Glutamine Metabolism Is Required for Alveolar Regeneration during Lung Injury
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Bleomycin-Induced Lung Injury
2.3. Mouse Lung Dissociation and Flow Cytometry
2.4. Organoid Culture
2.5. RNA Extraction and Real-Time PCR
2.6. Detection of Glutamate and α-Ketoglutarate
2.7. Determination of Hydroxyproline Content
2.8. Histopathological Staining of Mouse Lung Tissue
2.9. AT2 Cell Viability Assay
2.10. Single-Cell RNA-seq Analysis
2.11. Transcriptomic and Metabolomic Analyses
2.12. Statistical Analysis
3. Results
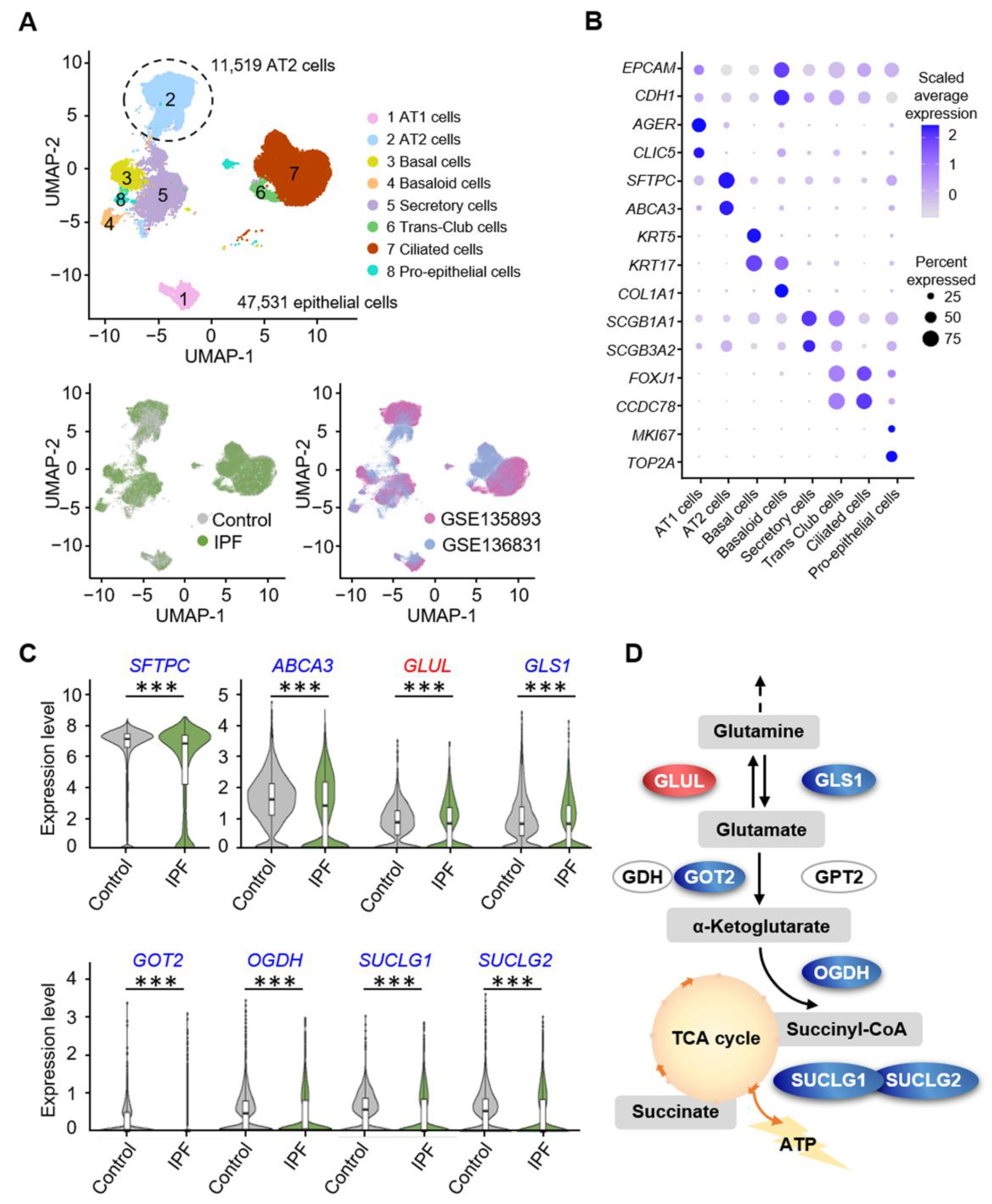
3.1. Alveolar Progenitor Cells in Patients with IPF Exhibit Downregulated Glutamine Catabolism
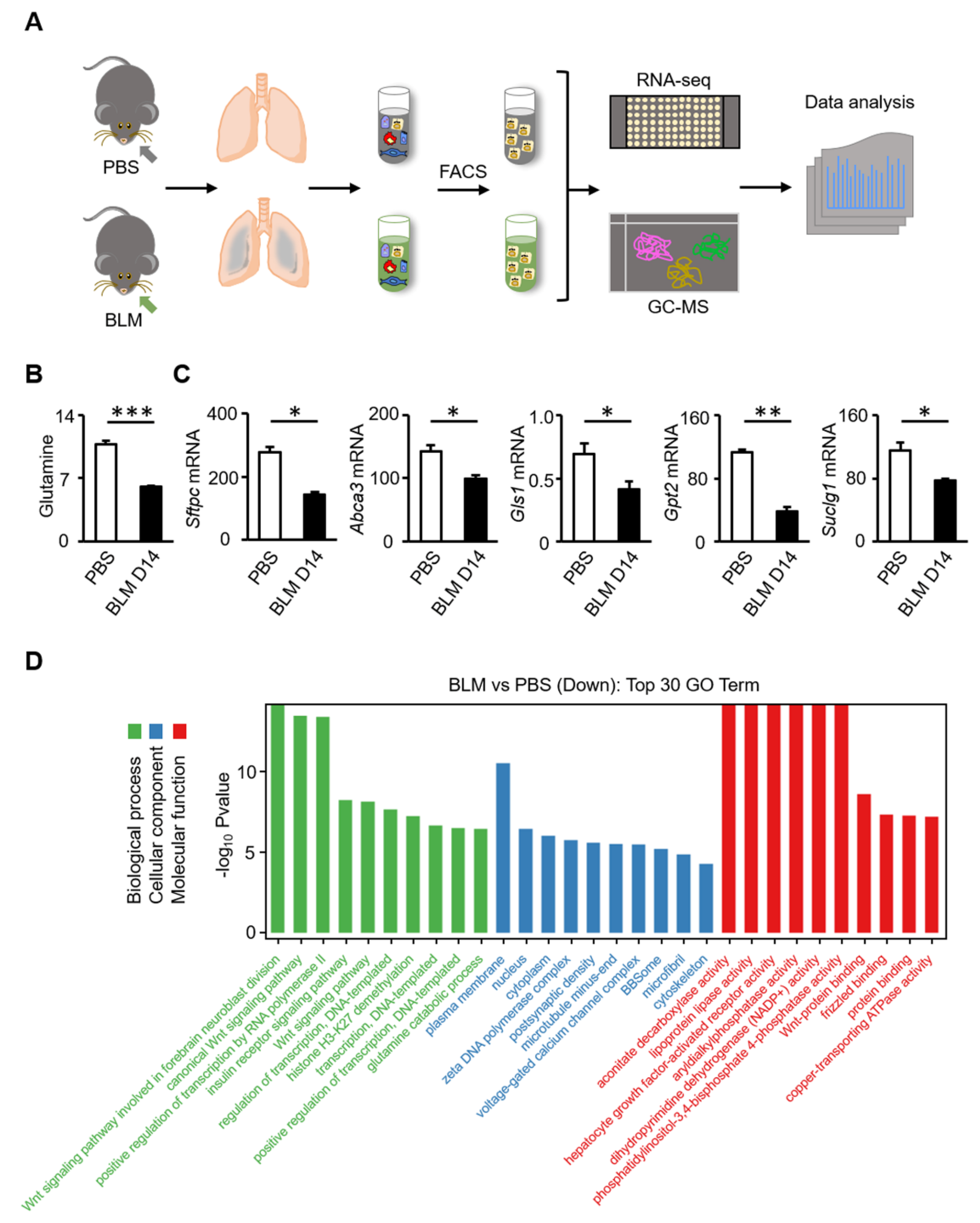
3.2. Glutamine Metabolic Pathway Is Inhibited in AT2 Cells of Bleomycin-Induced Fibrosis in Mice
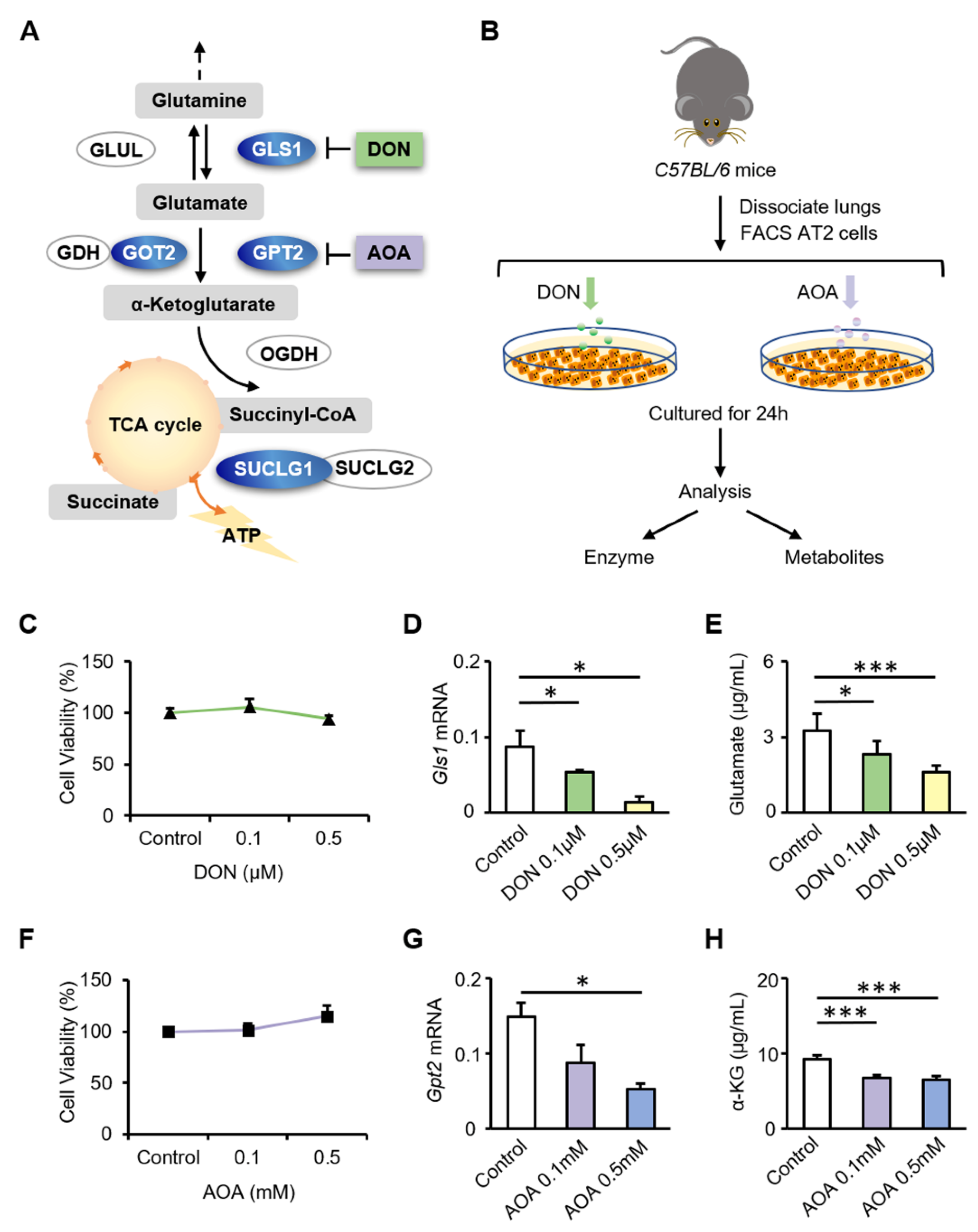
3.3. Inhibition of GLS1 and GPT2 in AT2 Cells In Vitro Leads to Decreased Glutamine Metabolism
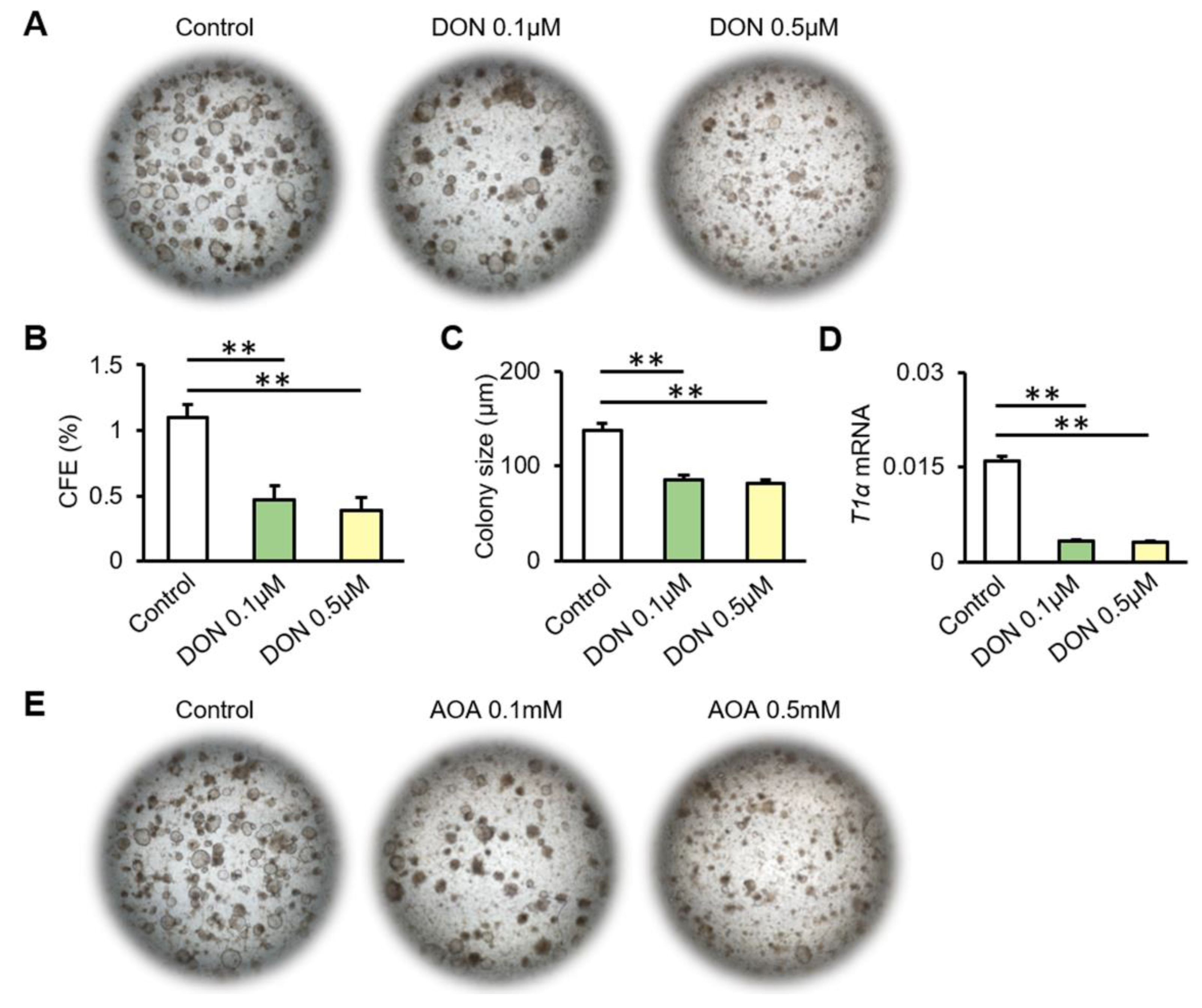
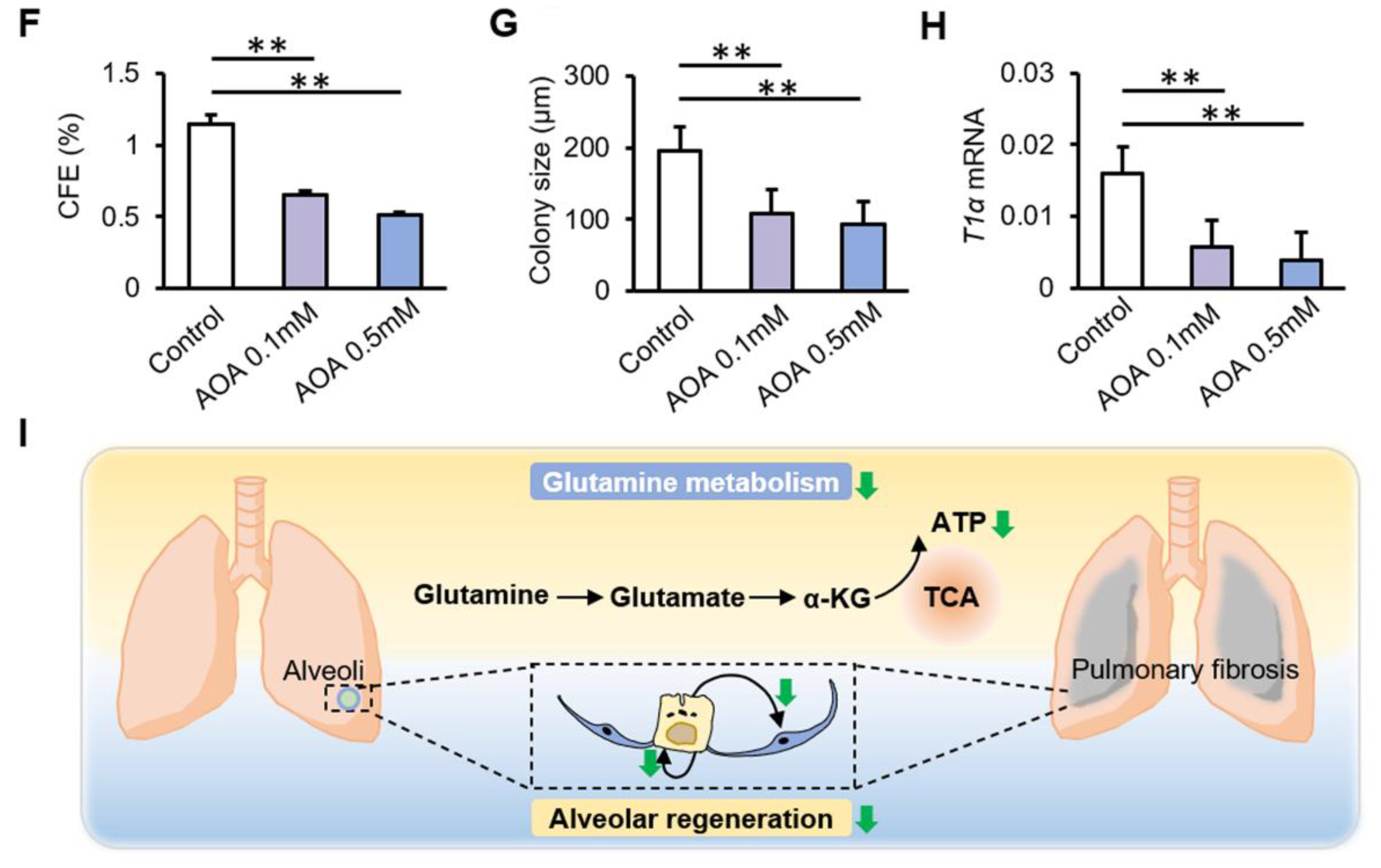
3.4. Inhibition of Glutamine Metabolism Reduces the Proliferation and Differentiation of AT2 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sousa-Victor, P.; García-Prat, L.; Muñoz-Cánoves, P. Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat. Rev. Mol. Cell Biol. 2021, 23, 204–226. [Google Scholar] [CrossRef] [PubMed]
- Benham-Pyle, B.W.; Brewster, C.E.; Kent, A.M.; Mann, F.G.; Chen, S.; Scott, A.R.; Box, A.C.; Alvarado, A.S. Identification of rare, transient post-mitotic cell states that are induced by injury and required for whole-body regeneration in Schmidtea mediterranea. Nat. Cell Biol. 2021, 23, 939–952. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Jimenez, S.; Barrera-Lopez, J.F.; Diaz-Chamorro, S.; Mateos-Quiros, C.M.; Rodriguez-Blanco, I.; Marquez-Perez, F.L.; Lorenzo, M.J.; Centeno, F.; Roman, A.C.; Carvajal-Gonzalez, J.M. p53 regulation by MDM2 contributes to self-renewal and differentiation of basal stem cells in mouse and human airway epithelium. FASEB J. 2021, 35, e21816. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, W.; Frank, D.B.; Zepp, J.A.; Morley, M.P.; Alkhaleel, F.A.; Kong, J.; Zhou, S.; Cantu, E.; Morrisey, E.E. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 2018, 555, 251–255. [Google Scholar] [CrossRef]
- Aspal, M.; Zemans, R.L. Mechanisms of ATII-to-ATI Cell Differentiation during Lung Regeneration. Int. J. Mol. Sci. 2020, 21, 3188. [Google Scholar] [CrossRef]
- Hamada, S.; Sato, A.; Hara-Chikuma, M.; Satooka, H.; Hasegawa, K.; Tanimura, K.; Tanizawa, K.; Inouchi, M.; Handa, T.; Oga, T.; et al. Role of mitochondrial hydrogen peroxide induced by intermittent hypoxia in airway epithelial wound repair in vitro. Exp. Cell Res. 2016, 344, 143–151. [Google Scholar] [CrossRef]
- McNulty, K.; Janes, S.M. Stem Cells and Pulmonary Fibrosis: Cause or Cure? Proc. Am. Thorac. Soc. 2012, 9, 164–171. [Google Scholar] [CrossRef]
- Wu, H.; Yu, Y.; Huang, H.; Hu, Y.; Fu, S.; Wang, Z.; Shi, M.; Zhao, X.; Yuan, J.; Li, J.; et al. Progressive Pulmonary Fibrosis Is Caused by Elevated Mechanical Tension on Alveolar Stem Cells. Cell 2020, 180, 107–121.e17. [Google Scholar] [CrossRef]
- Yin, L.; Zheng, D.; Limmon, G.V.; Leung, N.H.; Xu, S.; Rajapakse, J.C.; Yu, H.; Chow, V.T.; Chen, J. Aging exacerbates damage and delays repair of alveolar epithelia following influenza viral pneumonia. Respir. Res. 2014, 15, 116. [Google Scholar] [CrossRef]
- Olson, A.L.; Gifford, A.; Inase, N.; Pérez, E.F.; Suda, T. The epidemiology of idiopathic pulmonary fibrosis and interstitial lung diseases at risk of a progressive-fibrosing phenotype. Eur. Respir. Rev. 2018, 27, 180077. [Google Scholar] [CrossRef] [Green Version]
- Spagnolo, P.; Distler, O.; Ryerson, C.J.; Tzouvelekis, A.; Lee, J.S.; Bonella, F.; Bouros, D.; Hoffmann-Vold, A.-M.; Crestani, B.; Matteson, E.L. Mechanisms of progressive fibrosis in connective tissue disease (CTD)-associated interstitial lung diseases (ILDs). Ann. Rheum. Dis. 2020, 80, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Gross, T.J.; Hunninghake, G.W. Idiopathic Pulmonary Fibrosis. N. Engl. J. Med. 2001, 345, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Ni Wang, Z.; Tang, X.X. New Perspectives on the Aberrant Alveolar Repair of Idiopathic Pulmonary Fibrosis. Front. Cell Dev. Biol. 2020, 8, 580026. [Google Scholar] [CrossRef] [PubMed]
- Baratella, E.; Ruaro, B.; Giudici, F.; Wade, B.; Santagiuliana, M.; Salton, F.; Confalonieri, P.; Simbolo, M.; Scarpa, A.; Tollot, S.; et al. Evaluation of Correlations between Genetic Variants and High-Resolution Computed Tomography Patterns in Idiopathic Pulmonary Fibrosis. Diagnostics 2021, 11, 762. [Google Scholar] [CrossRef]
- Nabhan, A.N.; Brownfield, D.G.; Harbury, P.B.; Krasnow, M.A.; Desai, T.J. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 2018, 359, 1118–1123. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Li, M.; Li, W.; Yu, H.; Sun, X.; Zhang, Q.; Li, Y.; Li, X.; Li, Y.; Abel, E.D.; et al. Airway epithelial regeneration requires autophagy and glucose metabolism. Cell Death Dis. 2019, 10, 875. [Google Scholar] [CrossRef]
- Li, X.; Zhao, F.; Wang, A.; Cheng, P.; Chen, H. Role and mechanisms of autophagy in lung metabolism and repair. Cell. Mol. Life Sci. 2021, 78, 5051–5068. [Google Scholar] [CrossRef]
- Li, X.; Wu, J.; Sun, X.; Wu, Q.; Li, Y.; Li, K.; Zhang, Q.; Abel, E.D.; Chen, H. Autophagy Reprograms Alveolar Progenitor Cell Metabolism in Response to Lung Injury. Stem Cell Rep. 2020, 14, 420–432. [Google Scholar] [CrossRef] [Green Version]
- Hegab, A.E.; Ozaki, M.; Kagawa, S.; Fukunaga, K. Effect of High Fat Diet on the Severity and Repair of Lung Fibrosis in Mice. Stem Cells Dev. 2021, 30, 908–921. [Google Scholar] [CrossRef]
- Bergström, J.; Fürst, P.; O’Norée, L.; Vinnars, E. Intracellular free amino acid concentration in human muscle tissue. J. Appl. Physiol. 1974, 36, 693–697. [Google Scholar] [CrossRef]
- Tan, H.; Sim, A.Y.L.; Long, Y.C. Glutamine metabolism regulates autophagy-dependent mTORC1 reactivation during amino acid starvation. Nat. Commun. 2017, 8, 338. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wu, G.; Zhou, Z.; Dai, Z.; Sun, Y.; Ji, Y.; Li, W.; Wang, W.; Liu, C.; Han, F.; et al. Glutamine and intestinal barrier function. Amino Acids 2015, 47, 2143–2154. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Zeng, L.; Guoyao, W.; Kong, X.; Wu, G.; Yin, Y. The glutamine-alpha-ketoglutarate (AKG) metabolism and its nutritional implications. Amino Acids 2016, 48, 2067–2080. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Newman, H.; Shen, L.; Sharma, D.; Hu, G.; Mirando, A.J.; Zhang, H.; Knudsen, E.; Zhang, G.-F.; Hilton, M.; et al. Glutamine Metabolism Regulates Proliferation and Lineage Allocation in Skeletal Stem Cells. Cell Metab. 2019, 29, 966–978.e964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Tsai, Y.-H.; Tseng, B.-J.; Tseng, S.-H. Influence of Growth Hormone and Glutamine on Intestinal Stem Cells: A Narrative Review. Nutrients 2019, 11, 1941. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, A.; Deng, X.; Khuri, F.R.; Owonikoko, T.K. Altered Glutamine Metabolism and Therapeutic Opportunities for Lung Cancer. Clin. Lung Cancer 2014, 15, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Vanhove, K.; Derveaux, E.; Graulus, G.-J.; Mesotten, L.; Thomeer, M.; Noben, J.-P.; Guedens, W.; Adriaensens, P. Glutamine Addiction and Therapeutic Strategies in Lung Cancer. Int. J. Mol. Sci. 2019, 20, 252. [Google Scholar] [CrossRef] [Green Version]
- Barrios, J.; Kho, A.T.; Aven, L.; Mitchel, J.; Park, J.-A.; Randell, S.H.; Miller, L.A.; Tantisira, K.G.; Ai, X. Pulmonary Neuroendocrine Cells Secrete γ-Aminobutyric Acid to Induce Goblet Cell Hyperplasia in Primate Models. Am. J. Respir. Cell Mol. Biol. 2019, 60, 687–694. [Google Scholar] [CrossRef]
- Ge, J.; Cui, H.; Xie, N.; Banerjee, S.; Guo, S.; Dubey, S.; Barnes, S.; Liu, G. Glutaminolysis Promotes Collagen Translation and Stability via α-Ketoglutarate–mediated mTOR Activation and Proline Hydroxylation. Am. J. Respir. Cell Mol. Biol. 2018, 58, 378–390. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.-M.; Wang, A.; Li, Y.; Wu, Q.; Chen, H.-Y. Sex-determining region Y-box 2-positive alveolar cells are responsive to bleomycin-induced lung injury. Chin. Med. J. 2020, 134, 234–236. [Google Scholar] [CrossRef]
- Habermann, A.C.; Gutierrez, A.J.; Bui, L.T.; Yahn, S.L.; Winters, N.I.; Calvi, C.L.; Peter, L.; Chung, M.-I.; Taylor, C.J.; Jetter, C.; et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 2020, 6, eaba1972. [Google Scholar] [CrossRef] [PubMed]
- Travaglini, K.J.; Nabhan, A.N.; Penland, L.; Sinha, R.; Gillich, A.; Sit, R.V.; Chang, S.; Conley, S.D.; Mori, Y.; Seita, J.; et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 2020, 587, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Katzen, J.; Beers, M.F. Contributions of alveolar epithelial cell quality control to pulmonary fibrosis. J. Clin. Investig. 2020, 130, 5088–5099. [Google Scholar] [CrossRef] [PubMed]
- Adams, T.S.; Schupp, J.C.; Poli, S.; Ayaub, E.A.; Neumark, N.; Ahangari, F.; Chu, S.G.; Raby, B.A.; DeIuliis, G.; Januszyk, M.; et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 2020, 6, eaba1983. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M.; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Selman, M.; Pardo, A. The leading role of epithelial cells in the pathogenesis of idiopathic pulmonary fibrosis. Cell. Signal. 2020, 66, 109482. [Google Scholar] [CrossRef]
- Wolters, P.J.; Blackwell, T.S.; Eickelberg, O.; Loyd, J.; Kaminski, N.; Jenkins, G.; Maher, T.M.; Molina, M.M.; Noble, P.W.; Raghu, G.; et al. Time for a change: Is idiopathic pulmonary fibrosis still idiopathic and only fibrotic? Lancet Respir. Med. 2018, 6, 154–160. [Google Scholar] [CrossRef] [Green Version]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.-R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Tran, H.T.N.; Ang, K.S.; Chevrier, M.; Zhang, X.; Lee, N.Y.S.; Goh, M.; Chen, J. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020, 21, 12. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Risacher, S.L.; Shen, L.; Saykin, A.J. Network approaches to systems biology analysis of complex disease: Integrative methods for multi-omics data. Brief Bioinform. 2018, 19, 1370–1381. [Google Scholar] [CrossRef] [Green Version]
- Nachef, M.; Ali, A.K.; Almutairi, S.M.; Lee, S.-H. Targeting SLC1A5 and SLC3A2/SLC7A5 as a Potential Strategy to Strengthen Anti-Tumor Immunity in the Tumor Microenvironment. Front. Immunol. 2021, 12, 624324. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhang, N.; Tang, T.; Feng, F.; Sun, H.; Qu, W. Target the human Alanine/Serine/Cysteine Transporter 2(ASCT2): Achievement and Future for Novel Cancer Therapy. Pharmacol. Res. 2020, 158, 104844. [Google Scholar] [CrossRef] [PubMed]
- Clark, E.C.; Patel, S.D.; Chadwick, P.R.; Warhurst, G.; Curry, A.; Carlson, G.L. Glutamine deprivation facilitates tumour necrosis factor induced bacterial translocation in Caco-2 cells by depletion of enterocyte fuel substrate. Gut 2003, 52, 224–230. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, G.P.; Kitoko, J.Z.; Lima-Gomes, P.D.S.; Rochael, N.C.; de Araújo, C.C.; Lugon, P.N.; dos Santos, H.L.; Martins, E.G.L.; Ornellas, F.M.; de Oliveira, H.D.; et al. Glutamine Therapy Reduces Inflammation and Extracellular Trap Release in Experimental Acute Respiratory Distress Syndrome of Pulmonary Origin. Nutrients 2019, 11, 831. [Google Scholar] [CrossRef] [Green Version]
- Bharadwaj, S.; Singh, M.; Kirtipal, N.; Kang, S.G. SARS-CoV-2 and Glutamine: SARS-CoV-2 Triggered Pathogenesis via Metabolic Reprograming of Glutamine in Host Cells. Front. Mol. Biosci. 2020, 7, 627842. [Google Scholar] [CrossRef]
- Shaghaghi, H.; Para, R.; Tran, C.; Roman, J.; Ojeda-Lassalle, Y.; Sun, J.; Romero, F.; Summer, R. Glutamine restores mitochondrial respiration in bleomycin-injured epithelial cells. Free Radic. Biol. Med. 2021, 176, 335–344. [Google Scholar] [CrossRef]
- Hamanaka, R.B.; O’Leary, E.M.; Witt, L.J.; Tian, Y.; Gökalp, G.A.; Meliton, A.Y.; Dulin, N.O.; Mutlu, G.M. Glutamine Metabolism Is Required for Collagen Protein Synthesis in Lung Fibroblasts. Am. J. Respir. Cell Mol. Biol. 2019, 61, 597–606. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Li, X.; Ma, Q.; Wang, Q.; Wu, J.; Yu, H.; Li, K.; Li, Y.; Wang, J.; Zhang, Q.; et al. Glutamine Metabolism Is Required for Alveolar Regeneration during Lung Injury. Biomolecules 2022, 12, 728. https://doi.org/10.3390/biom12050728
Wang S, Li X, Ma Q, Wang Q, Wu J, Yu H, Li K, Li Y, Wang J, Zhang Q, et al. Glutamine Metabolism Is Required for Alveolar Regeneration during Lung Injury. Biomolecules. 2022; 12(5):728. https://doi.org/10.3390/biom12050728
Chicago/Turabian StyleWang, Sisi, Xue Li, Qingwen Ma, Qi Wang, Junping Wu, Hongzhi Yu, Kuan Li, Yu Li, Jianhai Wang, Qiuyang Zhang, and et al. 2022. "Glutamine Metabolism Is Required for Alveolar Regeneration during Lung Injury" Biomolecules 12, no. 5: 728. https://doi.org/10.3390/biom12050728
APA StyleWang, S., Li, X., Ma, Q., Wang, Q., Wu, J., Yu, H., Li, K., Li, Y., Wang, J., Zhang, Q., Wang, Y., Wu, Q., & Chen, H. (2022). Glutamine Metabolism Is Required for Alveolar Regeneration during Lung Injury. Biomolecules, 12(5), 728. https://doi.org/10.3390/biom12050728





