 732–741. [Google Scholar] [CrossRef]
- LaPorte, D.C. The isocitrate dehydrogenase phosphorylation cycle: Regulation and enzymology. J. Cell. Biochem. 1993, 51, 14–18. [Google Scholar] [CrossRef]
- LaPorte, D.C.; Chung, T. A single gene codes for the kinase and phosphatase which regulate isocitrate dehydrogenase. J. Biol. Chem. 1985, 260, 15291–15297. [Google Scholar] [CrossRef]
- Stueland, C.S.; Eck, K.R.; Stieglbauer, K.T.; LaPorte, D.C. Isocitrate dehydrogenase kinase/phosphatase exhibits an intrinsic adenosine triphosphatase activity. J. Biol. Chem. 1987, 262, 16095–16099. [Google Scholar] [CrossRef]
- Cozzone, A.J. Regulation of acetate metabolism by protein phosphorylation in enteric bacteria. Annu. Rev. Microbiol. 1998, 52, 127–164. [Google Scholar] [CrossRef]
- Zheng, J.; Jia, Z. Structure of the bifunctional isocitrate dehydrogenase kinase/phosphatase. Nature 2010, 465, 961–965. [Google Scholar] [CrossRef]
- Thorsness, P.E.; Koshland, D.E., Jr. Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J. Biol. Chem. 1987, 262, 10422–10425. [Google Scholar] [CrossRef]
- Zheng, J.; Yates, S.P.; Jia, Z. Structural and mechanistic insights into the bifunctional enzyme isocitrate dehydrogenase kinase/phosphatase AceK. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 2656–2668. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stueland, C.S.; Ikeda, T.P.; LaPorte, D.C. Mutation of the predicted ATP binding site inactivates both activities of isocitrate dehydrogenase kinase/phosphatase. J. Biol. Chem. 1989, 264, 13775–13779. [Google Scholar] [CrossRef]
- Yates, S.P.; Edwards, T.E.; Bryan, C.M.; Stein, A.J.; Van Voorhis, W.C.; Myler, P.J.; Stewart, L.J.; Zheng, J.; Jia, Z. Structural basis of the substrate specificity of bifunctional isocitrate dehydrogenase kinase/phosphatase. Biochemistry 2011, 50, 8103–8106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ikeda, T.; LaPorte, D.C. Isocitrate dehydrogenase kinase/phosphatase: AceK alleles that express kinase but not phosphatase activity. J. Bacteriol. 1991, 173, 1801–1806. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miller, S.P.; Karschnia, E.J.; Ikeda, T.P.; LaPorte, D.C. Isocitrate dehydrogenase kinase/phosphatase. Kinetic characteristics of the wild-type and two mutant proteins. J. Biol. Chem. 1996, 271, 19124–19128. [Google Scholar] [CrossRef]
- Deutscher, J.; Pevec, B.; Beyreuther, K.; Kiltz, H.H.; Hengstenberg, W. Streptococcal phosphoenolpyruvate-sugar phosphotransferase system: Amino acid sequence and site of ATP-dependent phosphorylation of HPr. Biochemistry 1986, 25, 6543–6551. [Google Scholar] [CrossRef]
- Deutscher, J.; Saier, M.H., Jr. ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 1983, 80, 6790–6794. [Google Scholar] [CrossRef]
- Kravanja, M.; Engelmann, R.; Dossonnet, V.; Bluggel, M.; Meyer, H.E.; Frank, R.; Galinier, A.; Deutscher, J.; Schnell, N.; Hengstenberg, W. The hprK gene of Enterococcus faecalis encodes a novel bifunctional enzyme: The HPr kinase/phosphatase. Mol. Microbiol. 1999, 31, 59–66. [Google Scholar] [CrossRef]
- Galinier, A.; Kravanja, M.; Engelmann, R.; Hengstenberg, W.; Kilhoffer, M.C.; Deutscher, J.; Haiech, J. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc. Natl. Acad. Sci. USA 1998, 95, 1823–1828. [Google Scholar] [CrossRef]
- Jones, B.E.; Dossonnet, V.; Kuster, E.; Hillen, W.; Deutscher, J.; Klevit, R.E. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J. Biol. Chem. 1997, 272, 26530–26535. [Google Scholar] [CrossRef]
- Boel, G.; Mijakovic, I.; Maze, A.; Poncet, S.; Taha, M.K.; Larribe, M.; Darbon, E.; Khemiri, A.; Galinier, A.; Deutscher, J. Transcription regulators potentially controlled by HPr kinase/phosphorylase in Gram-negative bacteria. J. Mol. Microbiol. Biotechnol. 2003, 5, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Herro, R.; Poncet, S.; Cossart, P.; Buchrieser, C.; Gouin, E.; Glaser, P.; Deutscher, J. How seryl-phosphorylated HPr inhibits PrfA, a transcription activator of Listeria monocytogenes virulence genes. J. Mol. Microbiol. Biotechnol. 2005, 9, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.S.; Steinhauer, K.; Hillen, W.; Stulke, J.; Brennan, R.G. Crystal structure of HPr kinase/phosphatase from Mycoplasma pneumoniae. J. Mol. Biol. 2003, 326, 1203–1217. [Google Scholar] [CrossRef]
- Fieulaine, S.; Morera, S.; Poncet, S.; Monedero, V.; Gueguen-Chaignon, V.; Galinier, A.; Janin, J.; Deutscher, J.; Nessler, S. X-ray structure of HPr kinase: A bacterial protein kinase with a P-loop nucleotide-binding domain. EMBO J. 2001, 20, 3917–3927. [Google Scholar] [CrossRef] [PubMed]
- Marquez, J.A.; Hasenbein, S.; Koch, B.; Fieulaine, S.; Nessler, S.; Russell, R.B.; Hengstenberg, W.; Scheffzek, K. Structure of the full-length HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution: Mimicking the product/substrate of the phospho transfer reactions. Proc. Natl. Acad. Sci. USA 2002, 99, 3458–3463. [Google Scholar] [CrossRef]
- Nessler, S. The bacterial HPr kinase/phosphorylase: A new type of Ser/Thr kinase as antimicrobial target. Biochim. Biophys. Acta 2005, 1754, 126–131. [Google Scholar] [CrossRef]
- Chaptal, V.; Vincent, F.; Gueguen-Chaignon, V.; Monedero, V.; Poncet, S.; Deutscher, J.; Nessler, S.; Morera, S. Structural analysis of the bacterial HPr kinase/phosphorylase V267F mutant gives insights into the allosteric regulation mechanism of this bifunctional enzyme. J. Biol. Chem. 2007, 282, 34952–34957. [Google Scholar] [CrossRef]
- Ramstrom, H.; Bourotte, M.; Philippe, C.; Schmitt, M.; Haiech, J.; Bourguignon, J.J. Heterocyclic bis-cations as starting hits for design of inhibitors of the bifunctional enzyme histidine-containing protein kinase/phosphatase from Bacillus subtilis. J. Med. Chem. 2004, 47, 2264–2275. [Google Scholar] [CrossRef]
- Sachdeva, P.; Narayan, A.; Misra, R.; Brahmachari, V.; Singh, Y. Loss of kinase activity in Mycobacterium tuberculosis multidomain protein Rv1364c. FEBS J. 2008, 275, 6295–6308. [Google Scholar] [CrossRef]
- Misra, R.; Menon, D.; Arora, G.; Virmani, R.; Gaur, M.; Naz, S.; Jaisinghani, N.; Bhaduri, A.; Bothra, A.; Maji, A.; et al. Tuning the Mycobacterium tuberculosis Alternative Sigma Factor SigF through the Multidomain Regulator Rv1364c and Osmosensory Kinase Protein Kinase D. J. Bacteriol. 2019, 201, e00725-18. [Google Scholar] [CrossRef]
- Greenstein, A.E.; Hammel, M.; Cavazos, A.; Alber, T. Interdomain communication in the Mycobacterium tuberculosis environmental phosphatase Rv1364c. J. Biol. Chem. 2009, 284, 29828–29835. [Google Scholar] [CrossRef] [PubMed]
- King-Scott, J.; Konarev, P.V.; Panjikar, S.; Jordanova, R.; Svergun, D.I.; Tucker, P.A. Structural characterization of the multidomain regulatory protein Rv1364c from Mycobacterium tuberculosis. Structure 2011, 19, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Hunter, T. Metabolic Kinases Moonlighting as Protein Kinases. Trends Biochem. Sci. 2018, 43, 301–310. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).