On the Study of Deubiquitinases: Using the Right Tools for the Job
Abstract
:1. Introduction
1.1. Deubiquitinase Enzymes: Classification and Activity Regulation
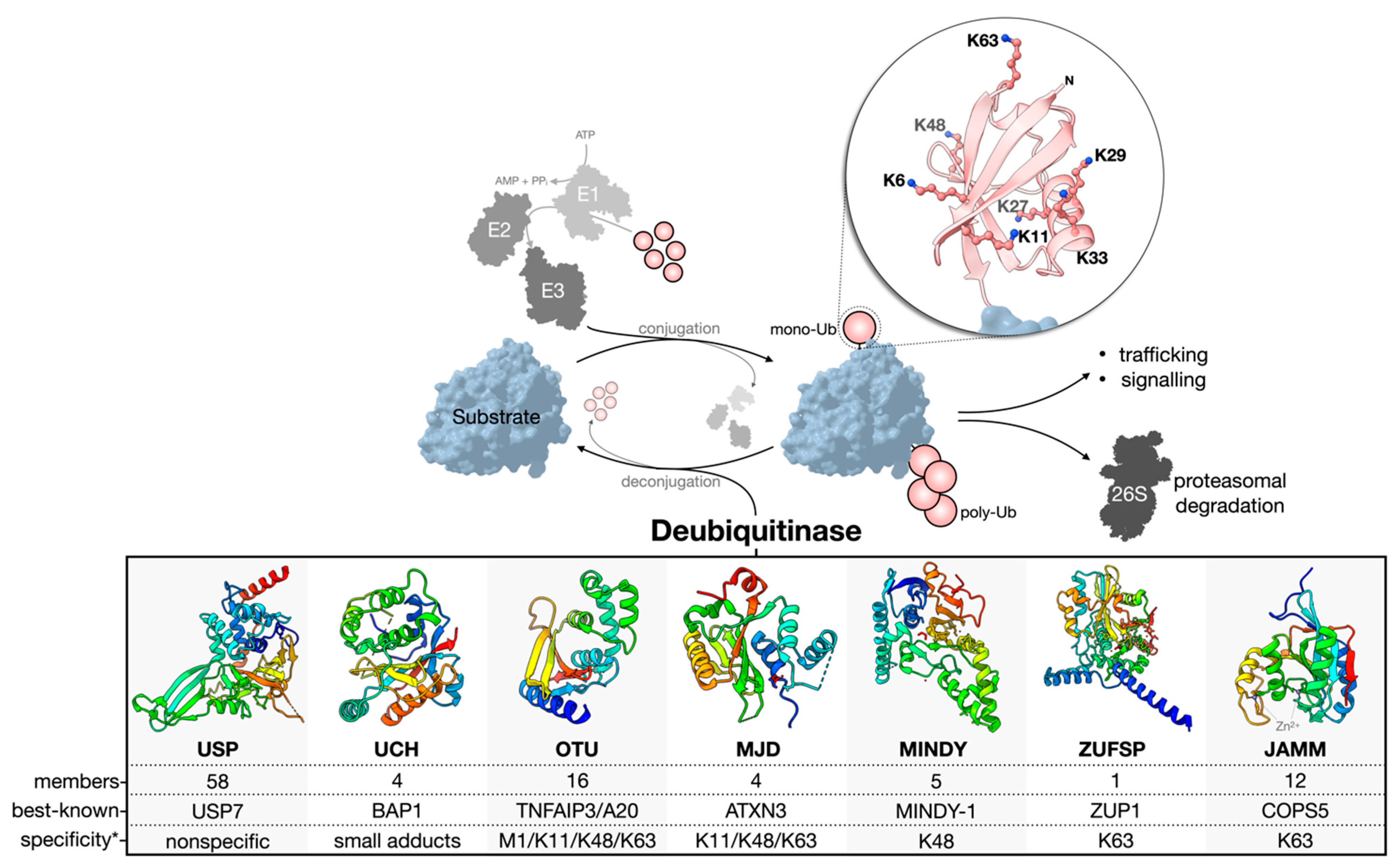
1.2. Challenges and Missteps When Studying DUBs at the Cellular Level
2. Small Molecule DUB Inhibitors
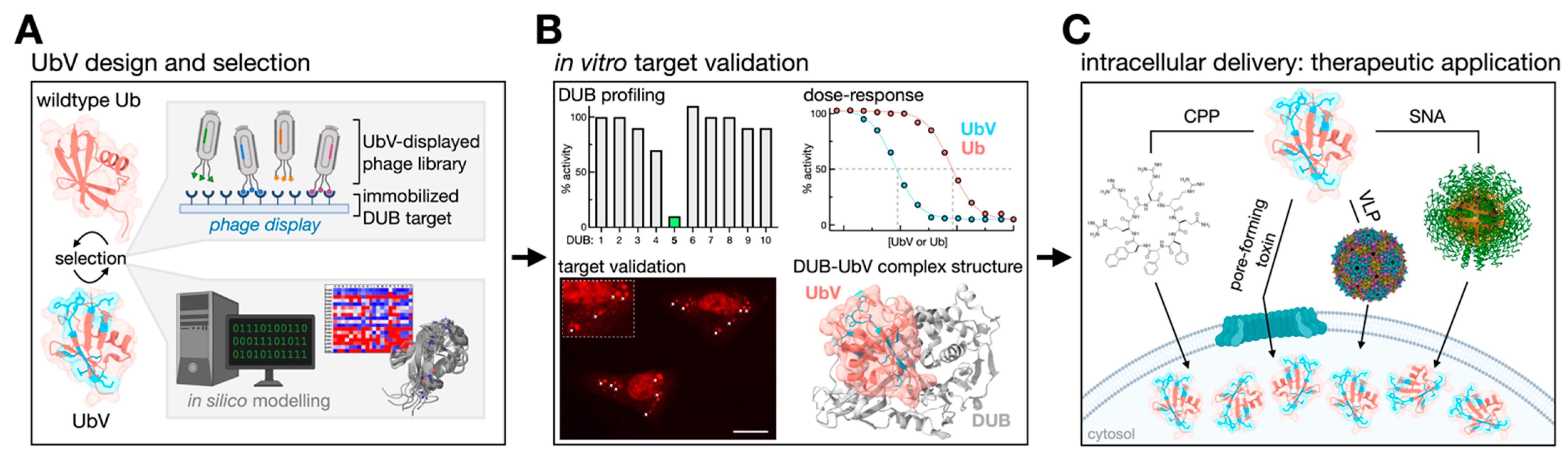
3. Ubiquitin Variants: A Combinatorial Approach to Selective DUB Targeting
3.1. Diversifying the UbV Portfolio
3.2. UbV Characterization
3.3. Intracellular Delivery of UbVs
4. Activity-Based Probes: Capturing DUBs in Action
ABP Specificity
5. Unifying Technologies to Study DUBs at the Cellular Level
5.1. DUB-Omics
5.2. Genetic and Pharmacological Modulation Reveals DUB Biology
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Varshavsky, A. The Early History of the Ubiquitin Field. Protein Sci. Publ. Protein Soc. 2006, 15, 647–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swatek, K.N.; Komander, D. Ubiquitin Modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mevissen, T.E.T.; Komander, D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef] [Green Version]
- Koyano, F.; Okatsu, K.; Kosako, H.; Tamura, Y.; Go, E.; Kimura, M.; Kimura, Y.; Tsuchiya, H.; Yoshihara, H.; Hirokawa, T.; et al. Ubiquitin Is Phosphorylated by PINK1 to Activate Parkin. Nature 2014, 510, 162–166. [Google Scholar] [CrossRef]
- Okatsu, K.; Sato, Y.; Yamano, K.; Matsuda, N.; Negishi, L.; Takahashi, A.; Yamagata, A.; Goto-Ito, S.; Mishima, M.; Ito, Y.; et al. Structural Insights into Ubiquitin Phosphorylation by PINK1. Sci. Rep. 2018, 8, 10382. [Google Scholar] [CrossRef]
- Ordureau, A.; Heo, J.-M.; Duda, D.M.; Paulo, J.A.; Olszewski, J.L.; Yanishevski, D.; Rinehart, J.; Schulman, B.A.; Harper, J.W. Defining Roles of PARKIN and Ubiquitin Phosphorylation by PINK1 in Mitochondrial Quality Control Using a Ubiquitin Replacement Strategy. Proc. Natl. Acad. Sci. USA 2015, 112, 6637–6642. [Google Scholar] [CrossRef] [Green Version]
- Wauer, T.; Swatek, K.N.; Wagstaff, J.L.; Gladkova, C.; Pruneda, J.N.; Michel, M.A.; Gersch, M.; Johnson, C.M.; Freund, S.M.; Komander, D. Ubiquitin Ser65 Phosphorylation Affects Ubiquitin Structure, Chain Assembly and Hydrolysis. EMBO J. 2015, 34, 307–325. [Google Scholar] [CrossRef]
- Hepowit, N.L.; Kolbe, C.-C.; Zelle, S.R.; Latz, E.; MacGurn, J.A. Regulation of Ubiquitin and Ubiquitin-like Modifiers by Phosphorylation. FEBS J. 2021. [Google Scholar] [CrossRef]
- Walser, F.; Mulder, M.P.C.; Bragantini, B.; Burger, S.; Gubser, T.; Gatti, M.; Botuyan, M.V.; Villa, A.; Altmeyer, M.; Neri, D.; et al. Ubiquitin Phosphorylation at Thr12 Modulates the DNA Damage Response. Mol. Cell 2020, 80, 423–436.e9. [Google Scholar] [CrossRef]
- Ohtake, F.; Saeki, Y.; Sakamoto, K.; Ohtake, K.; Nishikawa, H.; Tsuchiya, H.; Ohta, T.; Tanaka, K.; Kanno, J. Ubiquitin Acetylation Inhibits Polyubiquitin Chain Elongation. EMBO Rep. 2015, 16, 192–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez Berrocal, D.A.; Witting, K.F.; Ovaa, H.; Mulder, M.P.C. Hybrid Chains: A Collaboration of Ubiquitin and Ubiquitin-Like Modifiers Introducing Cross-Functionality to the Ubiquitin Code. Front. Chem. 2020, 7, 931. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Dixit, V.M. Drugging the Undruggables: Exploring the Ubiquitin System for Drug Development. Cell Res. 2016, 26, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Koyuncu, S.; Loureiro, R.; Lee, H.J.; Wagle, P.; Krueger, M.; Vilchez, D. Rewiring of the Ubiquitinated Proteome Determines Ageing in C. Elegans. Nature 2021, 596, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Kasherman, M.A.; Premarathne, S.; Burne, T.H.J.; Wood, S.A.; Piper, M. The Ubiquitin System: A Regulatory Hub for Intellectual Disability and Autism Spectrum Disorder. Mol. Neurobiol. 2020, 57, 2179–2193. [Google Scholar] [CrossRef]
- Clague, M.J.; Heride, C.; Urbé, S. The Demographics of the Ubiquitin System. Trends Cell Biol. 2015, 25, 417–426. [Google Scholar] [CrossRef]
- Clague, M.J.; Urbé, S.; Komander, D. Breaking the Chains: Deubiquitylating Enzyme Specificity Begets Function. Nat. Rev. Mol. Cell Biol. 2019, 20, 338–352. [Google Scholar] [CrossRef]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbé, S. Deubiquitylases from Genes to Organism. Physiol. Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef]
- Vlasschaert, C.; Cook, D.; Xia, X.; Gray, D.A. The Evolution and Functional Diversification of the Deubiquitinating Enzyme Superfamily. Genome Biol. Evol. 2017, 9, 558–573. [Google Scholar] [CrossRef] [Green Version]
- Grou, C.P.; Pinto, M.P.; Mendes, A.V.; Domingues, P.; Azevedo, J.E. The de Novo Synthesis of Ubiquitin: Identification of Deubiquitinases Acting on Ubiquitin Precursors. Sci. Rep. 2015, 5, 12836. [Google Scholar] [CrossRef] [Green Version]
- de Poot, S.A.H.; Tian, G.; Finley, D. Meddling with Fate: The Proteasomal Deubiquitinating Enzymes. J. Mol. Biol. 2017, 429, 3525–3545. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, C.P.; MacGurn, J.A. Coupling Conjugation and Deconjugation Activities to Achieve Cellular Ubiquitin Dynamics. Trends Biochem. Sci. 2020, 45, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Wertz, I.E.; O’Rourke, K.M.; Zhou, H.; Eby, M.; Aravind, L.; Seshagiri, S.; Wu, P.; Wiesmann, C.; Baker, R.; Boone, D.L.; et al. De-Ubiquitination and Ubiquitin Ligase Domains of A20 Downregulate NF-ΚB Signalling. Nature 2004, 430, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Faesen, A.C.; Luna-Vargas, M.P.A.; Geurink, P.P.; Clerici, M.; Merkx, R.; Van Dijk, W.J.; Hameed, D.S.; El Oualid, F.; Ovaa, H.; Sixma, T.K. The Differential Modulation of USP Activity by Internal Regulatory Domains, Interactors and Eight Ubiquitin Chain Types. Chem. Biol. 2011, 18, 1550–1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mevissen, T.E.T.; Hospenthal, M.K.; Geurink, P.P.; Elliott, P.R.; Akutsu, M.; Arnaudo, N.; Ekkebus, R.; Kulathu, Y.; Wauer, T.; El Oualid, F.; et al. OTU Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis. Cell 2013, 154, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Sowa, M.E.; Bennett, E.J.; Gygi, S.P.; Harper, J.W. Defining the Human Deubiquitinating Enzyme Interaction Landscape. Cell 2009, 138, 389–403. [Google Scholar] [CrossRef] [Green Version]
- Leznicki, P.; Natarajan, J.; Bader, G.; Spevak, W.; Schlattl, A.; Rehman, S.A.A.; Pathak, D.; Weidlich, S.; Zoephel, A.; Bordone, M.C.; et al. Expansion of DUB Functionality Generated by Alternative Isoforms-USP35, a Case Study. J. Cell Sci. 2018, 131, jcs212753. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and Cellular Roles of Ubiquitin-Specific Deubiquitinating Enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef] [Green Version]
- Gersch, M.; Wagstaff, J.L.; Toms, A.V.; Graves, B.; Freund, S.M.V.; Komander, D. Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity. Mol. Cell 2019, 74, 436–451.e7. [Google Scholar] [CrossRef]
- Sanchez-Bailon, M.P.; Choi, S.-Y.; Dufficy, E.R.; Sharma, K.; McNee, G.S.; Gunnell, E.; Chiang, K.; Sahay, D.; Maslen, S.; Stewart, G.S.; et al. Arginine Methylation and Ubiquitylation Crosstalk Controls DNA End-Resection and Homologous Recombination Repair. Nat. Commun. 2021, 12, 6313. [Google Scholar] [CrossRef]
- Walden, M.; Masandi, S.K.; Pawłowski, K.; Zeqiraj, E. Pseudo-DUBs as Allosteric Activators and Molecular Scaffolds of Protein Complexes. Biochem. Soc. Trans. 2018, 46, 453–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, T.; Cohen, R.E. A Cryptic Protease Couples Deubiquitination and Degradation by the Proteasome. Nature 2002, 419, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Pathare, G.R.; Nagy, I.; Śledź, P.; Anderson, D.J.; Zhou, H.-J.; Pardon, E.; Steyaert, J.; Förster, F.; Bracher, A.; Baumeister, W. Crystal Structure of the Proteasomal Deubiquitylation Module Rpn8-Rpn11. Proc. Natl. Acad. Sci. USA 2014, 111, 2984–2989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worden, E.J.; Padovani, C.; Martin, A. Structure of the Rpn11–Rpn8 Dimer Reveals Mechanisms of Substrate Deubiquitination during Proteasomal Degradation. Nat. Struct. Mol. Biol. 2014, 21, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Ronau, J.A.; Beckmann, J.F.; Hochstrasser, M. Substrate Specificity of the Ubiquitin and Ubl Proteases. Cell Res. 2016, 26, 441–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the Chains: Structure and Function of the Deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Ritorto, M.S.; Ewan, R.; Perez-Oliva, A.B.; Knebel, A.; Buhrlage, S.J.; Wightman, M.; Kelly, S.M.; Wood, N.T.; Virdee, S.; Gray, N.S.; et al. Screening of DUB Activity and Specificity by MALDI-TOF Mass Spectrometry. Nat. Commun. 2014, 5, 4763. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, H.; Yamanaka, S.; Kuwada, S.; Higaki, K.; Kido, K.; Sato, Y.; Fukai, S.; Tokunaga, F.; Sawasaki, T. A Human DUB Protein Array for Clarification of Linkage Specificity of Polyubiquitin Chain and Application to Evaluation of Its Inhibitors. Biomedicines 2020, 8, 152. [Google Scholar] [CrossRef]
- Paudel, P.; Zhang, Q.; Leung, C.; Greenberg, H.C.; Guo, Y.; Chern, Y.-H.; Dong, A.; Li, Y.; Vedadi, M.; Zhuang, Z.; et al. Crystal Structure and Activity-Based Labeling Reveal the Mechanisms for Linkage-Specific Substrate Recognition by Deubiquitinase USP9X. Proc. Natl. Acad. Sci. USA 2019, 116, 7288–7297. [Google Scholar] [CrossRef] [Green Version]
- Kategaya, L.; Di Lello, P.; Rougé, L.; Pastor, R.; Clark, K.R.; Drummond, J.; Kleinheinz, T.; Lin, E.; Upton, J.-P.; Prakash, S.; et al. USP7 Small-Molecule Inhibitors Interfere with Ubiquitin Binding. Nature 2017, 550, 534–538. [Google Scholar] [CrossRef] [Green Version]
- Abdul Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.-Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwasna, D.; Abdul Rehman, S.A.; Natarajan, J.; Matthews, S.; Madden, R.; De Cesare, V.; Weidlich, S.; Virdee, S.; Ahel, I.; Gibbs-Seymour, I.; et al. Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability. Mol. Cell 2018, 70, 150–164.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komander, D.; Reyes-Turcu, F.; Licchesi, J.D.F.; Odenwaelder, P.; Wilkinson, K.D.; Barford, D. Molecular Discrimination of Structurally Equivalent Lys 63-Linked and Linear Polyubiquitin Chains. EMBO Rep. 2009, 10, 466–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCullough, J.; Row, P.E.; Lorenzo, Ó.; Doherty, M.; Beynon, R.; Clague, M.J.; Urbé, S. Activation of the Endosome-Associated Ubiquitin Isopeptidase AMSH by STAM, a Component of the Multivesicular Body-Sorting Machinery. Curr. Biol. 2006, 16, 160–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermanns, T.; Pichlo, C.; Baumann, U.; Hofmann, K. A Structural Basis for the Diverse Linkage Specificities within the ZUFSP Deubiquitinase Family. Nat. Commun. 2022, 13, 401. [Google Scholar] [CrossRef] [PubMed]
- Abdul Rehman, S.A.; Armstrong, L.A.; Lange, S.M.; Kristariyanto, Y.A.; Gräwert, T.W.; Knebel, A.; Svergun, D.I.; Kulathu, Y. Mechanism of Activation and Regulation of Deubiquitinase Activity in MINDY1 and MINDY2. Mol. Cell 2021, 81, 4176–4190.e6. [Google Scholar] [CrossRef] [PubMed]
- Misaghi, S.; Galardy, P.J.; Meester, W.J.N.; Ovaa, H.; Ploegh, H.L.; Gaudet, R. Structure of the Ubiquitin Hydrolase UCH-L3 Complexed with a Suicide Substrate. J. Biol. Chem. 2005, 280, 1512–1520. [Google Scholar] [CrossRef] [Green Version]
- Molland, K.; Zhou, Q.; Mesecar, A.D. A 2.2 Å Resolution Structure of the USP7 Catalytic Domain in a New Space Group Elaborates upon Structural Rearrangements Resulting from Ubiquitin Binding. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 283–287. [Google Scholar] [CrossRef] [Green Version]
- Crystal Structure of a Deubiquitinating Enzyme (Human UCH-L3) at 1.8 å Resolution. EMBO J. 1997, 16, 3787–3796. [CrossRef]
- Grasty, K.C.; Weeks, S.D.; Loll, P.J. Structural Insights into the Activity and Regulation of Human Josephin-2. J. Struct. Biol. X 2019, 3, 100011. [Google Scholar] [CrossRef]
- Hermanns, T.; Pichlo, C.; Woiwode, I.; Klopffleisch, K.; Witting, K.F.; Ovaa, H.; Baumann, U.; Hofmann, K. A Family of Unconventional Deubiquitinases with Modular Chain Specificity Determinants. Nat. Commun. 2018, 9, 799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Liu, Q.; Mallette, E.; Caba, C.; Hou, F.; Fux, J.; LaPlante, G.; Dong, A.; Zhang, Q.; Zheng, H.; et al. Structural and Functional Characterization of Ubiquitin Variant Inhibitors for the JAMM-Family Deubiquitinases STAMBP and STAMBPL1. J. Biol. Chem. 2021, 297, 101107. [Google Scholar] [CrossRef] [PubMed]
- Snyder, N.A.; Silva, G.M. Deubiquitinating Enzymes (DUBs): Regulation, Homeostasis, and Oxidative Stress Response. J. Biol. Chem. 2021, 297, 101077. [Google Scholar] [CrossRef] [PubMed]
- Amer-Sarsour, F.; Kordonsky, A.; Berdichevsky, Y.; Prag, G.; Ashkenazi, A. Deubiquitylating Enzymes in Neuronal Health and Disease. Cell Death Dis. 2021, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Cai, W.; Ai, D.; Zhang, X.; Yao, L. The Role of Deubiquitinases in Vascular Diseases. J. Cardiovasc. Transl. Res. 2020, 13, 131–141. [Google Scholar] [CrossRef]
- Basar, M.A.; Beck, D.B.; Werner, A. Deubiquitylases in Developmental Ubiquitin Signaling and Congenital Diseases. Cell Death Differ. 2021, 28, 538–556. [Google Scholar] [CrossRef]
- Parihar, N.; Bhatt, L.K. Deubiquitylating Enzymes: Potential Target in Autoimmune Diseases. Inflammopharmacology 2021, 29, 1683–1699. [Google Scholar] [CrossRef]
- Cruz, L.; Soares, P.; Correia, M. Ubiquitin-Specific Proteases: Players in Cancer Cellular Processes. Pharmaceuticals 2021, 14, 848. [Google Scholar] [CrossRef]
- Kim, S.-H.; Baek, K.-H. Regulation of Cancer Metabolism by Deubiquitinating Enzymes: The Warburg Effect. Int. J. Mol. Sci. 2021, 22, 6173. [Google Scholar] [CrossRef]
- Errington, T.M.; Mathur, M.; Soderberg, C.K.; Denis, A.; Perfito, N.; Iorns, E.; Nosek, B.A. Investigating the Replicability of Preclinical Cancer Biology. eLife 2021, 10, e71601. [Google Scholar] [CrossRef]
- Errington, T.M.; Denis, A.; Perfito, N.; Iorns, E.; Nosek, B.A. Challenges for Assessing Replicability in Preclinical Cancer Biology. eLife 2021, 10, e67995. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, M.; Jolly, L.A.; Gecz, J.; Wood, S.A. La FAM Fatale: USP9X in Development and Disease. Cell. Mol. Life Sci. CMLS 2015, 72, 2075–2089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouchantaf, R.; Azakir, B.A.; McPherson, P.S.; Millard, S.M.; Wood, S.A.; Angers, A. The Ubiquitin Ligase Itch Is Auto-Ubiquitylated in Vivo and in Vitro but Is Protected from Degradation by Interacting with the Deubiquitylating Enzyme FAM/USP9X. J. Biol. Chem. 2006, 281, 38738–38747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, O.M.; Carvalho, J.; Spencer-Dene, B.; Mitter, R.; Frith, D.; Snijders, A.P.; Wood, S.A.; Behrens, A. The Deubiquitinase USP9X Regulates FBW7 Stability and Suppresses Colorectal Cancer. J. Clin. Investig. 2018, 128, 1326–1337. [Google Scholar] [CrossRef] [Green Version]
- Donato, N.J.; Talpaz, M.; Peterson, L.; Young, M.; Showalter, H.D.; Wobus, C.; O’Riordan, M.X.D.; Ermann, M. Deubiquitinase Inhibitors and Methods for Use of the Same. WO2015054555A1, 16 April 2015. [Google Scholar]
- Akiyama, H.; Umezawa, Y.; Watanabe, D.; Okada, K.; Ishida, S.; Nogami, A.; Miura, O. Inhibition of USP9X Downregulates JAK2-V617F and Induces Apoptosis Synergistically with BH3 Mimetics Preferentially in Ruxolitinib-Persistent JAK2-V617F-Positive Leukemic Cells. Cancers 2020, 12, 406. [Google Scholar] [CrossRef]
- Potu, H.; Kandarpa, M.; Peterson, L.F.; Durham, A.; Donato, N.J.; Talpaz, M. Downregulation of SOX2 by Inhibition of Usp9X Induces Apoptosis in Melanoma. Oncotarget 2021, 12, 160–172. [Google Scholar] [CrossRef]
- Jaiswal, A.; Murakami, K.; Elia, A.; Shibahara, Y.; Done, S.J.; Wood, S.A.; Donato, N.J.; Ohashi, P.S.; Reedijk, M. Therapeutic Inhibition of USP9x-Mediated Notch Signaling in Triple-Negative Breast Cancer. Proc. Natl. Acad. Sci. USA 2021, 118, e2101592118. [Google Scholar] [CrossRef]
- Zhang, Z.; Cui, Z.; Xie, Z.; Li, C.; Xu, C.; Guo, X.; Yu, J.; Chen, T.; Facchinetti, F.; Bohnenberger, H.; et al. Deubiquitinase USP5 Promotes Non-Small Cell Lung Cancer Cell Proliferation by Stabilizing Cyclin D1. Transl. Lung Cancer Res. 2021, 10, 3995–4011. [Google Scholar] [CrossRef]
- Potu, H.; Kandarpa, M.; Peterson, L.F.; Donato, N.J.; Talpaz, M. Tumor Necrosis Factor Related Apoptosis Inducing Ligand (TRAIL) Regulates Deubiquitinase USP5 in Tumor Cells. Oncotarget 2019, 10, 5745–5754. [Google Scholar] [CrossRef]
- Peterson, L.F.; Sun, H.; Liu, Y.; Potu, H.; Kandarpa, M.; Ermann, M.; Courtney, S.M.; Young, M.; Showalter, H.D.; Sun, D.; et al. Targeting Deubiquitinase Activity with a Novel Small-Molecule Inhibitor as Therapy for B-Cell Malignancies. Blood 2015, 125, 3588–3597. [Google Scholar] [CrossRef]
- Clancy, A.; Heride, C.; Pinto-Fernández, A.; Elcocks, H.; Kallinos, A.; Kayser-Bricker, K.J.; Wang, W.; Smith, V.; Davis, S.; Fessler, S.; et al. The Deubiquitylase USP9X Controls Ribosomal Stalling. J. Cell Biol. 2021, 220, e202004211. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xia, H.; Kim, M.; Xu, L.; Li, Y.; Zhang, L.; Cai, Y.; Norberg, H.V.; Zhang, T.; Furuya, T.; et al. Beclin1 Controls the Levels of P53 by Regulating the Deubiquitination Activity of USP10 and USP13. Cell 2011, 147, 223–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Sartori, M.A.; Makhnevych, T.; Federowicz, K.E.; Dong, X.; Liu, L.; Nim, S.; Dong, A.; Yang, J.; Li, Y.; et al. Generation and Validation of Intracellular Ubiquitin Variant Inhibitors for USP7 and USP10. J. Mol. Biol. 2017, 429, 3546–3560. [Google Scholar] [CrossRef] [PubMed]
- Bednash, J.S.; Weathington, N.; Londino, J.; Rojas, M.; Gulick, D.L.; Fort, R.; Han, S.; McKelvey, A.C.; Chen, B.B.; Mallampalli, R.K. Targeting the Deubiquitinase STAMBP Inhibits NALP7 Inflammasome Activity. Nat. Commun. 2017, 8, 15203. [Google Scholar] [CrossRef] [Green Version]
- Mulder, M.P.C.; Witting, K.F.; Ovaa, H. Cracking the Ubiquitin Code: The Ubiquitin Toolbox. Curr. Issues Mol. Biol. 2020, 37, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Lange, S.M.; Armstrong, L.A.; Kulathu, Y. Deubiquitinases: From Mechanisms to Their Inhibition by Small Molecules. Mol. Cell 2021, 82, 15–29. [Google Scholar] [CrossRef]
- Pruneda, J.N.; Komander, D. Evaluating Enzyme Activities and Structures of DUBs. Methods Enzymol. 2019, 618, 321–341. [Google Scholar] [CrossRef]
- Lundgren, S.; Albertella, M.; Belda, O.; Derbyshire, D.; Henderson, I.; Lindberg, J.; Odrzywol, E.; Stoor, C.; Strömberg, K.; Unnerståle, S. Comprehensive Profiling of DUB Inhibitors Using the Medivir DUB Platform; Discovery on Target: Boston, MA, USA, 2016. [Google Scholar]
- Dexheimer, T.S.; Rosenthal, A.S.; Liang, Q.; Chen, J.; Villamil, M.A.; Kerns, E.H.; Simeonov, A.; Jadhav, A.; Zhuang, Z.; Maloney, D.J. Discovery of ML323 as a Novel Inhibitor of the USP1/UAF1 Deubiquitinase Complex. In Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information: Bethesda, MD, USA, 2010. [Google Scholar]
- Liang, Q.; Dexheimer, T.S.; Zhang, P.; Rosenthal, A.S.; Villamil, M.A.; You, C.; Zhang, Q.; Chen, J.; Ott, C.A.; Sun, H.; et al. A Selective USP1-UAF1 Inhibitor Links Deubiquitination to DNA Damage Responses. Nat. Chem. Biol. 2014, 10, 298–304. [Google Scholar] [CrossRef] [Green Version]
- Arrowsmith, C.H.; Audia, J.E.; Austin, C.; Baell, J.; Bennett, J.; Blagg, J.; Bountra, C.; Brennan, P.E.; Brown, P.J.; Bunnage, M.E.; et al. The Promise and Peril of Chemical Probes. Nat. Chem. Biol. 2015, 11, 541. [Google Scholar] [CrossRef] [Green Version]
- Pozhidaeva, A.; Bezsonova, I. USP7: Structure, Substrate Specificity, and Inhibition. DNA Repair 2019, 76, 30–39. [Google Scholar] [CrossRef]
- Wertz, I.E.; Murray, J.M. Structurally-Defined Deubiquitinase Inhibitors Provide Opportunities to Investigate Disease Mechanisms. Drug Discov. Today Technol. 2019, 31, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Varca, A.C.; Casalena, D.; Chan, W.C.; Hu, B.; Magin, R.S.; Roberts, R.M.; Liu, X.; Zhu, H.; Seo, H.-S.; Dhe-Paganon, S.; et al. Identification and Validation of Selective Deubiquitinase Inhibitors. Cell Chem. Biol. 2021, 28, 1758–1771.e13. [Google Scholar] [CrossRef] [PubMed]
- Schauer, N.J.; Magin, R.S.; Liu, X.; Doherty, L.M.; Buhrlage, S.J. Advances in Discovering Deubiquitinating Enzyme (DUB) Inhibitors. J. Med. Chem. 2020, 63, 2731–2750. [Google Scholar] [CrossRef]
- Liu, L.; Damerell, D.R.; Koukouflis, L.; Tong, Y.; Marsden, B.D.; Schapira, M. UbiHub: A Data Hub for the Explorers of Ubiquitination Pathways. Bioinformatics 2019, 35, 2882–2884. [Google Scholar] [CrossRef] [PubMed]
- Rusilowicz-Jones, E.V.; Jardine, J.; Kallinos, A.; Pinto-Fernandez, A.; Guenther, F.; Giurrandino, M.; Barone, F.G.; McCarron, K.; Burke, C.J.; Murad, A.; et al. USP30 Sets a Trigger Threshold for PINK1–PARKIN Amplification of Mitochondrial Ubiquitylation. Life Sci. Alliance 2020, 3. [Google Scholar] [CrossRef] [PubMed]
- Panyain, N.; Godinat, A.; Thawani, A.R.; Lachiondo-Ortega, S.; Mason, K.; Elkhalifa, S.; Smith, L.M.; Harrigan, J.A.; Tate, E.W. Activity-Based Protein Profiling Reveals Deubiquitinase and Aldehyde Dehydrogenase Targets of a Cyanopyrrolidine Probe. RSC Med. Chem. 2021, 12, 1935–1943. [Google Scholar] [CrossRef]
- Panyain, N.; Godinat, A.; Lanyon-Hogg, T.; Lachiondo-Ortega, S.; Will, E.J.; Soudy, C.; Mondal, M.; Mason, K.; Elkhalifa, S.; Smith, L.M.; et al. Discovery of a Potent and Selective Covalent Inhibitor and Activity-Based Probe for the Deubiquitylating Enzyme UCHL1, with Antifibrotic Activity. J. Am. Chem. Soc. 2020, 142, 12020–12026. [Google Scholar] [CrossRef]
- Blagg, J.; Workman, P. Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology. Cancer Cell 2017, 32, 9–25. [Google Scholar] [CrossRef] [Green Version]
- Schlierf, A.; Altmann, E.; Quancard, J.; Jefferson, A.B.; Assenberg, R.; Renatus, M.; Jones, M.; Hassiepen, U.; Schaefer, M.; Kiffe, M.; et al. Targeted Inhibition of the COP9 Signalosome for Treatment of Cancer. Nat. Commun. 2016, 7, 13166. [Google Scholar] [CrossRef]
- Altmann, E.; Erbel, P.; Renatus, M.; Schaefer, M.; Schlierf, A.; Druet, A.; Kieffer, L.; Sorge, M.; Pfister, K.; Hassiepen, U.; et al. Azaindoles as Zinc-Binding Small-Molecule Inhibitors of the JAMM Protease CSN5. Angew. Chem. Int. Ed. Engl. 2017, 56, 1294–1297. [Google Scholar] [CrossRef]
- Turnbull, A.P.; Ioannidis, S.; Krajewski, W.W.; Pinto-Fernandez, A.; Heride, C.; Martin, A.C.L.; Tonkin, L.M.; Townsend, E.C.; Buker, S.M.; Lancia, D.R.; et al. Molecular Basis of USP7 Inhibition by Selective Small-Molecule Inhibitors. Nature 2017, 550, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Gavory, G.; O’Dowd, C.R.; Helm, M.D.; Flasz, J.; Arkoudis, E.; Dossang, A.; Hughes, C.; Cassidy, E.; McClelland, K.; Odrzywol, E.; et al. Discovery and Characterization of Highly Potent and Selective Allosteric USP7 Inhibitors. Nat. Chem. Biol. 2018, 14, 118–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Dowd, C.R.; Helm, M.D.; Rountree, J.S.S.; Flasz, J.T.; Arkoudis, E.; Miel, H.; Hewitt, P.R.; Jordan, L.; Barker, O.; Hughes, C.; et al. Identification and Structure-Guided Development of Pyrimidinone Based USP7 Inhibitors. ACS Med. Chem. Lett. 2018, 9, 238–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schauer, N.J.; Liu, X.; Magin, R.S.; Doherty, L.M.; Chan, W.C.; Ficarro, S.B.; Hu, W.; Roberts, R.M.; Iacob, R.E.; Stolte, B.; et al. Selective USP7 Inhibition Elicits Cancer Cell Killing through a P53-Dependent Mechanism. Sci. Rep. 2020, 10, 5324. [Google Scholar] [CrossRef] [Green Version]
- Lamberto, I.; Liu, X.; Seo, H.-S.; Schauer, N.J.; Iacob, R.E.; Hu, W.; Das, D.; Mikhailova, T.; Weisberg, E.L.; Engen, J.R.; et al. Structure-Guided Development of a Potent and Selective Non-Covalent Active-Site Inhibitor of USP7. Cell Chem. Biol. 2017, 24, 1490–1500.e11. [Google Scholar] [CrossRef] [Green Version]
- Kluge, A.F.; Lagu, B.R.; Maiti, P.; Jaleel, M.; Webb, M.; Malhotra, J.; Mallat, A.; Srinivas, P.A.; Thompson, J.E. Novel Highly Selective Inhibitors of Ubiquitin Specific Protease 30 (USP30) Accelerate Mitophagy. Bioorg. Med. Chem. Lett. 2018, 28, 2655–2659. [Google Scholar] [CrossRef]
- Colland, F.; Formstecher, E.; Jacq, X.; Reverdy, C.; Planquette, C.; Conrath, S.; Trouplin, V.; Bianchi, J.; Aushev, V.N.; Camonis, J.; et al. Small-Molecule Inhibitor of USP7/HAUSP Ubiquitin Protease Stabilizes and Activates P53 in Cells. Mol. Cancer Ther. 2009, 8, 2286–2295. [Google Scholar] [CrossRef] [Green Version]
- Harrigan, J.A.; Jacq, X.; Martin, N.M.; Jackson, S.P. Deubiquitylating Enzymes and Drug Discovery: Emerging Opportunities. Nat. Rev. Drug Discov. 2018, 17, 57–77. [Google Scholar] [CrossRef]
- Mevissen, T.E.T.; Kulathu, Y.; Mulder, M.P.C.; Geurink, P.P.; Maslen, S.L.; Gersch, M.; Elliott, P.R.; Burke, J.E.; van Tol, B.D.M.; Akutsu, M.; et al. Molecular Basis of Lys11-Polyubiquitin Specificity in the Deubiquitinase Cezanne. Nature 2016, 538, 402–405. [Google Scholar] [CrossRef]
- Ernst, A.; Avvakumov, G.; Tong, J.; Fan, Y.; Zhao, Y.; Alberts, P.; Persaud, A.; Walker, J.R.; Neculai, A.M.; Neculai, D.; et al. A Strategy for Modulation of Enzymes in the Ubiquitin System. Science 2013, 339, 590–595. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Sidhu, S.S. Generating Intracellular Modulators of E3 Ligases and Deubiquitinases from Phage-Displayed Ubiquitin Variant Libraries. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2018; Volume 1844, pp. 101–119. [Google Scholar]
- Leung, I.; Dekel, A.; Shifman, J.M.; Sidhu, S.S. Saturation Scanning of Ubiquitin Variants Reveals a Common Hot Spot for Binding to USP2 and USP21. Proc. Natl. Acad. Sci. USA 2016, 113, 8705–8710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roscoe, B.P.; Thayer, K.M.; Zeldovich, K.B.; Fushman, D.; Bolon, D.N.A. Analyses of the Effects of All Ubiquitin Point Mutants on Yeast Growth Rate. J. Mol. Biol. 2013, 425, 1363–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tencer, A.H.; Liang, Q.; Zhuang, Z. Divergence in Ubiquitin Interaction and Catalysis among the Ubiquitin-Specific Protease Family Deubiquitinating Enzymes. Biochemistry 2016, 55, 4708–4719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurley, J.H.; Lee, S.; Prag, G. Ubiquitin-Binding Domains. Biochem. J. 2006, 399, 361–372. [Google Scholar] [CrossRef]
- Renatus, M.; Parrado, S.G.; D’Arcy, A.; Eidhoff, U.; Gerhartz, B.; Hassiepen, U.; Pierrat, B.; Riedl, R.; Vinzenz, D.; Worpenberg, S.; et al. Structural Basis of Ubiquitin Recognition by the Deubiquitinating Protease USP2. Struct. Lond. Engl. 1993 2006, 14, 1293–1302. [Google Scholar] [CrossRef] [Green Version]
- Garg, P.; Ceccarelli, D.F.; Keszei, A.F.A.; Kurinov, I.; Sicheri, F.; Sidhu, S.S. Structural and Functional Analysis of Ubiquitin-Based Inhibitors That Target the Backsides of E2 Enzymes. J. Mol. Biol. 2020, 432, 952–966. [Google Scholar] [CrossRef]
- Middleton, A.J.; Teyra, J.; Zhu, J.; Sidhu, S.S.; Day, C.L. Identification of Ubiquitin Variants That Inhibit the E2 Ubiquitin Conjugating Enzyme, Ube2k. ACS Chem. Biol. 2021, 16, 1745–1756. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, K.P.; Sartori, M.A.; Kamadurai, H.B.; Ordureau, A.; Jiang, C.; Mercredi, P.Y.; Murchie, R.; Hu, J.; Persaud, A.; et al. System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes. Mol. Cell 2016, 62, 121–136. [Google Scholar] [CrossRef] [Green Version]
- Gabrielsen, M.; Buetow, L.; Nakasone, M.A.; Ahmed, S.F.; Sibbet, G.J.; Smith, B.O.; Zhang, W.; Sidhu, S.S.; Huang, D.T. A General Strategy for Discovery of Inhibitors and Activators of RING and U-Box E3 Ligases with Ubiquitin Variants. Mol. Cell 2017, 68, 456–470.e10. [Google Scholar] [CrossRef] [Green Version]
- Watson, E.R.; Grace, C.R.R.; Zhang, W.; Miller, D.J.; Davidson, I.F.; Prabu, J.R.; Yu, S.; Bolhuis, D.L.; Kulko, E.T.; Vollrath, R.; et al. Protein Engineering of a Ubiquitin-Variant Inhibitor of APC/C Identifies a Cryptic K48 Ubiquitin Chain Binding Site. Proc. Natl. Acad. Sci. USA 2019, 116, 17280–17289. [Google Scholar] [CrossRef] [Green Version]
- Gorelik, M.; Orlicky, S.; Sartori, M.A.; Tang, X.; Marcon, E.; Kurinov, I.; Greenblatt, J.F.; Tyers, M.; Moffat, J.; Sicheri, F.; et al. Inhibition of SCF Ubiquitin Ligases by Engineered Ubiquitin Variants That Target the Cul1 Binding Site on the Skp1–F-Box Interface. Proc. Natl. Acad. Sci. USA 2016, 113, 3527–3532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeBlanc, N.; Mallette, E.; Zhang, W. Targeted Modulation of E3 Ligases Using Engineered Ubiquitin Variants. FEBS J. 2021, 288, 2143–2165. [Google Scholar] [CrossRef] [PubMed]
- Veggiani, G.; Yates, B.P.; Martyn, G.D.; Manczyk, N.; Singer, A.U.; Kurinov, I.; Sicheri, F.; Sidhu, S.S. Panel of Engineered Ubiquitin Variants Targeting the Family of Human Ubiquitin Interacting Motifs. ACS Chem. Biol. 2022, 17, 941–956. [Google Scholar] [CrossRef]
- Manczyk, N.; Yates, B.P.; Veggiani, G.; Ernst, A.; Sicheri, F.; Sidhu, S.S. Structural and Functional Characterization of a Ubiquitin Variant Engineered for Tight and Specific Binding to an Alpha-helical Ubiquitin Interacting Motif. Protein Sci. Publ. Protein Soc. 2017, 26, 1060–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manczyk, N.; Veggiani, G.; Gish, G.D.; Yates, B.P.; Ernst, A.; Sidhu, S.S.; Sicheri, F. Dimerization of a Ubiquitin Variant Leads to High Affinity Interactions with a Ubiquitin Interacting Motif. Protein Sci. 2019, 28, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Teyra, J.; Singer, A.U.; Schmitges, F.W.; Jaynes, P.; Kit Leng Lui, S.; Polyak, M.J.; Fodil, N.; Krieger, J.R.; Tong, J.; Schwerdtfeger, C.; et al. Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15. Structure 2019, 27, 590–605.e5. [Google Scholar] [CrossRef] [Green Version]
- Manczyk, N.; Veggiani, G.; Teyra, J.; Strilchuk, A.W.; Sidhu, S.S.; Sicheri, F. The Ubiquitin Interacting Motifs of USP37 Act on the Proximal Ub of a Di-Ub Chain to Enhance Catalytic Efficiency. Sci. Rep. 2019, 9, 4119. [Google Scholar] [CrossRef]
- Gjonaj, L.; Sapmaz, A.; Flierman, D.; Janssen, G.M.C.; van Veelen, P.A.; Ovaa, H. Development of a DUB-Selective Fluorogenic Substrate. Chem. Sci. 2019, 10, 10290–10296. [Google Scholar] [CrossRef]
- Hewitt, C.S.; Krabill, A.D.; Das, C.; Flaherty, D.P. Development of Ubiquitin Variants with Selectivity for Ubiquitin C-Terminal Hydrolase Deubiquitinases. Biochemistry 2020, 59, 3447–3462. [Google Scholar] [CrossRef]
- Hewitt, C.S.; Das, C.; Flaherty, D.P. Rational Development and Characterization of a Ubiquitin Variant with Selectivity for Ubiquitin C-Terminal Hydrolase L3. Biomolecules 2022, 12, 62. [Google Scholar] [CrossRef]
- Zhang, W.; Bailey-Elkin, B.A.; Knaap, R.C.M.; Khare, B.; Dalebout, T.J.; Johnson, G.G.; van Kasteren, P.B.; McLeish, N.J.; Gu, J.; He, W.; et al. Potent and Selective Inhibition of Pathogenic Viruses by Engineered Ubiquitin Variants. PLoS Pathog. 2017, 13, e1006372. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Burroughs, A.M.; Aravind, L. The Prokaryotic Antecedents of the Ubiquitin-Signaling System and the Early Evolution of Ubiquitin-like Beta-Grasp Domains. Genome Biol. 2006, 7, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.Q.; Veggiani, G.; Singer, A.; Teyra, J.; Chung, J.; Sidhu, S.S. A Panel of Engineered Ubiquitin Variants Targeting the Family of Domains Found in Ubiquitin Specific Proteases (DUSPs). J. Mol. Biol. 2021, 433, 167300. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, L.; Rouge, L.; Phillips, A.H.; Lam, C.; Liu, P.; Sandoval, W.; Helgason, E.; Murray, J.M.; Wertz, I.E.; et al. Conformational Stabilization of Ubiquitin Yields Potent and Selective Inhibitors of USP7. Nat. Chem. Biol. 2013, 9, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.H.; Zhang, Y.; Cunningham, C.N.; Zhou, L.; Forrest, W.F.; Liu, P.S.; Steffek, M.; Lee, J.; Tam, C.; Helgason, E.; et al. Conformational Dynamics Control Ubiquitin-Deubiquitinase Interactions and Influence in Vivo Signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 11379–11384. [Google Scholar] [CrossRef] [Green Version]
- Faesen, A.C.; Dirac, A.M.G.; Shanmugham, A.; Ovaa, H.; Perrakis, A.; Sixma, T.K. Mechanism of USP7/HAUSP Activation by Its C-Terminal Ubiquitin-like Domain and Allosteric Regulation by GMP-Synthetase. Mol. Cell 2011, 44, 147–159. [Google Scholar] [CrossRef] [Green Version]
- Fottner, M.; Weyh, M.; Gaussmann, S.; Schwarz, D.; Sattler, M.; Lang, K. A Modular Toolbox to Generate Complex Polymeric Ubiquitin Architectures Using Orthogonal Sortase Enzymes. Nat. Commun. 2021, 12, 6515. [Google Scholar] [CrossRef]
- Huppelschoten, Y.; van der Heden van Noort, G.J. State of the Art in (Semi-)Synthesis of Ubiquitin- and Ubiquitin-like Tools. Semin. Cell Dev. Biol. 2021, 21. [Google Scholar] [CrossRef]
- Conole, D.; Mondal, M.; Majmudar, J.D.; Tate, E.W. Recent Developments in Cell Permeable Deubiquitinating Enzyme Activity-Based Probes. Front. Chem. 2019, 7, 876. [Google Scholar] [CrossRef] [Green Version]
- Bruno, B.J.; Miller, G.D.; Lim, C.S. Basics and Recent Advances in Peptide and Protein Drug Delivery. Ther. Deliv. 2013, 4, 1443–1467. [Google Scholar] [CrossRef] [Green Version]
- Fu, A.; Tang, R.; Hardie, J.; Farkas, M.E.; Rotello, V.M. Promises and Pitfalls of Intracellular Delivery of Proteins. Bioconjug. Chem. 2014, 25, 1602–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gui, W.; Ott, C.A.; Yang, K.; Chung, J.S.; Shen, S.; Zhuang, Z. Cell-Permeable Activity-Based Ubiquitin Probes Enable Intracellular Profiling of Human Deubiquitinases. J. Am. Chem. Soc. 2018, 140, 12424–12433. [Google Scholar] [CrossRef] [PubMed]
- Claessen, J.H.L.; Witte, M.D.; Yoder, N.C.; Zhu, A.Y.; Spooner, E.; Ploegh, H.L. Catch-and-Release Probes Applied to Semi-Intact Cells Reveal Ubiquitin-Specific Protease Expression in Chlamydia Trachomatis Infection. ChemBioChem 2013, 14, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaczmarczyk, S.J.; Sitaraman, K.; Young, H.A.; Hughes, S.H.; Chatterjee, D.K. Protein Delivery Using Engineered Virus-like Particles. Proc. Natl. Acad. Sci. USA 2011, 108, 16998–17003. [Google Scholar] [CrossRef] [Green Version]
- Cronin, J.; Zhang, X.-Y.; Reiser, J. Altering the Tropism of Lentiviral Vectors through Pseudotyping. Curr. Gene Ther. 2005, 5, 387–398. [Google Scholar] [CrossRef]
- Banskota, S.; Raguram, A.; Suh, S.; Du, S.W.; Davis, J.R.; Choi, E.H.; Wang, X.; Nielsen, S.C.; Newby, G.A.; Randolph, P.B.; et al. Engineered Virus-like Particles for Efficient in Vivo Delivery of Therapeutic Proteins. Cell 2022, 185, 250–265.e16. [Google Scholar] [CrossRef]
- Brodin, J.D.; Sprangers, A.J.; McMillan, J.R.; Mirkin, C.A. DNA-Mediated Cellular Delivery of Functional Enzymes. J. Am. Chem. Soc. 2015, 137, 14838–14841. [Google Scholar] [CrossRef] [Green Version]
- Fang, H.; Peng, B.; Ong, S.Y.; Wu, Q.; Li, L.; Yao, S.Q. Recent Advances in Activity-Based Probes (ABPs) and Affinity-Based Probes (AfBPs) for Profiling of Enzymes. Chem. Sci. 2021, 12, 8288–8310. [Google Scholar] [CrossRef]
- Hewings, D.S.; Flygare, J.A.; Bogyo, M.; Wertz, I.E. Activity-Based Probes for the Ubiquitin Conjugation–Deconjugation Machinery: New Chemistries, New Tools, and New Insights. FEBS J. 2017, 284, 1555–1576. [Google Scholar] [CrossRef] [Green Version]
- Sanman, L.E.; Bogyo, M. Activity-Based Profiling of Proteases. Annu. Rev. Biochem. 2014, 83, 249–273. [Google Scholar] [CrossRef] [Green Version]
- Saghatelian, A.; Jessani, N.; Joseph, A.; Humphrey, M.; Cravatt, B.F. Activity-Based Probes for the Proteomic Profiling of Metalloproteases. Proc. Natl. Acad. Sci. USA 2004, 101, 10000–10005. [Google Scholar] [CrossRef] [Green Version]
- Hameed, D.S.; Sapmaz, A.; Burggraaff, L.; Amore, A.; Slingerland, C.J.; van Westen, G.J.P.; Ovaa, H. Development of Ubiquitin-Based Probe for Metalloprotease Deubiquitinases. Angew. Chem. Int. Ed. 2019, 58, 14477–14482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGouran, J.F.; Gaertner, S.R.; Altun, M.; Kramer, H.B.; Kessler, B.M. Deubiquitinating Enzyme Specificity for Ubiquitin Chain Topology Profiled by Di-Ubiquitin Activity Probes. Chem. Biol. 2013, 20, 1447–1455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, H.B.L.; Heilig, R.; Fischer, R.; Kessler, B.M.; Pinto-Fernández, A. ABPP-HT-High-Throughput Activity-Based Profiling of Deubiquitylating Enzyme Inhibitors in a Cellular Context. Front. Chem. 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Altun, M.; Kramer, H.B.; Willems, L.I.; McDermott, J.L.; Leach, C.A.; Goldenberg, S.J.; Kumar, K.G.S.; Konietzny, R.; Fischer, R.; Kogan, E.; et al. Activity-Based Chemical Proteomics Accelerates Inhibitor Development for Deubiquitylating Enzymes. Chem. Biol. 2011, 18, 1401–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iphöfer, A.; Kummer, A.; Nimtz, M.; Ritter, A.; Arnold, T.; Frank, R.; van den Heuvel, J.; Kessler, B.M.; Jänsch, L.; Franke, R. Profiling Ubiquitin Linkage Specificities of Deubiquitinating Enzymes with Branched Ubiquitin Isopeptide Probes. ChemBioChem 2012, 13, 1416–1420. [Google Scholar] [CrossRef] [PubMed]
- Ábrányi-Balogh, P.; Petri, L.; Imre, T.; Szijj, P.; Scarpino, A.; Hrast, M.; Mitrović, A.; Fonovič, U.P.; Németh, K.; Barreteau, H.; et al. A Road Map for Prioritizing Warheads for Cysteine Targeting Covalent Inhibitors. Eur. J. Med. Chem. 2018, 160, 94–107. [Google Scholar] [CrossRef]
- Hewings, D.S.; Heideker, J.; Ma, T.P.; AhYoung, A.P.; El Oualid, F.; Amore, A.; Costakes, G.T.; Kirchhofer, D.; Brasher, B.; Pillow, T.; et al. Reactive-Site-Centric Chemoproteomics Identifies a Distinct Class of Deubiquitinase Enzymes. Nat. Commun. 2018, 9, 1162. [Google Scholar] [CrossRef]
- Wang, T.; Yin, L.; Cooper, E.M.; Lai, M.-Y.; Dickey, S.; Pickart, C.M.; Fushman, D.; Wilkinson, K.D.; Cohen, R.E.; Wolberger, C. Evidence for Bidentate Substrate Binding as the Basis for the K48 Linkage Specificity of Otubain 1. J. Mol. Biol. 2009, 386, 1011–1023. [Google Scholar] [CrossRef] [Green Version]
- Huttlin, E.L.; Bruckner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual Proteome-Scale Networks Reveal Cell-Specific Remodeling of the Human Interactome. Cell 2021, 184, 3022–3040.e28. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the Human Interactome Defines Protein Communities and Disease Networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Schweppe, D.K.; Huttlin, E.L.; Harper, J.W.; Gygi, S.P. BioPlex Display: An Interactive Suite for Large-Scale AP–MS Protein–Protein Interaction Data. J. Proteome Res. 2018, 17, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Kouranti, I.; McLean, J.R.; Feoktistova, A.; Liang, P.; Johnson, A.E.; Roberts-Galbraith, R.H.; Gould, K.L. A Global Census of Fission Yeast Deubiquitinating Enzyme Localization and Interaction Networks Reveals Distinct Compartmentalization Profiles and Overlapping Functions in Endocytosis and Polarity. PLoS Biol. 2010, 8, e1000471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multilevel Proteomics Reveals Host Perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, H.; Pei, R.; Mao, B.; Zhao, Z.; Li, H.; Lin, Y.; Lu, K. The SARS-CoV-2 Protein ORF3a Inhibits Fusion of Autophagosomes with Lysosomes. Cell Discov. 2021, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A Promiscuous Biotin Ligase Fusion Protein Identifies Proximal and Interacting Proteins in Mammalian Cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.I.; Jensen, S.C.; Noble, K.A.; Kc, B.; Roux, K.H.; Motamedchaboki, K.; Roux, K.J. An Improved Smaller Biotin Ligase for BioID Proximity Labeling. Mol. Biol. Cell 2016, 27, 1188–1196. [Google Scholar] [CrossRef]
- Lobingier, B.T.; Hüttenhain, R.; Eichel, K.; Miller, K.B.; Ting, A.Y.; von Zastrow, M.; Krogan, N.J. An Approach to Spatiotemporally Resolve Protein Interaction Networks in Living Cells. Cell 2017, 169, 350–360.e12. [Google Scholar] [CrossRef] [Green Version]
- Branon, T.C.; Bosch, J.A.; Sanchez, A.D.; Udeshi, N.D.; Svinkina, T.; Carr, S.A.; Feldman, J.L.; Perrimon, N.; Ting, A.Y. Efficient Proximity Labeling in Living Cells and Organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. [Google Scholar] [CrossRef]
- Gingras, A.C.; Abe, K.T.; Raught, B. Getting to Know the Neighborhood: Using Proximity-Dependent Biotinylation to Characterize Protein Complexes and Map Organelles. Curr. Opin. Chem. Biol. 2019, 48, 44–54. [Google Scholar] [CrossRef]
- Go, C.D.; Knight, J.D.R.; Rajasekharan, A.; Rathod, B.; Hesketh, G.G.; Abe, K.T.; Youn, J.-Y.; Samavarchi-Tehrani, P.; Zhang, H.; Zhu, L.Y.; et al. A Proximity-Dependent Biotinylation Map of a Human Cell. Nature 2021, 595, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Hassink, G.C.; Zhao, B.; Sompallae, R.; Altun, M.; Gastaldello, S.; Zinin, N.V.; Masucci, M.G.; Lindsten, K. The ER-Resident Ubiquitin-Specific Protease 19 Participates in the UPR and Rescues ERAD Substrates. EMBO Rep. 2009, 10, 755–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.-Y.; Varthi, M.; Sykes, S.M.; Phillips, C.; Warzecha, C.; Zhu, W.; Wyce, A.; Thorne, A.W.; Berger, S.L.; McMahon, S.B. The Putative Cancer Stem Cell Marker USP22 Is a Subunit of the Human SAGA Complex Required for Activator-Driven Transcription and Cell Cycle Progression. Mol. Cell 2008, 29, 102–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwok, S.F.; Solano, R.; Tsuge, T.; Chamovitz, D.A.; Ecker, J.R.; Matsui, M.; Deng, X.W. Arabidopsis Homologs of a C-Jun Coactivator Are Present Both in Monomeric Form and in the COP9 Complex, and Their Abundance Is Differentially Affected by the Pleiotropic Cop/Det/Fus Mutations. Plant Cell 1998, 10, 1779–1790. [Google Scholar] [CrossRef] [Green Version]
- Velasco, K.; Zhao, B.; Callegari, S.; Altun, M.; Liu, H.; Hassink, G.; Masucci, M.G.; Lindsten, K. An N-Terminal SIAH-Interacting Motif Regulates the Stability of the Ubiquitin Specific Protease (USP)-19. Biochem. Biophys. Res. Commun. 2013, 433, 390–395. [Google Scholar] [CrossRef]
- Wu, X.; Yen, L.; Irwin, L.; Sweeney, C.; Carraway, K.L. Stabilization of the E3 Ubiquitin Ligase Nrdp1 by the Deubiquitinating Enzyme USP8. Mol. Cell. Biol. 2004, 24, 7748–7757. [Google Scholar] [CrossRef] [Green Version]
- Deng, M.; Yang, X.; Qin, B.; Liu, T.; Zhang, H.; Guo, W.; Lee, S.B.; Kim, J.J.; Yuan, J.; Pei, H.; et al. Deubiquitination and Activation of AMPK by USP10. Mol. Cell 2016, 61, 614–624. [Google Scholar] [CrossRef] [Green Version]
- Bushman, J.W.; Donovan, K.A.; Schauer, N.J.; Liu, X.; Hu, W.; Varca, A.C.; Buhrlage, S.J.; Fischer, E.S. Proteomics-Based Identification of DUB Substrates Using Selective Inhibitors. Cell Chem. Biol. 2021, 28, 78–87.e3. [Google Scholar] [CrossRef]
- Doudna, J.A.; Gersbach, C.A. Genome Editing: The End of the Beginning. Genome Biol. 2015, 16, 292. [Google Scholar] [CrossRef] [Green Version]
- Doudna, J.A.; Charpentier, E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Artegiani, B.; van Voorthuijsen, L.; Lindeboom, R.G.H.; Seinstra, D.; Heo, I.; Tapia, P.; López-Iglesias, C.; Postrach, D.; Dayton, T.; Oka, R.; et al. Probing the Tumor Suppressor Function of BAP1 in CRISPR-Engineered Human Liver Organoids. Cell Stem Cell 2019, 24, 927–943.e6. [Google Scholar] [CrossRef] [PubMed]
- Gažová, I.; Lefevre, L.; Bush, S.J.; Rojo, R.; Hume, D.A.; Lengeling, A.; Summers, K.M. CRISPR-Cas9 Editing of Human Histone Deubiquitinase Gene USP16 in Human Monocytic Leukemia Cell Line THP-1. Front. Cell Dev. Biol. 2021, 9, 679544. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Lee, Y.-H.; Jones, A.E.; Woolnough, J.L.; Zhou, D.; Dai, Q.; Wu, Q.; Giles, K.E.; Townes, T.M.; Wang, H. The Histone H2A Deubiquitinase Usp16 Regulates Embryonic Stem Cell Gene Expression and Lineage Commitment. Nat. Commun. 2014, 5, 3818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacLeod, G.; Bozek, D.A.; Rajakulendran, N.; Monteiro, V.; Ahmadi, M.; Steinhart, Z.; Kushida, M.M.; Yu, H.; Coutinho, F.J.; Cavalli, F.M.G.; et al. Genome-Wide CRISPR-Cas9 Screens Expose Genetic Vulnerabilities and Mechanisms of Temozolomide Sensitivity in Glioblastoma Stem Cells. Cell Rep. 2019, 27, 971–986.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Characterization | |||||||
|---|---|---|---|---|---|---|---|
| Compound | Target | IC50/Ki/Kd (nM) | MoA | Structure (pdb) | Negative Control | Validation | Ref. |
| IMP-1710 | UCHL1 | 38 | covalent, slowly reversible | IMP-1711 | cell-based assays; dose-response; in vivo target engagement; chemical proteomics | [90] | |
| CSN5i-3 | CSN5 | 5.8 | non-covalent | 5jog | R,R-CSN5i-2e | enzyme assays; cell-based assays; dose-response; XRD | [92] |
| Azaindole derivatives cmpds 4, 6 1 | CSN5 | 90, 60 | non-covalent | 5m5q 2 | enzyme assays; NMR; SPR; CellTiter-glo; XRD | [93] | |
| FT671 | USP7 | 52 | non-covalent | 5nge | mutagenesis; cell-based assays; SPR; MS | [94] | |
| GNE6640 | USP7 | 750 | non-covalent | 5uqv | GNE-6641 | enzyme assays; cell-based assays; NMR; MS; XRD | [40] |
| ALM-4, ALM-5 | USP7 | 1.5, 22 | non-covalent | 5n9t 3 | ent-ALM-4 | cell-based assays; SPR; target engagement assay; XRD | [95] |
| Pyrimidinone derivatives 4 | USP7 | 6−87 4 | non-covalent | 6f5h 5 | enzyme assays; cell-based assays; in vitro ADME assay; XRD | [96] | |
| XL177A | USP7 | 0.34 | covalent | XL177B | enzyme assays; cell-based assays; ABPP-MS; HDX | [97] | |
| XL188 | USP7 | 90 | non-covalent | 5vs6 | XL203C | enzyme assays; cell-based assays; ITC; XRD | [98] |
| FT709 | USP9X | 82 | non-covalent | cell-based assays; proteomics; SPR; ELISA | [72] | ||
| ML323 | USP1/UAF1 | 76 | non-covalent | NCGC-959 | enzyme assays; cell-based assays; orthogonal gel-based assays | [81] | |
| MF-094 | USP30 | 120 | non-covalent | MF-095 | enzyme assays; mitophagy assay | [99] | |
| FT385 | USP30 | 1 | covalent | enzyme assays; cell-based assays; BLI; ABPP | [88] | ||
| Characterization | ||||||||
|---|---|---|---|---|---|---|---|---|
| UbV | Target | IC50 (nM) | EC50 (nM) | Kd (nM) | Validation | Specificity 1 | Co-Structure (pdb) | Ref. |
| 2.3 | USP2a | 25 | phage ELISA; enzyme assays; XRD | ++ | 3v6e | [103] | ||
| 7.2 | USP7 | 0.91 | 10.9 | phage ELISA; enzyme assays; cell-based assays; AP-MS; GF; XRD | +++ | [74] | ||
| 8.2 | USP8 | 4.8 | phage ELISA; enzyme assays; cell-based assays; AP-MS; XRD | +++ | 3n3k | [103] | ||
| 10.1 | USP10 | 46.2 | 39.9 | phage ELISA; enzyme assays; cell-based assays | ++ | [74] | ||
| 15.1/D | USP15 | 6.6 | ELISA; enzyme assays; cell-based assays; AP-MS | +++ | 6dj9 2 | [120] 3 | ||
| M20 | USP16 | in silico, enzyme assays; cell-based assays; AP-MS; ABP profiling | +++ | [122] | ||||
| 21.4 | USP21 | 2.4 | phage ELISA; enzyme assays; cell-based assays; AP-MS; XRD | +++ | 3mtn | [103] | ||
| 25.1 | USP25 | 130 | competitive phage ELISA | ++ | [117] | |||
| 28 | USP28 | 15 | competitive phage ELISA; cell-based assays | ++ | [117] | |||
| 37.3 | USP37 | 190 | competitive phage ELISA | ++ | [117] | |||
| T9F/T66K | UCHL1 | 2200 | 1300 | in silico; BLI; enzyme assays; cell-based assays | + | [123] | ||
| Q40V/T66K/V70F | UCHL3 | 4 | 49 | in silico; BLI; enzyme assays; cell-based assays | + | [124] | ||
| SP.3 | STAMBP | 9.8 | 60 | phage ELISA; enzyme assays; ITC; XRD | + | 7l97 4 | [52] | |
| BR.1 | BRISC | phage ELISA | ++ | [103] | ||||
| B1.1 | OTUB1 | 11,100 | 20 | phage ELISA; enzyme assays; XRD | ++ | 4i6l | [103] | |
| OTUD1 | OTUD1 | 10 | competitive phage ELISA | ++ | [117] | |||
| Ataxin3.2 & 3.3 | ATXN3 | 20 | competitive phage ELISA | ++ | [117] | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caba, C.; Mohammadzadeh, A.; Tong, Y. On the Study of Deubiquitinases: Using the Right Tools for the Job. Biomolecules 2022, 12, 703. https://doi.org/10.3390/biom12050703
Caba C, Mohammadzadeh A, Tong Y. On the Study of Deubiquitinases: Using the Right Tools for the Job. Biomolecules. 2022; 12(5):703. https://doi.org/10.3390/biom12050703
Chicago/Turabian StyleCaba, Cody, Azam Mohammadzadeh, and Yufeng Tong. 2022. "On the Study of Deubiquitinases: Using the Right Tools for the Job" Biomolecules 12, no. 5: 703. https://doi.org/10.3390/biom12050703
APA StyleCaba, C., Mohammadzadeh, A., & Tong, Y. (2022). On the Study of Deubiquitinases: Using the Right Tools for the Job. Biomolecules, 12(5), 703. https://doi.org/10.3390/biom12050703






